Abstract
Many molecules decode not only the concentration of cellular signals, but also their temporal dynamics. However, little is known about the mechanisms that underlie the detection and discrimination of dynamic signals. We used computational modelling of the interaction of a ligand with multiple targets to investigate how kinetic and thermodynamic parameters regulate their capabilities to respond to dynamic signals. Our results demonstrated that the detection and discrimination of temporal features of signal inputs occur for reactions proceeding outside mass-action equilibrium. For these reactions, thermodynamic parameters such as affinity do not predict their outcomes. Additionally, we showed that, at non-equilibrium, the association rate constants determine the amount of product formed in reversible reactions. In contrast, the dissociation rate constants regulate the time interval required for reversible reactions to achieve equilibrium and, consequently, control their ability to detect and discriminate dynamic features of cellular signals.
Introduction
Cells detect endogenous signals through changes in the activities of biomolecules that integrate signalling pathways and networks1–3. Similarly, many drugs exert their effects by regulating components of signalling networks4,5. In recent decades, the advances of molecular biology and proteomics promoted a rapid growth in the understanding of the topological organization of signalling networks and pathways2,6,7. However, despite the wealth of data, the comprehension of the dynamics of interconnected biomolecules and how they underlie specific cellular processes in response to a vast variety of signals remain a challenge2,5,6,8.
Signalling pathways and networks typically show high numbers of cross-talks and redundancies2,5,6,9. Often, networks that share mutual components execute opposite cellular responses10,11. Moreover, common intracellular signals trigger several competing processes9,10,12,13.
To ensure the appropriate response to different signals, the activities of the biomolecules must be tailored to detect only the correct information14. Historically, the law of mass action extensively influenced our understanding of signalling transduction and the mechanisms of drug action15,16. In consequence, we tend to explain the activation of a molecule by a cellular signal or the effect of a drug as dose/concentration-dependent15. Thus, putative differences in the affinities for common activators is the typical explanation for the differential activations of competing signalling pathways17,18. Affinity is also a core concept in pharmacology, commonly used to predict the efficacy of drugs and lead compounds15,16,19,20. However, the concentrations of drugs and endogenous signals fluctuate constantly in the biological systems and often with faster time scales than the rates of binding and unbinding from their cellular targets16,18,20. Frequently the rate constants of the reactions play a more decisive role to their outcomes than thermodynamic parameters such as binding affinity16,18,19,21. Cumulating evidences have showed that the lifetime of a drug on its target is often more important for its physiological effects than the affinity of the drug/target complex16,19. Similarly, several biomolecules and signalling pathways detect the temporal dynamics of intracellular signals13,14,18,22,23, which implies that the concentrations of their activators are not the only property carrying information2,13,14,22. Therefore, one of the most important aspects of cellular signalling transduction that still needs to be addressed is the identification of the mechanisms that underlie the detection and discrimination of the dynamic features of cellular signals.
In this work, we used computational models that simulate the interactions between a ligand and different targets to characterize the role of kinetic and thermodynamic parameters in the detection and discrimination of dynamic signals. Our results indicated that only reactions outside mass-action equilibrium are sensitive to the temporal features of signal inputs. Consequently, their outcomes are not predicted by thermodynamic parameters such as binding affinities and dissociation constants. We also demonstrated that, outside mass-action equilibrium, the association rate constants regulate the amount of product formed in reversible reactions. The dissociation rate constants control the time required for reversible reactions to achieve equilibrium and determine their ability to detect and discriminate dynamic features of cellular signals. Moreover, in sequential reactions, fast dissociation rate constants act as bottlenecks for the propagation of dynamic signals.
Results
Mechanisms for the detection and discrimination of the durations of signals
Thermodynamic and kinetic parameters regulate chemical reactions, but their individual contributions vary18,24. For a reversible reaction of binding and unbinding (reaction 1) between a molecule M and a ligand L forming the complex LM:
| chemical equation 1 |
the dissociation constant (KD) quantifies the binding affinity of the complex LM formed at equilibrium, which is mathematically defined by equation 1:
| 1 |
where the brackets indicate concentrations.
According to equation 1, the KD of a reversible reaction specifies which species are more abundant at equilibrium (the reactants L and M or the product LM). The KD of a reversible reaction is related with its Gibbs free energy (ΔG), which designates the stability of the product LM relative to the reactants L and M (Fig. 1A)24,25. As thermodynamic quantities, KD and ΔG define the relative concentrations of its components at equilibrium, but do not indicate whether the reversible reaction occurs in a feasible time24. It is the energy barrier (energy of activation, EA) that must be overcome during a reaction that determines its velocity24. A low-energy barrier corresponds to a fast reaction and a high-energy barrier corresponds to a slow reaction (Fig. 1A). EA regulates the rate constant (k) of a reaction, but not whether it is thermodynamically favourable24. When reactions occur at equilibrium, they are under thermodynamic control and regulated by thermodynamic parameters such as KD24,25. When they proceed outside equilibrium, they are under kinetic control and their rate constants determine their outcomes24,25.
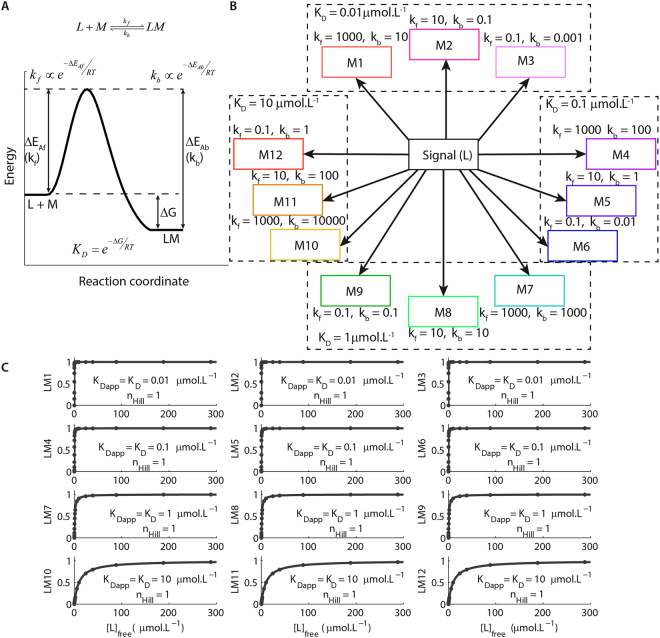
Figure 1.
