Abstract
Bacteria and archaea possess adaptive immunity against foreign genetic materials through clustered regularly interspaced short palindromic repeat (CRISPR) systems. The discovery of this intriguing bacterial system heralded a revolutionary change in the field of medical science. The CRISPR and CRISPR-associated protein 9 (Cas9) based molecular mechanism has been applied to genome editing. This CRISPR-Cas9 technique is now able to mediate precise genetic corrections or disruptions in in vitro and in vivo environments. The accuracy and versatility of CRISPR-Cas have been capitalized upon in biological and medical research and bring new hope to cancer research. Cancer involves complex alterations and multiple mutations, translocations and chromosomal losses and gains. The ability to identify and correct such mutations is an important goal in cancer treatment. In the context of this complex cancer genomic landscape, there is a need for a simple and flexible genetic tool that can easily identify functional cancer driver genes within a comparatively short time. The CRISPR-Cas system shows promising potential for modeling, repairing and correcting genetic events in different types of cancer. This article reviews the concept of CRISPR-Cas, its application and related advantages in oncology.
Keywords: cancer, CRISPR-Cas9, genetics, immunity, medical research
Introduction
Over the centuries, new technology has changed the field of medical science. Medical technology is essential for healthcare and improves quality of life. Diagnostics and treatments have become easier and more accurate due to advancements in areas like genetic engineering, biotechnology and nuclear medicine.1 In the current decade, there have been tremendous developments in the field of genetic engineering and related technologies. For several years, scientists have been using ‘gene targeting’ to introduce new changes into a specific site in the genome by removing or adding single bases or whole genes. Furthermore, researchers have used technologies derived from the prokaryotic immune system.2,3 Systems involving the clustered regularly interspaced short palindromic repeat (CRISPR) and its associated proteins (Cas) have become the most reliable tools for gene editing. The idea of the CRISPR-Cas technique has been adapted from the bacterial immune system. The CRISPR-Cas9 system has been widely adopted all over the world and successfully applied to target essential genes in different organisms and cell lines, including bacteria, zebrafish, monkeys, rabbits, mice and even humans.4
Cancer is one of the most significant public health problems around the world. It is the second leading cause of death around the globe, with about 8.8 million deaths due to cancer in 2015. The number of expected new cases will increase globally by about 70% over the next two decades.5 Current tools are insufficient to fight cancer, and scientists are always looking for helpful new technologies. In this regard, the CRISPR-Cas9 system brings new hope. Rewriting of the genetic code in humans is possible through CRISPR. A patient has been injected with cells that contain edited genes using the revolutionary CRISPR-Cas9 technique.6 The induction of CRISPR is simpler and more efficient than other technologies and will probably accelerate gene-editing procedures across the world. To date, it has been applied in several different in vivo and in vitro cancer models. Oncologists are very excited about the efficacy, accuracy and potential of CRISPR in the field of cancer research. Peking University in Beijing has started three clinical trials using the CRISPR technique against urinary bladder, prostate and renal cancers.6
Time line of CRISPR
CRISPR-Cas9 has brought revolutionary change to the field of medical and biological sciences in recent years, as summarized in Table 1. In 1987, a group of Japanese scientists reported a series of short direct repeats interspaced with short sequences in the genome of Escherichia coli.7 Two years later, Francisco Mojica (University of Alicante) found a curious structure that was roughly palindromic, with the repeated sequence of 30 bases separated by spacers of around 36 bases.8 He discovered similar repeats in the closely related H. volcanii. These intriguing discoveries motivated him to study the phenomena further; in 2000, he named these structures short regularly spaced repeats (SRSRs). Later, the name was changed to clustered regularly interspaced palindromic repeats (CRISPR).9 This vital finding helped scientists to hypothesize, correctly, that CRISPR is an adaptive immune system; Vergnaud and his group published similar findings in 2005.10
Table 1.
Timeline of CRISPR.
| The milestones of CRISPR technology | Contribution | Time Period(s) | Reference |
|---|---|---|---|
| A series of short direct repeats interspaced with short sequences: genome of Escherichia coli | Osaka University, Japan | 1987 | Ishino and colleagues7 |
| Discovery of CRISPR and its function | Francisco Mojica, University of Alicante, Spain | 1993–2005 | Mojica11 |
| Discovery of Cas9 and protospacer adjacent motif (PAM) | Alexander Bolotin, French National Institute for Agricultural Research (INRA), France | May 2005 | Bolotin and colleagues12 |
| Hypothetical scheme of adaptive immunity | Eugene Koonin, US National Center for Biotechnology Information, NIH, Maryland | March 2006 | Mojica and colleagues13 |
| Experimental demonstration of adaptive immunity | Philippe Horvath, Danisco France SAS, France | March 2007 | Barrangou and colleagues2 |
| Spacer sequences are transcribed into guide RNAs | John van der Oost, University of Wageningen, Netherlands | August 2008 | Brouns and colleagues14 |
| CRISPR acts on DNA targets | Luciano Marraffini and Erik Sontheimer, Northwestern University, Illinois | December 2008 | Marraffini and Sontheimer15 |
| Cas9 cleaves target DNA | Sylvain Moineau, University of Laval, Quebec City, Canada | December 2010 | Garneau and colleagues16 |
| Discovery of trace RNA for Cas9 system | Emmanuelle Charpentier, Umea University, Sweden and University of Vienna, Austria | March 2011 | Deltcheva and colleagues17 |
| DNA interference mechanism: CRISPR systems | Howard Hughes Medical Institute (HHMI), University of California, Berkeley, California University of Vienna, A-1030 Vienna, Austria, Department of Molecular Biology, Umea University, Sweden |