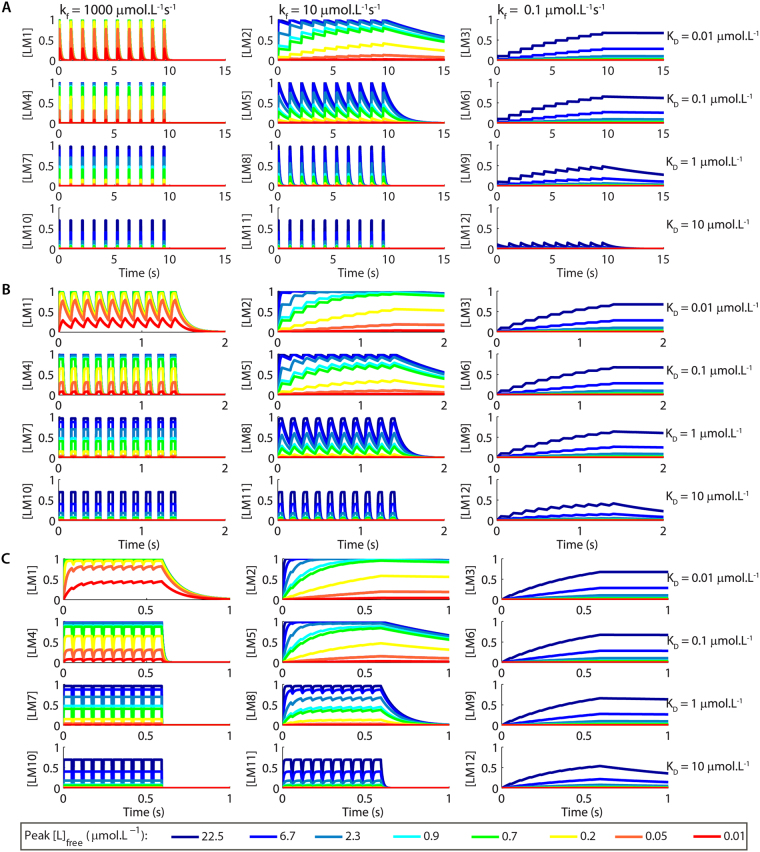
Thermodynamic and kinetic parameter of chemical reactions. (A) The energy profile for a simple reversible reaction of complex formation. The species L and M are the initial reactants, LM is the complex formed, kf and kb are the rate constants for the association (forward) and dissociation (backward) reactions, respectively. ΔEAf and ΔEAb state for the energy of activation for the forward and backward reaction, respectively, R is the ideal gas constant, and T is the temperature in Kelvin. (B) Diagram of the simulated system, which consists of twelve different molecules (M1-12) interacting with a ligand (L) with different affinities (KDs) and rate constants (kfs are given in µmol.L−1.s−1 and kbs in s−1). (C) Dose-response curves for the formation of the complex LM1-LM12 as functions of [L]free. The KDapps estimated with these curves were set as the control KDs for the formation of LM1-LM12 in our simulations.
In biological systems, the concentrations of drugs and endogenous signals vary often with time scales faster than the rates of binding and unbinding from their cellular targets16,18,20. In consequence, many cellular reactions do not achieve equilibrium or steady-state16,18,20. We hypothesised that only reactions that proceed outside mass-action equilibrium detect and discriminate dynamic cellular signals. To test this hypothesis, we simulated the interactions of twelve different molecules (M1-M12) with a ligand L to form the corresponding complexes LM1-LM12 (Fig. 1B). We simulated the formation of three complexes (LM1-LM3) with high affinity at equilibrium (KD = 0.01 µmol.L−1), six with moderate affinity (LM4-LM6 with KD = 0.1 µmol.L−1 and LM7-LM9 with KD = 1 µmol.L−1) and three (LM10-LM12) with low affinity (KD = 10 µmol.L−1). For each KD, we implemented three different sets of rate constants of association (kf) and dissociation (kb) to simulate reactions with varied velocities (Fig. 1B). We then obtained the dose-response curves for the formations of LM1-LM12 as functions of free concentrations of L ([L]free) at equilibrium (Fig. 1C) to ensure that the values of KD used in the model matched the concentration of free ligand ([L]free) required to promote the half-maximum activation of each complex implemented, which we verified by fitting the equation (2):
| 2 |
where A is the activity (i.e. normalized concentration) of the complexes LM1-LM12, Amax corresponds to their maximum activity (=1), the term nhill is the Hill coefficient and KDapp is the apparent KD. As expected, independently of the rate constants used for the reactions simulated, the KDapps of the dose-responses corresponded exactly to the KDs implemented (Fig. 1C). We set these KDapps as the control KDs of LM1-LM12 hereafter. All curves presented nHill equal to 1.
Next, we used square pulses of [L]free with different durations and peak concentrations to verify how the thermodynamic and kinetic parameters used regulate the detection and discrimination of dynamical signals, which we defined as the ability of molecules to respond and display different levels of activation to changes in the temporal properties of their signal activators. The durations and amplitudes of the pulses of [L]free were set in the simulations in a non-conservative manner. Thus, the concentrations of L used in the pulses were buffered. Consequently, all molecules M1-M12 were exposed to the same signals and there was no competition among them.
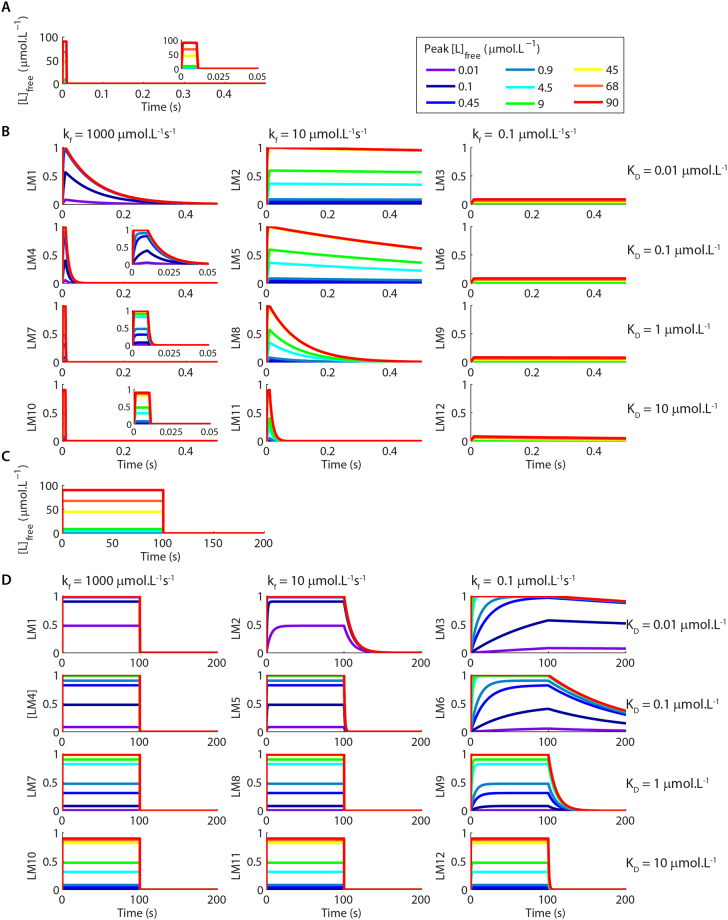
The association and dissociation of complexes that have identical affinities at equilibrium proceeded with different time courses when we used square pulses of [L]free as input signals (Fig. 2). Moreover, complexes that have the same KD at equilibrium displayed different levels of activation (Fig. 2). These differences were strongly pronounced for short pulses, which possess durations within the range of pivotal cellular signals (varying from milliseconds to few seconds)26–28, and gradually disappeared as we stimulated the model with pulses that were long enough to allow the reactions to achieve equilibrium. For instance, LM10, LM11 and LM12 presented very different levels of activation when stimulated by brief pulses of [L]free (10 ms) (Fig. 2A,B), but equivalent activations for pulses of [L]free of 100 s (Fig. 2C,D).
Figure 2.
Examples of time courses of LM1-LM12 for pulses of [L]free with different durations and concentrations. (A,B) Pulses of [L]free of 10 ms of duration and varying concentrations (A) and the corresponding activations of LM1-LM12 (B). The insets show the results with different scales for better visualization. (C,D) Pulses of L with 100 s of duration and varying concentrations (C) and the activations of LM1-LM12 (D). The KDs for the formation of the complexes at equilibrium are showed on the right, and the association rate constants of complex formation (kf) on the top of the panels showed in B and D. The kb for each reaction is calculated by: . The legend indicates the colour code used to represent each concentration of [L]free.
To analyse these data, we used equation 2 to fit dose-responses curves of the peak concentrations of LM1-LM12 obtained as functions of the peak amplitudes of the pulses of [L]free with different durations, and estimated the values of their KDapp and nHill for comparisons with the results of the system at equilibrium.
The dose-response curves of most complexes showed that the durations of pulses of [L]free modulated their formations (Fig. 3A) by changing the values of KDapp in comparison to their control KDs in a duration-dependent manner. Figure 3B shows the KDapp/KD ratios to facilitate their comparisons, we listed the exact values of KDapp in Suppl. Table S1. As we increased the durations of pulses of [L]free, the values of KDapp decreased until they matched the control KDs (KDapp/KD = 1) indicating that the reversible reactions had reached equilibrium, which happened at different pulse durations for the molecules simulated (Fig. 3B). The dynamic changes of KDapps showed that the molecules detected the durations and the peak concentrations of the pulses of [L]free by temporally integrating these signals over time. Once the durations of pulses of [L]free were sufficiently long for the reactions to achieve mass-action equilibrium, they became insensitive to time and detected only the variations in the concentrations of L.
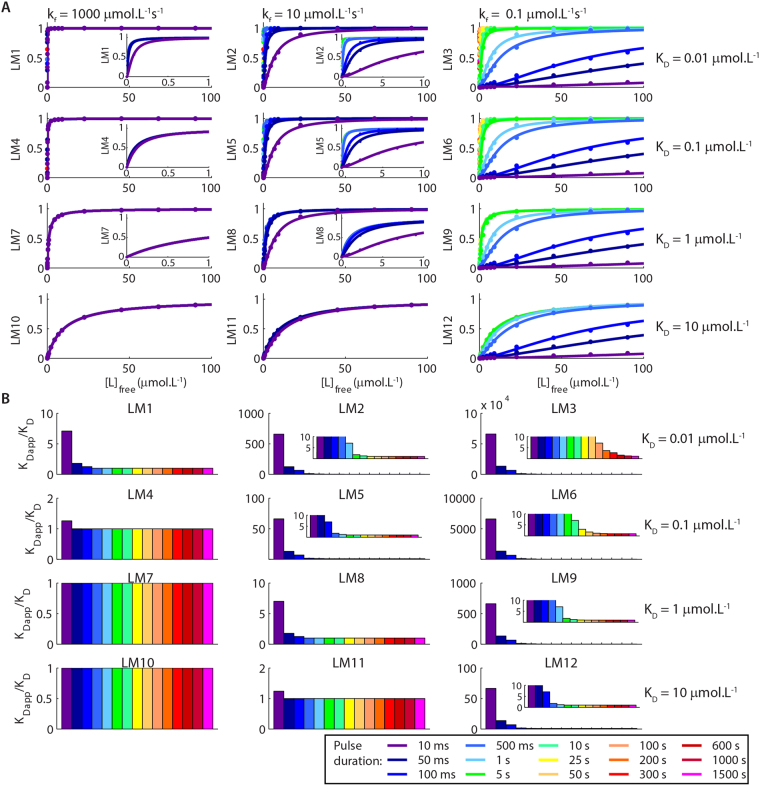
Figure 3.
Activations of LM1-LM12 for pulses of [L]free with different durations and concentrations. (A) Dose-response curves for the formations of LM1-LM12 as functions of pulses of [L]free with different durations. (B) KDapp/KD ratios calculated using the KDs showed in Fig. 1C and the KDapps (Suppl. Table S1) obtained from the curves showed in A. The control KDs for the interaction of each molecule with L are showed on the right of the panels and the kfs for the association reactions are indicated on the top of A. The kb for each reaction is calculated by: . The insets show the same results in a different scale for better visualization. The legend indicates the colour code used to represent each duration of [L]free.
Our results demonstrated that the key point for the temporal discrimination of cellular signals relies on the different time scales in which each reversible reaction reach thermodynamic equilibrium. The longer it takes for a reaction to reach equilibrium, the larger is the range of durations of signals it can detect and discriminate by dynamically changing its KDapp (Fig. 3B).
The results of Fig. 3B also revealed that the dissociation rate constants (kbs) used in our simulations played a pivotal role in determining the time required for each reversible reaction to reach equilibrium (Fig. 3B). In a reversible reaction, the slower is the kb the longer it takes for the activation of a given molecule to peak29. Our results indicated that, for the conditions that we simulated, the slower was the kb the longer was the time interval required for the reactions to reach equilibrium independently of the kf used. For instance, the reactions of formations of LM7 and LM10 occurred with very fast kbs in our simulations. Their formations proceeded at equilibrium for all pulse durations tested, consequently, they only detected the concentrations of L (Fig. 3B). However, the formation of LM1, which happened with the same kf used for the formations of LM7 and LM10 but a slower kb, required pulses of 500 ms to exhibit KDapp compatible to its control KD. Reactions that have same kbs required identical durations of pulses of [L]free to reach equilibrium independently of their kfs (Fig. 3B and Suppl. Table S1, compare the pairs LM1 and LM8, LM2 and LM9, LM5 and LM12). The complexes that dissociated with identical values of kb also presented similar KDapp/KD ratios (Fig. 3B).
Outside the mass-action equilibrium, reversible reactions with identical values of kf had equivalent numerical values of KDapp independently of their kbs and of their control KDs (Suppl. Table S1). For instance, the values of KDapp obtained for the formation of LM3 were much more similar to the KDapps of LM9 for most pulse durations tested than the KDapps of LM1 (Suppl. Table S1), even though LM1 and LM3 have identical KDs at equilibrium and the KD of LM9 is 100-fold weaker. The larger were their kfs, the lower were their KDapps observed at non-equilibrium. This result indicates that complexes with faster kfs activate preferentially outside mass-action equilibrium. However, the closer the reversible reactions got to reaching equilibrium, the lesser their outcomes depended on their kfs and the more they depended on their thermodynamic affinities as expected30.