June 2012 | Jinek and colleagues18 |
| CRISPR systems can function heterologously in other species | Virginijus Siksnys, Vilnius University, Lithuania | September 2012 | Gasiunas and colleagues19 |
| RNA-guided human genome engineering | Harvard Medical School, Boston, Department of Biomedical Engineering, Boston University, Boston, Massachusetts |
January 2013 | Mali and colleagues4 |
| CRISPR-Cas9 harnessed for genome editing | Feng Zhang, Broad Institute of MIT and Harvard, McGovern Institute for Brain Research at MIT, Massachusetts | January 2013 | Ding and colleagues20 |
While studying Streptococcus thermophiles in 2005, Bolotin and his team revealed an unusual CRISPR locus in the sequence of genes, and successfully identified the novel Cas9 gene. They also noted the protospacer adjacent motif (PAM), which is required for target recognition in the CRISPR-Cas9 system.12 The following year, Koonin and his team proposed a hypothetical mechanism scheme for CRISPR cascades of bacterial immune system based on inserts homologous to phage DNA in the natural spacer array; they inferred that the Cas proteins might comprise a novel DNA repair system.21,22 In 2007, Horvath and his team showed experimentally that the CRISPR system is an adaptive immune system. They fused new phage DNA into the CRISPR array and showed successful defense against the next attacking phage.2 Then, in August 2008, critical information was reported by John van der Oost and his colleagues. In E. coli, they determined the phage-derived spacer sequences that were transcribed into small RNAs, named CRISPR RNAs (crRNAs). Those crRNAs guide the Cas proteins to the target-specific part of the DNA.14 In December of the same year, Marraffini and Sontheimer found that the target molecule is DNA, not RNA.15 They demonstrated that this system could also be applied in non-bacterial systems. Around 2 years later, Moineau and his colleagues confirmed that Cas9 is the only protein required for cleavage of DNA.16 In July 2011, Siksnys and his team cloned the entire CRISPR-Cas locus from S. thermophiles and expressed it in E. coli. From this experiment, they confirmed that it was capable of providing plasmid resistance.23 They showed that the RuvC domain could cleave the non-complementary strand, and the HNH domain cleaves the complementary site. They also proved that Cas9 could be reprogrammed to target a site of choice by changing the sequence of the crRNA.19
At the same time, another puzzle was solved by the group of Emmanuelle Charpentier. She reported that tracrRNA forms a duplex with crRNA, and this duplex guides Cas9 to its targets. They also enumerated a DNA interference mechanism system involving a dual-RNA structure that directs a Cas9 endonuclease to induce site-specific double-stranded breaks in target DNA.17 Later in 2012, Charpentier and Doudna revealed that crRNA and tracrRNA could be fused together to form a single simplified system.18 In 2013, Feng Zhang and his team from Broad Institute of MIT and Harvard, engineered two varieties of Cas9 orthologs from S. thermophilus and S. pyogenes and demonstrated targeted genome cleavage in mouse and human cells.24 In the same year, George McDonald Church and his team engineered the type II bacterial CRISPR system with customized guide RNA (gRNA) in human cells.4
CRISPR-Cas9 system
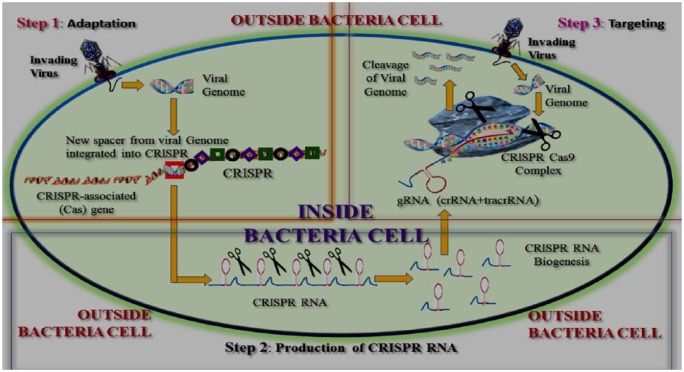
Bacterial CRISPR spacers are short, variable sequences derived from the genomes of viruses that previously invaded the bacteria. Such sequences provide ‘genetic memory’. During viral attacks, the CRISPR defense mechanism of bacteria shears viral genome sequences analogous to spacer sequences (Figure 1). If the invading virus is new, a new spacer is formed and archived into the sequences of spacers.3
Figure 1.
Graphical representation of the CRISPR-Cas9 system.
Step 1. Adaptation – DNA from the invading virus is processed into short segments. These segments are inserted into the CRISPR sequence to function as new spacers.
Step 2. Production of CRISPR RNA – the DNA undergoes a transcription process that copies DNA into RNA. The single-stranded RNA is cut into short pieces called CRISPR RNAs.
Step 3. Targeting – CRISPR RNAs are programmed to destroy the viral material. Here, the ‘RNA sequences’ are copied from the viral DNA sequences.
Adaptation in mammalian cells: role of gRNA in CRISPR-Cas9
The specificity of CRISPR-based immunity is not only useful for bacteria. The system was not only adapted in mammalian cells, but also applied as a potential gene-editing weapon. The modular design is a unique feature of the CRISPR-Cas9 system. The CRISPR complex consists of two modules: a CRISPR-associated (Cas) endonuclease module and a CRISPR RNA (crRNA) module. The Cas endonuclease module initiates the double-stranded DNA breaks and a crRNA module that specifies the target DNA sequence. The target module (sgRNA) and the endonuclease module (Cas9), which are encoded separately, can be optimized without altering the function of each other. The regulation of gene expression can be controlled through CRISPR-Cas9.4 Especially in mammalian cells, the co-expression of Cas9 and sgRNA is sufficient to incite sequence-specific DNA cuts.25 gRNAs can guide the insertion or deletion of uridine residues into the mitochondrial mRNAs in kinetoplastid protists during RNA editing. The gRNA is a short, synthetic RNA composed of a ‘scaffold’ sequence necessary for Cas9-binding and a user-defined ∼20 nucleotide ‘spacer’ or ‘targeting’ sequence that defines the genomic target to be modified.26 A double-strand DNA cut is possible at a particular genomic region where Cas9 nuclease can be programmed by a gRNA. It is also important to consider various factors for designing a gRNA for gene knockout, including the location of the CRISPR-Cas9 targeted sequence within the gene.27