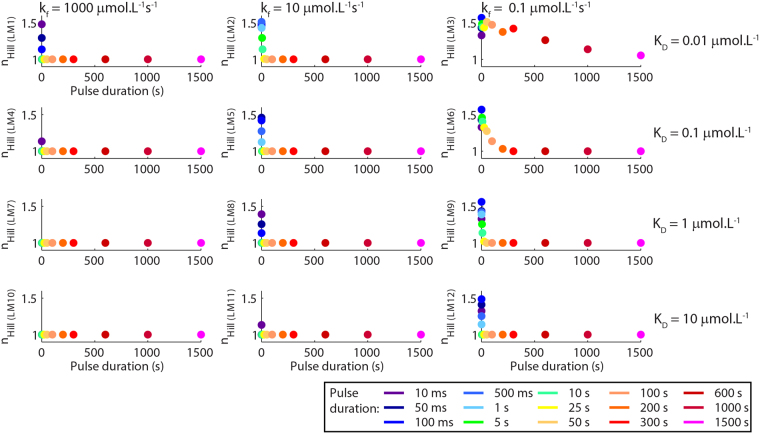
In addition to the changes in KDapp, we verified that durations of the pulses of [L]free promoted variations in the nHill for the reactions that proceeded outside mass-action equilibrium, which displayed nHill larger than 1 even though the components of our system have no allosteric cooperativity (Fig. 4). The parameter nHill is commonly defined as an “interacting-coefficient” that reflects the cooperative binding of ligands to multiple sites of a molecule31. Nevertheless, it is important to note that, in addition to allosteric cooperativity, nHill larger than 1 can indicate ultrasensitivity. Multiple mechanisms promote ultrasensitivity including feedback loops, small changes in reactions near saturating conditions, distributive phosphorylations, among others32–36. In a ultrasensitivity system, nHill designates the degree of bistability33,34,36. In our results, we verified that the values of nHill became larger than 1 only for reactions happening outside mass-action equilibrium. For these reactions, the values of nHill increased as we reduced the durations of pulses of [L]free. The changes of nHill resulted from ultrasensitivity promoted by the filtering of fast signals with low amplitudes as if they were noise. Previously, it was proposed that biology evolved to use non-equilibrium to efficiently discriminate signals from noise16, which is consistent with our results. We had observed similar changes of nHill previously18.
Figure 4.
Values of nHill estimated from the dose-response curves showed in Fig. 3A.
Detection and discrimination of frequencies and number of pulses of dynamic signals
Next, we investigated how the kinetic and thermodynamic parameters underlie the discrimination of interpulse intervals and number of pulses of trains of signals of L, a property displayed by several enzymes and signalling pathways18,22,23,37. We stimulated the formation of LM1-LM12 with trains of ten pulses of [L]free delivered at 1 Hz (1 s of interpulse interval), 10 Hz (100 ms of interpulse interval), or 100 Hz (10 ms of interpulse interval). Each pulse had duration of 50 ms (Suppl. Fig. S1A–C) or 100 ms (Suppl. Fig. S1D–F). Figure 5 and Suppl. Fig. S2 show examples of the time courses of LM1-LM12 observed. To verify whether the formation of LM1-LM12 detected the interpulse interval and the number of pulses of L simulated, we measured the peak amplitude of LM1-LM12 formed as functions of the peak of each pulse of [L]free within a train (Suppl. Fig. S3). We used these data to fit ten dose-response curves for each frequency tested using equation 2 (Suppl. Figs S4 and S5). Each curve corresponded to the formations of LM1-LM12 observed for a specific pulse number (Suppl. Figs S4 and S5). With these curves, we investigated whether the values of KDapp and nHill varied during each train and quantified their discrepancies from the control KDs (Fig. 1C).
Figure 5.
Examples of the time courses for the formation of LM1-LM12 obtained using trains of ten pulses of [L]free with different peak amplitudes released at 1 Hz (A), 10 Hz (B), or 100 Hz (C). Each pulse of [L]free had 50 ms of duration. The time courses of [L]free are showed in Suppl. Fig. S1A–C. The control KDs for the interactions of M1-M12 with L are showed on the right of the panels and the kfs for the association reactions are indicated in A. The legend indicates the colour code used to represent each concentration of [L]free.
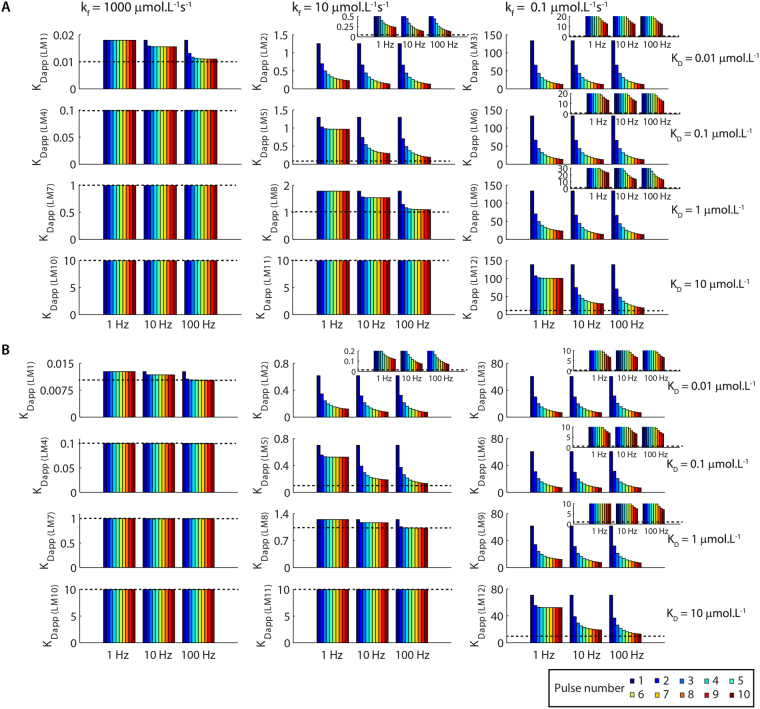
The KDapps obtained (Fig. 6A and B) revealed different types of dynamic signal discriminations that relied heavily on the kbs used in the reactions of the different complexes. The KDapps for the formations of LM4 (kb = 100 s−1), LM7 (kb = 1000 s−1), LM10 (kb = 10000 s−1) and LM11 (kb = 100 s−1) corresponded to the control KDs to all situations tested and demonstrated that these complexes were insensitive to the number of pulses, the interpulse intervals and the durations of pulses used in the simulations. However, as we decreased the kbs, this scenario changed.
Figure 6.
Variations of the KDapps observed for the stimulations of the model with trains of pulses of [L]free. We estimated the KDapps (in µmol.L−1) from dose-responses curves showed in Suppl. Figs S4 and S5. The results showed in (A) and (B) were obtained for pulses of 50 ms and 100 ms, respectively. The dashed lines correspond to the control KDs, also indicated on the right of the panels, the kfs for the association reactions are indicated in A. The insets show the same results in different scales for better visualization. The legend shows the colour code used to represent the pulse number of [L]free.
The KDapps of LM1 and LM8, which had kb = 10 s−1, detected and discriminated mainly the interpulse interval between the signals of [L]free used. Moreover, we observed changes of their KDapps during the initial pulses of [L]free released at 10 Hz and 100 Hz. Thus, LM1 and LM8 also discriminated a limited number of pulses released at intermediary or high frequencies, because their interpulse intervals were shorter than the time required for the inactivation of both complexes. Consequently, there were summations of their activations for the initial pulses of trains released at 10 Hz and 100 Hz (Figs 5 and S2), which promoted alterations in their KDapps (Fig. 6A and B). In contrast, pulses released at 1 Hz had a long interpulse interval (1 s) that prevented the accumulation of LM1 and LM8 from one pulse to another (Figs 5 and S2). The KDapps of LM1 and LM8 matched the control KDs when stimulated with 3 or more pulses of L with 100 ms of duration released at 100 Hz.
LM5 and LM12 (kb = 1 s−1) exhibited KDapps that changed as functions of both the interpulse interval and the number of pulses of [L]free released at 10 Hz and 100 Hz. For signals of [L]free released at 1 Hz, the KDapps of LM5 and LM12 changed only during the initial four pulses (Fig. 6A and B). Thus, LM5 and LM12 acted as good detectors and discriminators of the interpulse intervals for all frequencies tested and of the number of pulses of [L]free released at moderate to high frequencies, but poor detectors of the number of pulses released at a low frequency (Fig. 6A and B).
The KDapps of LM2 and LM9 (kb = 0.1 s−1) detected and discriminated the number of pulses of [L]free for all frequencies tested. In addition, LM2 and LM9 discriminated the interpulse interval of pulses released at 1 Hz from pulses released at 10 Hz or 100 Hz. However, their KDapps did not discriminate the interpulse interval of pulses released at 10 Hz from pulses released at 100 Hz (Fig. 6A and B).
LM3 and LM6, the two complexes with the slowest kbs implemented (kb = 0.001 s−1 and 0.01 s−1, respectively), presented KDapps that discriminated the number of pulses of [L]free, but were insensitive to their interpulse intervals (Fig. 6A and B).
None of the KDapps of LM2, LM3, LM5, LM6, LM9 and LM12 matched their control KDs (Fig. 6A and B, black dashed lines) evidencing that they did not reach mass-action equilibrium in the situations simulated. Moreover, the values of nHill for all the complexes that did not present values of KDapp compatible with the control KDs were larger than 1, which indicated that their activations had a bistability not observed at equilibrium (Suppl. Fig. S6). The kbs used in the simulations regulated the KDapp/KD ratio, as observed in the previous session. Hence, molecules with identical kbs (LM1/LM8, LM2/LM9, LM5/LM12) exhibit KDapps that diverged from KD with equivalent magnitudes (Suppl. Fig. S7).
Detection and discrimination of dynamic signals in sequential reactions
Next, we explored the detection and discrimination of dynamic signals in sequential reactions. Firstly, we implemented the reactions of association/dissociation of LM1-LM12 with the targets T1-T12 using one set of rate constants (kf = 10 µmol−1.L.s−1, kb = 0.1 s−1, KD = 0.01 µmol.L−1). Specifically, LM1 reacted with Tf1, LM2 with Tf2, LM3 with Tf3, and so on, which resulted in twelve ternary complexes LM1T1-LM12T12 formed according to the sequential reactions:
| chemical equation 2 |
where n = 1, 2, 3, …, 12. The parameters kf and kb refer to the rate constants used for the association/dissociation of LM1-LM12 (Fig. 1B).
In a different set of simulations, we implemented the interactions of LM1-LM12 with the targets T′1-T′12 to form LM1T′1-LM12T′12 using a different set of rate constants (kf = 0.1 µmol−1.L.s−1, kb = 0.001 s−1, KD = 0.01 µmol.L−1), but equivalent sequential reactions:
| chemical equation 3 |
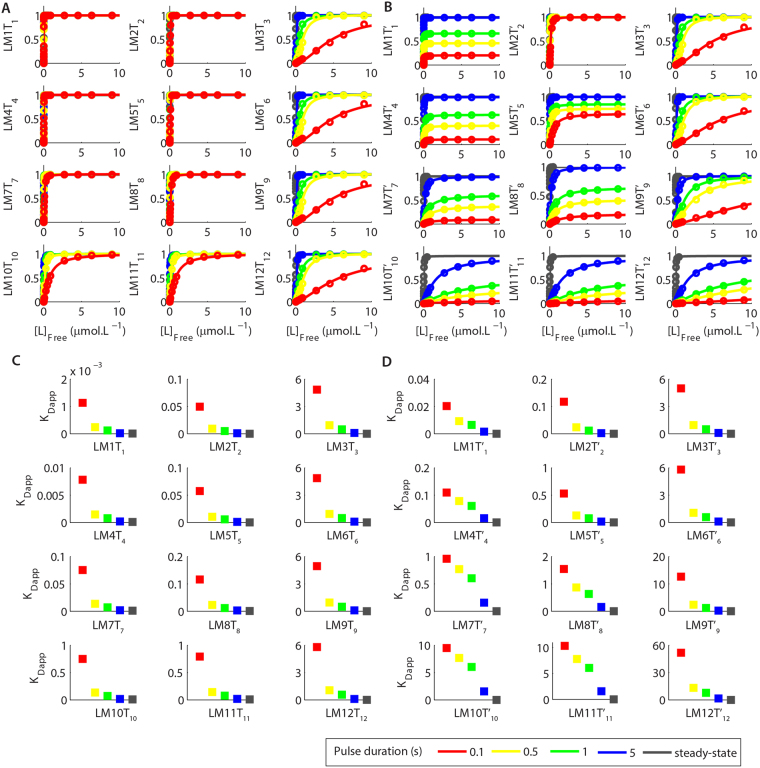
Initially, we simulated the formations of LM1T1-LM12T12 and LM1T′1-LM12T′12 at steady-state as functions of different [L]free to obtain dose-response curves fitted with equation 2 and estimate the control KDs and nhill. Note that the KD used for the interactions of LM1-LM12 with T1-T12 and with T′1-T′12 are identical (KD = 0.01 µmol.L−1). Nevertheless, the control KDs for the formations of LM1T1-LM12T12 and LM1T′1-LM12T′12 as functions of [L]free varied according to the KD of their binary precursors. Thus, the ternary complexes formed by LM1, LM2 and LM3 (LM1T1, LM2T2, LM3T3, LM1T′1, LM2T′2 and LM3T′3) exhibited control KDs as functions of [L]free of approximately 0.00001 µmol.L−1 (Fig. 7, Suppl. Table S2), which is 1000-fold lower than the control KDs of their binary precursors (Suppl. Table S1). The ternary complexes formed by LM4, LM5 and LM6 (LM4T4, LM5T5, LM6T6, LM4T′4, LM5T′5 and LM6T′6) had control KDs as a function of [L]free of 0.0001 µmol.L−1, which is higher than the KDs of the ternary complexes LM1T1-LM3T3 and LM1T′1-LM3T′3, but is also 1000-lower than the KDs for the formations of their precursors LM4, LM5 and LM6 (Fig. 7, Suppl. Table S2). The same pattern was also observed for the other ternary complexes simulated. Consequently, all the ternary complexes exhibited control KDs for their activations as functions of [L]free at steady-state approximately 1000-fold lower than the KDs of their binary precursors, which demonstrated that the binding of each binary complex to a target affected its interaction with L. This type of alteration is commonly observed in biological systems18,38.