In mammalian cells, knocking out a gene is highly effective when Cas9 is targeted to the exon regions of a specified gene. In this regard, cutting by Cas9 and subsequent repairing by NHEJ (nonhomologous end joining) result in indel mutations. Usually, such an indel causes a frameshift in the target gene and leads to production of a nonfunctional, truncated protein or the degradation of mutant mRNA through nonsense-mediated mRNA decay. RNA interference (RNAi) typically does not achieve complete silencing. CRISPR interference (CRISPRi), an alternative gene-silencing approach, uses the high binding affinity of Cas9 for its target sequence. Mutation in the nuclease domains of Cas9 produces a nuclease-dead protein (dCas9), which binds to the target DNA without cleaving it.28 After binding to the target DNA, dCas9 can suppress its expression by interfering with the transcription machinery.29
Application of CRISPR-Cas9 in cancer drug development
Identifying a target gene or protein is one of the most crucial parts of the drug discovery process.30 Screening the target in the mammalian cell, before CRISPR, depends on RNAi-based gene knockdown libraries and cDNA-based gene overexpression libraries. The first was used to detect gain-of-function (GOF), and the latter was used to detect high-throughput loss of function (LOF). However, RNAi often produces false-negative rates for incomplete gene silencing and a high false-positive rate due to the effects of notable off-targets. To overcome these drawbacks, shRNA/siRNA libraries are used simultaneously, and the overall procedures related to this methodology eventually increase the cost and/or size of the libraries. Moreover, lentiviral insertion-dependent LOF is restricted to cell lines with a haploid genome.31 The advent of CRISPR-Cas9 has opened a new door for functional genomics studies. This technique offers precise editing of a genome, enabling genetic research of defective genes and their behavior. It can be applied for the systematic identification of genes that support cancer cell viability and regulate cancer drug sensitivity.29 The LOF CRISPR libraries can be applied for both positive and negative selection assays. Again, GOF CRISPR libraries are simple to build and deploy compared to cDNA libraries, and the libraries also allow access to large genes that are not available in cDNA libraries. In comparison to cDNA libraries, however, the magnitude of CRISPR activation (CRISPRa) gene overexpression might be more variable.32,33 After screening to identify the target for drug development, the thorough archiving of these cancer genes is vital. This catalog will help to appraise the quality of future members of LOF CRISPR libraries in lethality screening and will also fine-tune the discovery of the target by avoiding cell-essential genes. CRISPR knockout and CRISPRi approaches have already been applied to screen very large sgRNA library pools and to identify essential cancer-lethal genes.34–36 These genes reveal the functional dependencies in individual cancer cell lines, which could be potential components for drug targets. The retroviral libraries of gRNAs in CRISPR help in targeting every gene within the genome. Around 3000 human genes have been identified to be associated with different genetic diseases, and another ~500 genes have been identified to be associated with complex diseases or various infections. Identification of various genes is rapidly increasing, and it is believed that 4000–7000 additional disease-associated genes will be defined in the next few years.37 BCR and ABL are examples of genes related to lethality of the chronic myelogenous leukemia cell line KBM7, which harbors BCR–ABL translocation.38 KRAS and PIK3CA have also been identified as lethal hits for the colorectal cancer cell lines DLD-1 and HCT116.39 These genes play pivotal roles in cellular processes including DNA replication, RNA processing and proteolysis.34–36 An important finding is that the CRISPR system is not totally free of the off-target effect. Researchers are studying different methods to overcome the obstacle and hence pave the way for developing cancer drugs using the CRISPR system.29,40,41
Delivery vehicle for CRISPR-Cas9
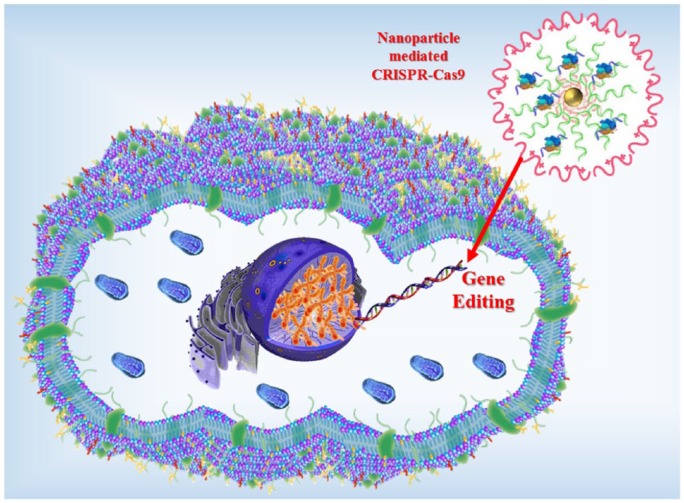
Often, the therapeutic translation of the CRISPR-Cas9 system is impeded due to lack of an appropriate delivery carrier. The high molecular weight and complexity of the CRISPR-Cas9 system make delivery difficult.42 Interestingly, researchers have developed a new nanoscale vehicle termed nanoclews, coated with a positively charged material such as lipid or polymers,37 that can disrupt the endosomal membrane and remain free inside the cell. When the nanoclew enters into a cell, it is absorbed by the cellular endocytic mechanisms. Then, the CRISPR-Cas9 complexes are separated from the nanoclew structure and make their way to the nucleus. Finally, the CRISPR-Cas9 complex reaches the nucleus and starts gene editing. The nanoclews are made of a single strand of DNA and help in delivering the CRISPR-Cas9 gene-editing complex into cells both in vivo and in vitro.43 The DNA nanoclew-based delivery method of CRISPR-Cas9 is shown in Figure 2.
Figure 2.
Graphical representation of a nanoscale delivery vehicle for CRISPR-Cas9 (not to scale).
CRISPR-Cas9 in cancer modeling
Genetic mutations and epigenetic alterations play significant roles in tumorigenesis. Cancer is a multiple-hit disease that is a result of mutations in genes involved in the control of cellular function, growth and division. That is why epigenetic modulation and genome editing are important for cancer modeling and therapeutic efficacy. The whole-genome sequencing data of human cancer cells indicated the complexity of the cancer genome, including numerous point mutations and large genome rearrangements. Cellular and animal models can be established using these data in order to understand the molecular mechanisms underlying tumorigenic responses. New candidate genes to be considered as target genes for cancer therapy can also be validated through these models.44,45
Figure 3 presents an overview of cancer modeling with the CRISPR-Cas9 system. In recent years, the CRISPR-Cas9 system has produced a revolution in the field of cancer modeling. While the other genome-editing tools – ZFNs and TALENs – are based on sequence recognition via protein–DNA interactions, the CRISPR-Cas9 method is known to target specific genomic loci with sgRNA. In addition, since RNA is much easier to synthesize and introduce into a cell than protein domains, the CRISPR-Cas9 tool is regarded as a more promising way to facilitate targeted genome modifications.46
Figure 3.
A schematic overview of cancer modeling using the CRISPR-Cas9 technique.