Figure 7.
Detection and discrimination of dynamic signals by sequential reactions. (A) Dose-response curves of the complexes LM1Tf1-LM12Tf12 as functions of [L]free with different durations. (B) Dose-response curves of the complexes LM1T′1-LM12T′12 as functions of [L]free with different durations. (A,B) Values of KDapp estimated for the activation of LM1Tf1-LM12Tf12 (C) and LM1T′1-LM12T′12 (D) for pulses of [L]free with different durations. The legend indicates the colour code used to represent durations of the pulses of [L]free.
Next, we used square pulses of [L]free with different durations (100 ms, 500 ms, 1 s, and 5 s) and peak concentrations to investigate how they regulated the formations of the ternary complexes LM1T1-LM12T12 and LM1T′1-LM12T′12. The results obtained were used to trace dose-responses curves of the peak concentrations of LM1T1-LM12T12 and LM1T′1-LM12T′12 as functions of the peak [L]free using equation 2. The curves were used to verify whether the ternary complexes simulated detected and discriminated the durations of the signals of [L]free by changing their values of KDapp, nHill and maximum activation (Amax) in comparisons to the values observed at steady-state (Fig. 7A,B).
In the previous sessions, we demonstrated that the rate constants used for the interactions of L with its targets M1-M12 modulated their KDapp and nHill. The results showed in Fig. 7A,B revealed that the rate constants used in the reactions of M1-M12 with L can also modulate the values of Amax obtained for the dose-response curves of the formations of their respective ternary complexes. Thus, binary precursors that dissociated with fast kbs (≥1 s−1) from L impaired the Amax observed for the activation of their corresponding ternary complexes. However, such impairment only occurred for the ternary complexes formed with slow kf (0.1 µmol−1.L.s−1), which indicates that it is the combination of the kb of the precursor with the kf for its interaction with its target that regulates Amax (Fig. 7B) and, in addition, also affected the values of KDapp and nHill (Fig. 7C,D and Suppl. Fig. S8).
Our results demonstrated that all ternary complexes simulated decoded the pulses duration tested and exhibited changes in their KDapp values in comparisons to their control KD values observed at steady-state (Fig. 7C,D). Yet, all the ternary complexes that exhibited impairments of Amax for the durations of pulses of [L]free also showed higher shifts in their values of KDapp in comparison to the values observed at steady-state (Fig. 7C,D). Thus, the combination of short half-lives of fast dissociating binary precursors greatly impaired the formation of ternary complexes that associate with slow kf. For instance, the binary complexes LM4, LM5 and LM6, which share the same control KD, dissociated with kb of 100 s−1, 1 s−1 and 0.01 s−1, respectively. Due to the fast inactivation rate of LM4, the dose-response curves of activation of LM4T′4 as functions of pulses of [L]free with different durations exhibited strong modulations of Amax, but a similar modulation was not observed for the activation of LM4T4, which reacted faster with its precursor. Moreover, LM4T′4 exhibited much higher values of KDapp than LM4T4 for the pulses of [L]free tested though both species have the same binary precursor and identical control KD. The curves for the activations of LM5T′5 showed a similar pattern observed for the curves of LM4T′4, but with less pronounced modulations as its binary precursor had a slower kb. LM5T′5 also exhibited higher values of KDapp than LM5T5, though both complexes have identical control KD values and interact with the same binary precursor. In contrast, the dose-response curves of LM6T′6 had no variation in their Amax because its precursor, LM6, had a slow kb. In addition, the values of KDapp verified for LM6T6 were very similar to the values observed for LM6T′6. Thus, the slow time course for the inactivation of LM6 allow it to act as a “molecular memory” and propagate the transient signals of L for longer periods in comparison to LM4 and LM5.
In Suppl. Fig. S8A and B we plotted the nHill obtained for each dose-response curve showed in Fig. 7A and B. Our results demonstrated that, outside mass-action equilibrium, the formations of LM1T1-LM12T12 showed higher values of nHill in comparison to their binary precursors LM1-LM12, which indicated an increase in bistability along the sequential reactions simulated caused exclusively by kinetic factors (Suppl. Fig. S8A). Nevertheless, for the ternary complexes LM1T′1-LM12T′12, the curves that presented impairments of Amax exhibited values of nHill close to 1 and often lower than the values observed for their binary precursors (Suppl. Fig. S8B). These results indicated that, when we used pulses of [L]free with different durations to promote the formation of LM1T′1-LM12T′12, the short half-life of binary precursors with fast kbs impaired their activations and affected not only their Amax and KDapp, but also their values of nHill.
Detection and discrimination of dynamic signals in competing systems
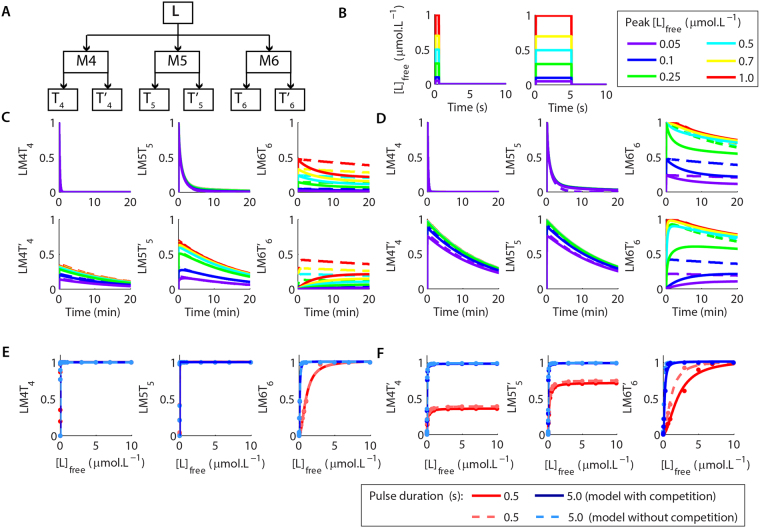
The last stage of our work consisted in investigating how competition among different molecules shapes their response. For this analysis, we used a simplified version of our system containing only the formation of LM4, LM5 and LM6, which exhibit very distinct patterns of activation even though they share the same control KD. Each one of this species were responsible for the activation of two targets simulated as described in the previous session. However, for this stage of the work, we simulated the two targets activated by each LM complex competing for their activators (Fig. 8A). We then used pulses of L with different durations (0.5 s and 5 s) to verify the consequences of competition in the results previously described (Fig. 8B–D).
Figure 8.
The role of competition in the detection and discrimination of dynamic signals. (A) The simulated system consisted of 3 targets activated by L (M4, M5, and M6) forming the complex LM4, LM5 and LM6. Each complex activated two substrates in a competitive manner. (B) We used pulses of [L]free with different durations (0.5 and 5 s) and peak concentrations to verify the role of competition in the activation of the targets of LM4, LM5, and LM6. (C,D) The results obtained showed that competition did not affected the peak activation of fast-reacting targets (LM4T4, LM5T5, LM6T6) for pulses of [L]free with 0.5 s (C) or 5 s (D) of duration in comparison to the results without competition (dashed lines). However, the presence of competition impaired the peak activation of slow reacting-targets (LM4T′4, LM5T′5, LM6T′6), promoting a slightly reduction in the peak amplitude of their dose-response curves of activation and a shift in their values of KDapp (E-F), which became larger (Suppl. Table S3).