Tyler Jacks and his team recently elucidated the feasibility of using this powerful tool for modeling liver cancer by directly mutating cancer genes in an in vivo study.47 They showed that this technology can be adapted into a cancer model with lung cancer mutations using genetically engineered mouse models.48 Scott W. Lowe and his team explained that an inducible CRISPR (iCRISPR) system could effectively be used for creating biallelic mutations in multiple target loci, implying that this platform provides a new position from which to study the LOF phenotypes in vivo.49 It was also expected that the CRISPR-Cas9 system will help to generate a mimic displaying cancer chromosomal translocations in human cells. This system allows the generation of cellular models that can demonstrate the primary oncogenic events driving tumorigenesis. This technique depends on a pair of plasmids expressing Cas9 and two sgRNAs to actually target the breakpoints of a cancer translocation. In acute myeloid leukemia (AML), t(8;21)/RUNX1-ETO chromosomal translocation is observed in HEK293 and CD34+ human hematopoietic progenitor cells. Choi and Meyerson reported that the CRISPR-Cas9 system could generate a chromosomal translocation mimic (see Table 2).50
Table 2.
Overview of the application of the CRISPR-Cas9 system in cancer modeling.
| Cancer | Mouse strain and genotype | Alteration | Delivery | Target cell | Approach | Reference |
|---|---|---|---|---|---|---|
| Lung adenocarcinoma | CD1 and C57BL/6J (B6), p53+/– or p53−/−, KrasLSL-G12D/+ |
Chromosomal rearrangement | Adenoviral, lentiviral |
HEK293 (human) | In vitro | Choi and Meyerson50 |
| Maddalo and colleagues51 | ||||||
| Blasco and colleagues52 | ||||||
| Sánchez-Rivera and colleagues48 | ||||||
| Liver cancer | FVB/NJ mice | Loss-of-function and directed mutation | Hydrodynamic injection | Liver cells (mouse) | In vivo | Xue and colleagues47 |
| Pancreatic ductal adenocarcinoma | KrasLSL-G12D/+; R26LSL-Tom; H11LSL- Cas9/+ | Loss-of-function | Retrograde ductal injection of adeno/lentivirus | Somatic pancreatic cells (mouse) | In vivo | Chiou and colleagues53 |
| Burkitt lymphoma | Arf/– EμMyc | Translocation | Lentiviral and retroviral | 293T cells | In vivo | Malina and colleagues54 |
| Colon cancer | ApcMin/+ | Loss-of-function and directed mutation | Plasmid transfection | DLD1 and HCT116 cell lines (human) | In vitro | Antal and colleagues55 |
| Acute myeloid leukemia (AML) | p53 null HSPC | Loss-of-function | Plasmid transfection | mHSPC (mouse) | In vitro | Chen and colleagues56 |
| C57Bl/6 mice or heterozygous Flt3-ITD knock-in mice | Loss-of-function | Lentiviral | mHSPC (mouse) | In vitro | Heckl and colleagues57 | |
| Lung metastasis | KrasG12D/+; p53−/−; Dicer1+/− | Multiple hits from screen | Lentiviral | Human ES and iPS cells | In vivo metastases screen | Urnov and colleagues58 |
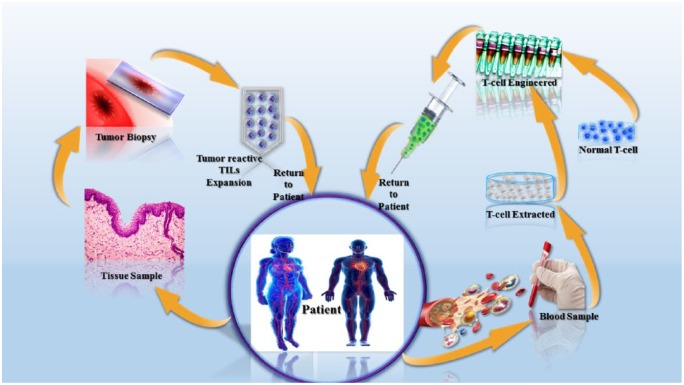
The CRISPR-Cas9 system shows many advantages over conventional gene-targeting technology; one is that it can directly modify the zygote genome. Wang and his team applied the system to modify mouse embryos by injecting Cas9 mRNA and sgRNAs into a fertilized egg.52 This type of genetically engineered modified mouse that carries multiple alterations at loci might have a significant role in cancer research. Moreover, researchers have developed genetically modified immune cells (e.g. T-cells) that showed the ability to kill cancer cells in mice (Figure 4). The cells were modified in such a way that they can express chimeric antigen receptors (CARs) on their surfaces. The CARs can recognize and attack cancer cells, because cancer cells express the corresponding antigen.59
Figure 4.
Cells are collected from the patient, edited by CRISPR-Cas9, and returned to the patient.
Application of CRISPR-Cas9 in cancer treatment
Scientists all over the world are using the CRISPR-Cas9 system to address cancer treatment from different research perspectives. In Table 3, that research is listed for quick reference. Again, the following text presents an overview of that research.
Table 3.
Previous studies and edited genes for different carcinomas.
| Type of cancer | Genes edited | Target | Author(s), publication year | Citation |
|---|---|---|---|---|
| Glioblastoma and medulloblastoma |
Trp53, Pten, Nf1 and Ptch1 | Patient-derived xenograft (PDX), cell-derived xenograft (CDX) and genetically engineered mouse model (GEMMs). | Monje and colleagues 2011 | Zhen and colleagues60 |
| Faria and colleagues 2015 | Li-Kuo and Kinzler61 | |||
| Becher and colleagues 2010 | Roper and colleagues62 | |||
| Bladder cancer | TP53, urothelial carcinoma-associated 1 (UCA1), long non-coding RNA-related nuclear protein (ncRAN) | 5637 and T24 bladder cancer cell lines | Mei Xue and colleagues 2014 | Yoshino and colleagues63 |
| Colorectal cancer | APC, TP53, KRAS, SMAD4 | GEMMs | Roper and colleagues 2017 | Roper and colleagues62 |
| Hepatocellular carcinoma | Pten and p53 genes | Embryonic stem cell targeting | Xue and colleagues 2014 | Xue and colleagues47 |
| Renal cell carcinoma | miR-210-3p | In vivo xenograft study in which Twist-related protein 1 (TWIST1) was the key target of miR-210-3p. | Yoshino and colleagues 2017 | Yoshino and colleagues63 |
| Von Hippel-Lindau (VHL) | Knockdown of VHL | Schokrpur and colleagues 2016 | Schokrpur and colleagues64 | |
| Breast cancer | Brahma (BRM) and Brahma-related Gene 1 (BRG1) | GEMMs | Wu and colleagues 2015 | Wu and colleagues65 |
| CDH1 | Annunziato and colleagues 2016 | Annunziato and colleagues66 | ||
| Human cervical cancer cells | HPV16 E6 gene | SiHa and CaSki cells | Yu and colleagues 2015 | Yu and colleagues67 |
| In vivo experiments targeting promoter + E6 + E7 transcript | Zhen and colleagues 2014 | Zhen and colleagues68 | ||
| Acute myeloid leukemia | miRNAs | Mammalian cell phenotypes | Wallace and colleagues 2016 | Wallace and colleagues69 |
| Heckl and colleagues 2014 | Heckl and colleagues57 | |||
| Ovarian cancer | Snail1 | Human ovarian adenocarcinoma (RMG-1) cells | Haraguchi and colleagues 2015 | Haraguchi and colleagues70 |
| HE4 | HE4-overexpressing SKOV3 cells | Ribeiro and colleagues 2016 | Ribeiro and colleagues71 | |
| LY75 | SKOV3 and TOV112, and A2780s and OV2008 | Faddaoui and colleagues 2016 | Faddaoui and colleagues72 | |
| OCIAD1 | BJNhem20-OCIAD1-CRISPR-39 line | Shetty and colleagues 2016 | Shetty and colleagues73 |
Brain cancer
Brain cancer is the most lethal among all cancers, regardless of gender and age. The therapies used against brain cancers such as gliomas have been more or less the same for the last five decades.74 There are also technical difficulties in the clinical management of brain cancer. For these reasons, researchers are trying to find solutions at the genetic level. In this context, CRISPR-Cas9 can be an efficient, convenient and less time-consuming technique.75 There are four types of animal models used in the study of gliomas and medulloblastomas of human brain cancer: patient-derived xenograft (PDX), cell-derived xenograft (CDX), genetically engineered mouse and in vivo mouse model.76 The Ptch1 gene responsible for medulloblastoma and the Trp53, Pten and Nf1 genes accountable for glioblastoma can be knocked out in the mouse brain using the CRISPR-Cas9 technique. This technique can also be used in further investigations against other regulatory genes found to be responsible for brain tumors.
Urinary bladder cancer
Long non-coding RNA (lncRNA) acts as a critical regulator of the development and progression of tumors and is a potential diagnostic biomarker. Upregulation of the long non-coding RNA PANDAR is associated with poor prognosis and promotes tumorigenesis in bladder cancer. Although the mechanism is not understood, PANDAR plays an effective role in the progression of bladder cancer (BCa).77 Several lncRNA genes are involved in bladder carcinoma, such as TP53,78 urothelial carcinoma-associated 1 (UCA1) and long non-coding RNAs-related nuclear protein (ncRAN).79 Upregulation of UCA1, which is expressed in both 5637 and T24 bladder cancer cell lines, fosters the propagation of BCa cells.80 LncRNA can be manipulated by the versatile gene-editing tool CRISPR-Cas9. Isolated genomic DNA from 5637 and T24 bladder cancer cells were transfected with CRISPR-Cas 9–UCA1 and then analyzed by T7 endonuclease 1 assays and DNA sequencing. A previous study showed that the CRISPR-Cas9 system efficiently knocked out the lncRNA–UCA1,60 indicating that this technique might be effective against other bladder cancer genes, both related or unrelated to lncRNAs.
Colorectal cancer
Colorectal cancer (CRC) arises from the colon or rectum. Tumor sequencing studies have revealed a significant number of candidate genes that are mutated in CRCs. These genes contribute to carcinogenesis, tumor phenotype and tumor progression.81 Functional assessment of putative cancer-associated genes usually requires in vivo experiments; in this context, genetically engineered mouse models (GEMMs) are being used.61 In studies using GEMMs, application of the CRISPR-Cas9-based editing system in orthotopic organoid transplantation of mice without cancer-predisposing mutations corrected the Apc and Trp53 tumor suppressor genes in colon epithelial cells. For engraftment at ectopic sites in mice, mutations in APC, TP53, KRAS, SMAD4 and PIK3CA were needed.62 This approach can be applied in different methods and/or different arrangements, depending on the goal of the experiment, and can be a crucial tool for determining the types of mutations that are most potent in transforming cells, thereby conferring a growth advantage in a multiclonal tumor.
Hepatocellular carcinoma
The CRISPR-Cas9 technique has versatile application potential and is being used in a variety of in vivo and ex vivo experiments. In hepatocellular carcinoma, this technique has already been used in different ways against tumor suppressor candidate genes in the liver.47,82 One approach was to target the tumor suppressors Pten and p53 genes alone or in combination via hydrodynamic tail vein injection. Moreover, sgRNA can create indels that disrupt the respective gene. A combination of Pten and p53 sgRNAs is capable of forming liver tumors similar to those in transgenic animals with CRE-loxP-deleted Pten and p53. This study highlights several points. First, it demonstrates the advantages of bypassing the lengthy process of genetically engineered strains with CRE-loxP technology. The low rate of indel formation is not limiting in a positive selection setting such as propagation of liver cancer. Second, it validates use of the CRISPR-Cas9 genome-editing system to edit the Pten and p53 tumor suppressor genes.47 This result offers the promise of success in further studies against hepatocellular carcinoma using the CRISPR-Cas9 technique, depending on the role, goal and overall model of the experiments.
Renal cell carcinoma
About 80% of renal cell carcinomas (RCCs) arise from tubular cells of the kidney and are a tumor type called clear cell RCC (ccRCC).83 Studies have revealed that five miRNAs in particular – miR-885-5p, miR-1274, miR-210-3p, miR-224 and miR-1290 – are unregulated in ccRCC.84 In an in vivo xenograft study with the CRISPR-Cas9 technique, depletion of miR-210-3p in RCC cell lines such as 786-O, A498 and Caki2 significantly augmented tumorigenesis together with a change in the morphology of A498 and Caki2 cells. Twist-related protein 1 (TWIST1) was the key target of miR-210-3p. The negative correlation between miR-210-3p and TWIST1 expression suggests that RCC progression is promoted by the suppression of TWIST1 mediated by miR-210-3p.63 This technique was also used in an effective disease model of metastatic renal cell carcinoma (mRCC), which showed that knockdown of the tumor suppressor Von Hippel-Lindau (VHL) might be the cause of RCC tumor cell proliferation. The success of these studies gives rise to the hope that this technique can be applied to detect the gene levels that cause RCC, in addition to activity against the progression of RCC.