The competition for a common activator had two consequences in our system. For the formation of fast-reacting ternary complexes (LM4T4, LM5T5, and LM6T6), independently of the pulse duration tested, the competition with slow-reacting ternary complexes (LM4T′4, LM5T′5, and LM6T′6) did not affect the maximum amplitude of activation for each pulse tested, but accelerated the inactivation of LM6T6 (Fig. 8B–D). In contrast, for the slow-binding complexes (LM4T′4, LM5T′5, and LM6T′6), the presence of competition affected their maximum activation, which reduced slightly the Amax of their dose-response curves of activation (8E-F, Suppl. Table S3). Moreover, for LM6T′6 competition promoted a delay in its activation curves (Fig. 8B–D) and shifted the KDapp of its dose-response curves (8E-F). Taken together, these results indicated that the effects of competition vary depending on the combination of the half-life of the initial precursor (LM4, LM5, and LM6) with the rates of activation of the subsequent molecules.
Discussion
Time is an important variable in the biological environments. The temporal dynamics of cellular signals regulate many molecules and signalling networks13,14,18,22,23,37. However, the comprehension of the molecular mechanisms that underlie the temporal regulation of cellular processes remains a challenge because much of our understanding of signalling processes results from data obtained at equilibrium or steady-state conditions. In this work, we focused on the identification of the molecular mechanisms that underlie the detection and discrimination of the temporal features of signals. For that, we simulated reactions of association and dissociation between molecules and a ligand. Previously, we used realistic models of different biomolecules with intricate interactions with endogenous ligands to investigate their modes of activation18. However, the level of complexity of our previous work made the comparison among different molecules difficult. Thus, in this work we have opted to use a generic and simpler system.
Biological systems are open systems in constant change16,18,19. The concentrations and levels of activation of biomolecules fluctuate continually, which sets perfect conditions for several reactions to proceed at non-equilibrium16,18,19. Consequently, many reactions in biological systems detect and discriminate dynamic signals and use temporal properties to display differential patterns of activation13,14,22,23,37. Because these reactions are sensitive to the temporal features of their components, they are under kinetic control and thermodynamic parameters such as KD do not predict their outcomes, which is a conclusion fully supported by our results.
Several data have revealed that the association and dissociation rate constants (kf and kb, respectively) for the interaction between biomolecules or of a drug with its targets are often more important than the binding affinity of the resulting complexes19,21,39,40. However, though in some systems the values of kf are crucial21, especially for the interaction of drugs with their targets the kb appears to play a more fundamental role19,40. Our results indicated that both rate constants are important in the detection of dynamical signals because they play different roles. At non-equilibrium, the kfs used in our simulations played a predominate role in determining the levels of activation of the ligand/molecule complexes simulated. The faster were kfs, the lower were the KDapps obtained, which indicate that molecules with fast kfs activate better at non-equilibrium. Similar results were observed previously21. Our results showed that the affinities observed at equilibrium do not ensure which molecules will activate in larger amounts18,19,21. A high affinity complex with slow rate constants can display a KDapp equivalent to the KDapp of a weak affinity complex when their reactions occur outside mass-action equilibrium. Only when the reactions approach mass-action equilibrium their rate constants become less important and their outcomes gradually become defined by their control KDs30.
In our simulations, the kbs played a pivotal role determining the time required for each reaction to achieve equilibrium. The slower was the kb used, the larger was the range of signal durations that a reaction detected and discriminated and the longer was the time required for it to reach equilibrium. Slow kbs also promoted reactions sensitive to the frequencies and number of pulses of reacting signals. Nevertheless, the slower was the kb used in our simulations, the better the reactions detected the number of pulses despite of their interpulse interval, which indicates that reactions with slow kbs integrate the signals over time more efficiently. These results contribute to explain why some molecules are sensitive to the interpulse interval of their signals, while others count pulses of signals regardless of their frequencies18,23. In addition, this type of information is crucial for the design of artificial signalling systems and probes41,42. We also demonstrated that the kbs played a crucial role in the propagation of dynamic signals. Thus, ligand/molecule complexes that dissociate with fast kbs do not propagate efficiently fast signals for slow-interacting targets. Consequently, in this scenario, complexes that dissociate slowly propagate dynamic signals better. Several observations have demonstrated that often drugs that dissociate slowly from their endogenous targets are more efficient, though the reasons for this process are not totally understood19,40. In this work, we have not explored this process specifically. The time intervals of the dynamic signals that we investigated are more compatible with physiological signals. However, our observations are not restricted to endogenous molecules and might explain the role of rate constants on the efficacy of drugs as well.
Methods
We implemented the computational models using BioNetGen43, a rule-based software for modelling signaling networks and pathways. All simulations were solved deterministically.
To define the parameters of the model, we used KDs (0.01 µmol.L−1, 0.1 µmol.L−1, 1 µmol.L−1 and 10 µmol.L−1) commonly found for the interactions between biomolecules10,18,38,44,45. We defined the kinetic parameters of the model by setting a kf of 1000 µmol−1.L.s−1 as our upper limit, which is consistent with the second order rate constant of a diffusion limited reaction in the cellular milieu. The other two kfs used in the model (10 µmol−1.L.s−1 and 0.1 µmol−1.L.s−1) were defined by dividing 1000 µmol−1.L.s−1 by 100 and 10000, respectively, in order to simulate reactions that cover a large range of velocities. All the kfs used are within the range of values observed for the interactions of biomolecules, which typically vary from 0.001 µmol−1.L.s−1 to 1000 µmol−1.L.s−1 46,47. For instance, calcium ions interact with many calcium-binding proteins with rates typically in the range of diffusion-limited reactions48. The complex calcium/calmodulin activates many targets with rate constants of binding around 1–10 µmol−1.L.s−1 18,49. In contrast, protein kinase A, a tetrameric enzyme involved in several signaling processes, has rate constants for the binding of its catalytic and regulatory subunits that vary around 0.5 to 0.05 µmol−1.L.s−1 44. We estimated the kbs for the reactions of the model using equation 3:
| 3 |
The concentration of M1-M12 was set to 1 µmol.L−1 initially (Figs 1–6). In the simulations showed in Figs 7 and 8, we set the initial concentrations of M1-M12 (M4-M6 in Fig. 8) to 10 µmol.L−1 and the concentrations of T1-T12 and T′1-T′12 to 1 µmol.L−1 (T4-T6 and T4′-T6′ in Fig. 8). The interaction of LM1-LM12 with T1-T12 and T′1-T′12 were simulated separately.