Breast cancer
Breast cancer (BC) is one of the most common causes of cancer death among women worldwide. Basal-like or triple-negative breast cancer (TNBC), a subtype of molecular BC lacking expression of estrogen receptor, progesterone receptor85 and HER2/neu tyrosine kinase receptor, shows the poorest prognosis among the BC subtypes. The CRISPR-Cas9 system was used to inhibit tumor growth and pulmonary metastasis by knocking out Cripto-1, an embryonic stem cell marker whose promoter showed activity in primary tumors. This study revealed that Cripto-1 could be an alternative therapeutic target for TNBC. The Brahma (BRM) and Brahma-related gene 1 (BRG1) are both overexpressed in primary BCs. Activity of the multi-subunit human SWI/SNF chromatin remodeling enzymes is catalyzed by BRM and BRG1.86 Use of the CRISPR-Cas9 technique to knockout the BRG1 or BRM gene showed that these two genes have at least some non-overlapping roles in promoting BC cell proliferation. Thus, both BRG1 and BRM are potential targets for BC therapy. Invasive lobular carcinoma (ILC) is another common type of human BC. In most cases, this carcinoma exhibits loss of cell–cell adhesion protein and methylation of the CDH1 gene promoter.87 A CRISPR-Cas9-mediated somatic gene-editing technique was used with an intraductal injection of lentiviral vectors that encode Cre recombinase into female mice carrying conditional alleles of the Cdh1 gene encoding E-cadherin. This newly developed platform can be utilized for rapid in vivo testing of putative tumor suppressor genes implicated in ILC. Similar platforms can be utilized for the development of novel in vivo models for the detection and cure of different BC subtypes.66
Cervical cancer
Cervical cancer is another common cancer in women worldwide. Human papillomavirus (HPV) is considered a major causative agent of cervical cancer. During HPV infection, the viral oncoprotein E6 promotes degradation of the host tumor suppressor protein p53, promoting malignant transformation of normal cervical cells.88 The CRISPR-Cas9 system was used to disrupt the HPV16 E6 gene. HPV16 E6 deoxyribonucleic acid was cleaved at particular sites, leading to apoptosis of HPV16-positive SiHa and CaSki cells. The HPV16 E6 ribonucleic acid-guided CRISPR-Cas system will be an effective therapeutic agent in cases of cervical malignancy related to HPV infection.67 Along these lines, in vivo experiments showed significant inhibition of tumorigenesis. Here, cells were engineered with CRISPR-Cas9 to target the promoter+E6+E7 transcript and edit the HPV16 E6 gene. The cells were inoculated into nude mice for establishment of transplanted tumor animal models.68 These in vivo and in vitro results demonstrate the potential of the CRISPR-Cas9 technique in detecting, diagnosing, preventing and curing human cervical cancer.
Acute myeloid leukemia
AML is a hematologic malignancy that carries a bad prognosis. miRNA expression is dysregulated in AML.89 In particular, miR-155 is regarded as a top miRNA candidate for promoting cellular fitness. The CRISPR-Cas9 technique can screen the functions of individual miRNAs and protein-coding genes during the growth of myeloid leukemia cell lines. This technique can be used to identify novel functional miRNAs in mammalian cell phenotypes and can also identify putative target proteins with opposing function. This provides a crucial tool for describing the effects of individual miRNAs and protein-coding genes in leukemic cells.69 Again, this technique can be used to generate mouse models of AML and consequently develop a broad range of in vivo models to understand the complexity of human diseases.57
Ovarian cancer
The epithelial-to-mesenchymal transition (EMT) is a common phenomenon during cancer metastasis.90 During this process, epithelial cells lose their junctions, and gene expression is reprogrammed. This transition is induced by several master regulators, which include several transcription factors such as Snail1, TWIST and zinc-finger E-box binding (ZEB). Knockdown of the Snail1 gene by application of the CRISPR-Cas9 technique demonstrated that loss of Snail1 changes the actin cytoskeleton. This technique was also used to determine the functions of Snail1 and block its expression in human ovarian adenocarcinoma (RMG-1) cells.70 The reasons for chemoresistance of epithelial ovarian cancer can also be determined at the genetic level using the CRISPR-Cas9 technique. Knockdown of the ovarian cancer biomarker HE4 reversed the chemoresistance.71 Knockdown of LY75 reduced migration as well as the invasiveness of the tumor cells in vitro and decreased the metastatic potential of EOC cell lines in in vivo environments.72 Ovarian carcinoma immunoreactive antigen domain containing 1 (OCIAD1) was also knocked out in a BJNhem20-OCIAD1-CRISPR-39 line using CRISPR-Cas9 as a strategy against ovarian cancer.73 This ex vivo experiment, along with other experiments based on CRISPR-Cas9 strategies, paves a new way for the treatment and/or prevention of ovarian cancer in the future.
Conclusion
Cancer presents a long-standing problem in the history of human health and so far has no holistic solution. Researchers from various parts of the world are searching relentlessly for an appropriate and efficient approach based on genetic technology that can provide a sustainable solution to this disease. The CRISPR-Cas9 system is cutting-edge gene-editing technology with wide potential that stands alone among other cytogenetic techniques of gene editing in cancer-related diseases. This technique also has the potential to be used in every field of medical and biological sciences. However, a significant number of ethical questions have arisen regarding the application of this technique without any protection against inappropriate usage. Misuse of this technique might create lethal conditions that could destroy human civilization. Regarding the use of CRISPR-Cas9 technology around the world, we found little use to date of this technique in developing countries due to the lack of expertise and proper infrastructure. CRISPR-Cas9 should be the preferred approach in deciphering the complex components of gene expression leading to any cancer.
Footnotes
Author contributions: ZAR, YJS, JHK, LAB and JYC designed the format of this paper, interpreted the data and wrote the manuscript. MFH, BM, MU and SBZ edited this paper.
Funding: This research was also supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2017R1A6A1A03015642), Korea.
Conflict of interest statement: The authors declare that there is no conflict of interest.
ORCID iDs: Mohammad Faisal Haidere  https://orcid.org/0000-0002-2041-7087
https://orcid.org/0000-0002-2041-7087
Jae Youl Cho  https://orcid.org/0000-0001-8141-9927
https://orcid.org/0000-0001-8141-9927
Contributor Information
Zubair Ahmed Ratan, Department of Biomedical Engineering, Khulna University of Engineering and Technology, Khulna, Bangladesh.
Young-Jin Son, Department of Pharmacy, Sunchon National University, Suncheon, Korea.
Mohammad Faisal Haidere, Centre for Advanced Research in Sciences (CARS), University of Dhaka, Dhaka, Bangladesh.
Bhuiyan Mohammad Mahtab Uddin, Department of Microbiology, Enam Medical College, Savar, Dhaka, Bangladesh.
Md. Abdullah Yusuf, Department of Microbiology, National Institute of Neurosciences & Hospital, Dhaka, Bangladesh.
Sojib Bin Zaman, International Centre for Diarrhoeal Disease Research, Dhaka, Bangladesh.
Jong-Hoon Kim, Department of Physiology, College of Veterinary Medicine, Chonbuk National University, Iksan 54596, Korea.
Laila Anjuman Banu, Department of Anatomy, Bangabandhu Sheikh Mujib Medical University (BSMMU), Shahbag, Dhaka 1000, Bangladesh.
Jae Youl Cho, Department of Genetic Engineering, Sungkyunkwan University, 2066 Seobu-ro, Suwon 16419, Korea.
References
- 1. Zaman SB, Hossain N, Ahammed S, et al. Contexts and opportunities of e-health technology in medical care. J Med Res and Innov 2017; 1: AV1–AV4. [Google Scholar]
- 2. Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315: 1709–1712. [DOI] [PubMed] [Google Scholar]
- 3. Ratan ZA, Zaman SB, Mehta V, et al. Application of fluorescence in situ hybridization (FISH) technique for the detection of genetic aberration in medical science. Cureus 2017; 9: e1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stewart B, Wild CP. World cancer report 2014. Lyon: IARC Press, 2014. [Google Scholar]
- 6. Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature 2016; 539: 479. [DOI] [PubMed] [Google Scholar]
- 7. Ishino Y, Shinagawa H, Makino K, et al. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987; 169: 5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mojica F, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol 1993; 9: 613–621. [DOI] [PubMed] [Google Scholar]
- 9. Mojica FJ, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evo 2005; 60: 174–182. [DOI] [PubMed] [Google Scholar]
- 10. Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005; 151: 653–663. [DOI] [PubMed] [Google Scholar]
- 11. Mojica FJ. La construcción del futuro. Concepto y modelo de prospectiva estratégica, territorial y tecnológica. Universidad Externado de Colombia, Bogotá, Colombia 2005. [Google Scholar]
- 12. Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005; 151: 2551–2561. [DOI] [PubMed] [Google Scholar]
- 13. Mojica F, Diez-Villasenor C, Garcia-Martinez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 2009; 155: 733–740. [DOI] [PubMed] [Google Scholar]
- 14. Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321: 960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008; 322: 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garneau JE, Dupuis M-E, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468: 67–71. [DOI] [PubMed] [Google Scholar]
- 17. Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011; 471: 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gasiunas G, Barrangou R, Horvath P, et al. Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 2012; 109: E2579–E2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ding Q, Regan SN, Xia Y, et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 2013; 12: 393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Makarova K, Slesarev A, Wolf Y, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 2006; 103: 15611–15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Makarova KS, Grishin NV, Shabalina SA, et al. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 2006; 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sapranauskas R, Gasiunas G, Fremaux C, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 2011; 39: 9275–9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cho SW, Kim S, Kim JM, et al. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol 2013; 31: 230–232. [DOI] [PubMed] [Google Scholar]
- 26. Lee CM, Cradick TJ, Fine EJ, et al. Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol Ther 2016; 24: 475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157: 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013; 154: 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Luo J. CRISPR/Cas9: from genome engineering to cancer drug discovery. Trends Cancer 2016; 2: 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pulley JM, Shirey-Rice JK, Lavieri RR, et al. Accelerating precision drug development and drug repurposing by leveraging human genetics. Assay Drug Dev Technol 2017; 15: 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bassik MC, Lebbink RJ, Churchman LS, et al. Rapid creation and quantitative monitoring of high coverage shRNA libraries. Nat Methods 2009; 6: 443–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shi J, Wang E, Milazzo JP, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 2015; 33: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Koike-Yusa H, Li Y, Tan E-P, et al. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol 2014; 32: 267–273. [DOI] [PubMed] [Google Scholar]
- 34. Wang T, Wei JJ, Sabatini DM, et al. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014; 343: 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shalem O, Sanjana NE, Hartenian E, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014; 343: 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gilbert LA, Horlbeck MA, Adamson B, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014; 159: 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yin H, Kauffman KJ, Anderson DG. Delivery technologies for genome editing. Nat Rev Drug Discov 2017; 16: 387–399. [DOI] [PubMed] [Google Scholar]
- 38. Golemovic M, Verstovsek S, Giles F, et al. AMN107, a novel aminopyrimidine inhibitor of Bcr-Abl, has in vitro activity against imatinib-resistant chronic myeloid leukemia. Clin Cancer Res 2005; 11: 4941–4947. [DOI] [PubMed] [Google Scholar]
- 39. Luo J, Emanuele MJ, Li D, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 2009; 137: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nissim L, Perli SD, Fridkin A, et al. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell 2014; 54: 698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu X, Scott DA, Kriz AJ, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 2014; 32: 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun W, Gu Z. Tailoring non-viral delivery vehicles for transporting genome-editing tools. Sci China Mater 2016; 60: 1–5. [Google Scholar]
- 43. Sun W, Ji W, Hall JM, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl 2015; 54: 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646–674. [DOI] [PubMed] [Google Scholar]
- 45. Lawrence L, Sincan M, Markello T, et al. The implications of familial incidental findings from exome sequencing: the NIH Undiagnosed Diseases Program experience. Genet Med 2014; 16: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wyman C, Kanaar R. DNA double-strand break repair: all’s well that ends well. Annu Rev Genet 2006; 40: 363–383. [DOI] [PubMed] [Google Scholar]
- 47. Xue W, Chen S, Yin H, et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 2014; 514: 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 2014; 516: 428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dow LE, Fisher J, O’Rourke KP, et al. Inducible in vivo genome editing with CRISPR-Cas9. Nat Biotechnol 2015; 33: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun 2014; 5: 3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Maddalo D, Manchado E, Concepcion CP, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014; 516: 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Blasco RB, Karaca E, Ambrogio C, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell Rep 2014; 9: 1219–1227. [DOI] [PubMed] [Google Scholar]
- 53. Chiou S-H, Winters IP, Wang J, et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 2015; 29: 1576–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malina A, Mills JR, Cencic R, et al. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev 2013; 27: 2602–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Antal CE, Hudson AM, Kang E, et al. Protein kinase C loss-of-function mutations in cancer reveal role as tumor suppressor. Cancer Res 2015; 75(Suppl. 15): 125. [Google Scholar]
- 56. Chen C, Liu Y, Rappaport AR, et al. MLL3 is a haploinsufficient 7q tumor suppressor in acute myeloid leukemia. Cancer Cell 2014; 25: 652–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heckl D, Kowalczyk MS, Yudovich D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol 2014; 32: 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet 2010; 11: 636–646. [DOI] [PubMed] [Google Scholar]
- 59. Eyquem J, Mansilla-Soto J, Giavridis T, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 2017; 543: 113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhen S, Hua L, Liu Y-H, et al. Inhibition of long non-coding RNA UCA1 by CRISPR/Cas9 attenuated malignant phenotypes of bladder cancer. Oncotarget 2017; 8: 9634–9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li-Kuo S, Kinzler KW. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992; 256: 668–670. [DOI] [PubMed] [Google Scholar]
- 62. Roper J, Tammela T, Cetinbas NM, et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 2017; 35: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoshino H, Yonemori M, Miyamoto K, et al. microRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017; 8: 20881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schokrpur S, Hu J, Moughon DL, et al. CRISPR-mediated VHL knockout generates an improved model for metastatic renal cell carcinoma. Sci Rep 2016; 6: 29032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wu Q, Madany P, Akech J, et al. The SWI/SNF ATPases are required for triple negative breast cancer cell proliferation. J Cell Physiol 2015; 230: 2683–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Annunziato S, Kas SM, Nethe M, et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev 2016; 30: 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yu L, Wang X, Zhu D, et al. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. Onco Targets Ther 2015; 8: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Zhen S, Hua L, Takahashi Y, et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun 2014; 450: 1422–1426. [DOI] [PubMed] [Google Scholar]
- 69. Wallace J, Hu R, Mosbruger TL, et al. Genome-wide CRISPR-Cas9 screen identifies microRNAs that regulate myeloid leukemia cell growth. PLoS One 2016; 11: e0153689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Haraguchi M, Sato M, Ozawa M. CRISPR/Cas9n-mediated deletion of the snail1 gene (SNAI1) reveals its role in regulating cell morphology, cell–cell interactions, and gene expression in ovarian cancer (RMG-1) cells. PLoS One 2015; 10: e0132260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ribeiro J, Schorl C, Yano N, et al. HE4 promotes collateral resistance to cisplatin and paclitaxel in ovarian cancer cells. J Ovarian Res 2016; 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Faddaoui A, Bachvarova M, Plante M, et al. The mannose receptor LY75 (DEC205/CD205) modulates cellular phenotype and metastatic potential of ovarian cancer cells. Oncotarget 2016; 7: 14125–14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shetty DK, Inamdar MS. Generation of a heterozygous knockout human embryonic stem cell line for the OCIAD1 locus using CRISPR/CAS9 mediated targeting: BJNhem20-OCIAD1-CRISPR-39. Stem Cell Res 2016; 16: 308–310. [DOI] [PubMed] [Google Scholar]
- 74. Fisher PG, Buffler PA. Malignant gliomas in 2005: where to GO from here? JAMA 2005; 293: 615–617. [DOI] [PubMed] [Google Scholar]
- 75. Zuckermann M, Kawauchi D, Gronych J. ‘CRISPR’ validation of recessive brain cancer genes in vivo. Oncotarget 2015; 6: 17865–17866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun 2015; 6: 7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen C, Huang J, Sun F, et al. MP48-14 long noncoding RNA lncRNA-BNCA promotes the progression of bladder cancer via regulating translation of P53. J Urol 2017; 197: e642. [Google Scholar]
- 78. Fujimoto K, Yamada Y, Okajima E, et al. Frequent association of p53 gene mutation in invasive bladder cancer. Cancer Res 1992; 52: 1393–1398. [PubMed] [Google Scholar]
- 79. Zhu Y, Yu M, Li Z, et al. ncRAN, a newly identified long noncoding RNA, enhances human bladder tumor growth, invasion, and survival. Urology 2011; 77: 510.e1–510e5. [DOI] [PubMed] [Google Scholar]
- 80. Xue M, Li X, Li Z, et al. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumour Biol 2014; 35: 6901–6912. [DOI] [PubMed] [Google Scholar]
- 81. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Weber J, Öllinger R, Friedrich M, et al. CRISPR/Cas9 somatic multiplex-mutagenesis for high-throughput functional cancer genomics in mice. Proc Natl Acad Sci U S A 2015; 112: 13982–13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Hadoux J, Vignot S, Rouge TDLM. Renal cell carcinoma: focus on safety and efficacy of temsirolimus. Clin Med Insights Oncol 2010; 4: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet 2008; 9: 102–114. [DOI] [PubMed] [Google Scholar]
- 85. Petridou E, Karpathios T, Dessypris N, et al. The role of dairy products and non alcoholic beverages in bone fractures among schoolage children. Scand J Soc Med 1997; 25: 119–125. [DOI] [PubMed] [Google Scholar]
- 86. Bakshi R, Hassan MQ, Pratap J, et al. The human SWI/SNF complex associates with RUNX1 to control transcription of hematopoietic target genes. J Cell Physiol 2010; 225: 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015; 163: 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer 2002; 2: 342–350. [DOI] [PubMed] [Google Scholar]
- 89. Garzon R, Garofalo M, Martelli MP, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A 2008; 105: 3945–3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196. [DOI] [PMC free article] [PubMed] [Google Scholar]