To obtain the dose-response curves at steady-state (Figs 1 and 7A,B), we performed the simulations until the reactions had reached steady-state. Then, we annotated the final concentrations of the complexes investigated and the concentration of free L ([L]free). To trace the dose-responses curves using square pulses of L, we simulated non-conservative signals of [L]free. The durations and peak concentrations of the pulses were set by the simulations and were not changed due to interactions with M1-M12, which prevented the competition among them. We defined the durations of the pulses of L setting 10 ms as our lower limit, which corresponds to fast calcium ion signals28. We then systematically increased the durations of the pulses until all complexes LM1-LM12 exhibited KDapps compatible with their control KDs. The data used in the dose-responses curves showed in Figs 3 and 6 corresponded to the peak activations of LM1-LM12 obtained as functions of the peak [L]free. In Figs 3 and 6, we varied the peak concentrations of the pulses of L from 0 µmol.L−1 to ~450 µmol.L−1 to achieve saturation of all complexes LM1-LM12. In Fig. 7, we varied the peak concentrations of the pulses of L from 0 µmol.L−1 to ~200 µmol.L−1 to saturate the complexes LM1T1-LM12T12 and LM1T′1-LM12T′12. Nevertheless, we opted to plot the curves of Figs 3, 6 and 7 with a smaller range of concentrations of [L]free for better visualization. We fitted the dose-response curves showed in Figs 3, 7 and 8 and Suppl. Figs S4, S5 and S8 using Matlab curve fitting tool with 95% of confidence interval. The full description of the reactions and the parameters used in the models are listed in Suppl. Table S4.
Electronic supplementary material
Acknowledgements
Research supported by Sao Paulo Research Foundation (FAPESP) grant #2015/50122–0 and DFG-IRTG 1740/2, FAPESP grant #2014/08481-0, and IBM/FAPESP grant #2016/18825-4.
Author Contributions
G. Antunes, designed research, built the model, performed the simulations, analysed the data and wrote the manuscript. F.M. Simoes-de-Souza designed research, analysed the data and wrote the manuscript, A.C. Roque wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
A correction to this article is available online at https://doi.org/10.1038/s41598-018-24192-7.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20842-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Koshland DE, Goldbeter A, Stock JB. Amplification and adaptation in regulatory and sensory systems. Science. 1982;217:220–5. doi: 10.1126/science.7089556. [DOI] [PubMed] [Google Scholar]
- 2.Kholodenko BN, Hancock JF, Kolch W. Signalling ballet in space and time. Nat Rev Mol Cell Biol. 2010;11:414–26. doi: 10.1038/nrm2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo RP, Liotta LA, Petricoin EF. Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nature Reviews Drug Discovery. 2007;6:871–880. doi: 10.1038/nrd2381. [DOI] [PubMed] [Google Scholar]
- 4.Abdel-Rahman SM, Kauffman RE. The Integration Of Pharmacokinetics And Pharmacodynamics: Understanding Dose-Response. Annual Review of Pharmacology and Toxicology. 2004;44:111–136. doi: 10.1146/annurev.pharmtox.44.101802.121347. [DOI] [PubMed] [Google Scholar]
- 5.Kolch W, Halasz M, Granovskaya M, Kholodenko BN. The dynamic control of signal transduction networks in cancer cells. Nature Reviews Cancer. 2015;15:515–527. doi: 10.1038/nrc3983. [DOI] [PubMed] [Google Scholar]
- 6.Kiel C, Serrano L. Challenges ahead in signal transduction: MAPK as an example. Curr Opin Biotechnol. 2012;23:305–14. doi: 10.1016/j.copbio.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Androulakis IP, Kamisoglu K, Mattick JS. Topology and Dynamics of Signaling Networks: In Search of Transcriptional Control of the Inflammatory Response. Annual Review of Biomedical Engineering. 2013;15:1–28. doi: 10.1146/annurev-bioeng-071812-152425. [DOI] [PubMed] [Google Scholar]
- 8.Behar M, Barken D, Werner SL, Hoffmann A. The dynamics of signaling as a pharmacological target. Cell. 2013;155:448–61. doi: 10.1016/j.cell.2013.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy MB, Beale HC, Carlisle HJ, Washburn LR. Integration of biochemical signalling in spines. Nat Rev Neurosci. 2005;6:423–34. doi: 10.1038/nrn1685. [DOI] [PubMed] [Google Scholar]
- 10.Antunes G, Roque AC, Simoes-de-Souza FM. Stochastic Induction of Long-Term Potentiation and Long-Term Depression. Scientific Reports. 2016;6:30899. doi: 10.1038/srep30899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coultrap SJ, et al. Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Rep. 2014;6:431–7. doi: 10.1016/j.celrep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neveu D, Zucker RS. Postsynaptic levels of [Ca2+]i needed to trigger LTD and LTP. Neuron. 1996;16:619–29. doi: 10.1016/S0896-6273(00)80081-1. [DOI] [PubMed] [Google Scholar]
- 13.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–6. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 14.Behar M, Hoffmann A. Understanding the temporal codes of intra-cellular signals. Curr Opin Genet Dev. 2010;20:684–93. doi: 10.1016/j.gde.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winquist RJ, Mullane K, Williams M. The fall and rise of pharmacology – (Re-)defining the discipline? Biochemical Pharmacology. 2014;87:4–24. doi: 10.1016/j.bcp.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Swinney DC. Opinion: Biochemical mechanisms of drug action: what does it take for success? Nature Reviews Drug Discovery. 2004;3:801–808. doi: 10.1038/nrd1500. [DOI] [PubMed] [Google Scholar]
- 17.Lisman J. A mechanism for the Hebb and the anti-Hebb processes underlying learning and memory. Proc Natl Acad Sci USA. 1989;86:9574–8. doi: 10.1073/pnas.86.23.9574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antunes G, Roque AC, Simoes de Souza FM. Modelling intracellular competition for calcium: kinetic and thermodynamic control of different molecular modes of signal decoding. Scientific Reports. 2016;6:23730. doi: 10.1038/srep23730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copeland RA. The drug–target residence time model: a 10-year retrospective. Nature Reviews Drug Discovery. 2015;15:87–95. doi: 10.1038/nrd.2015.18. [DOI] [PubMed] [Google Scholar]
- 20.Pan AC, Borhani DW, Dror RO, Shaw DE. Molecular determinants of drug–receptor binding kinetics. Drug Discovery Today. 2013;18:667–673. doi: 10.1016/j.drudis.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Kiel C, Serrano L. Cell Type-Specific Importance of Ras-c-Raf Complex Association Rate Constants for MAPK Signaling. Science Signaling. 2009;2:ra38–ra38. doi: 10.1126/scisignal.2000397. [DOI] [PubMed] [Google Scholar]
- 22.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–8. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 23.Fujii H, et al. Nonlinear decoding and asymmetric representation of neuronal input information by CaMKIIα and calcineurin. Cell Rep. 2013;3:978–87. doi: 10.1016/j.celrep.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Carey, F. A. & Sundberg, R. J. Advanced organic chemistry. (Springer, 2007).
- 25.Alberty RA. A Short History of the Thermodynamics of Enzyme-catalyzed Reactions. Journal of Biological Chemistry. 2004;279:27831–27836. doi: 10.1074/jbc.X400003200. [DOI] [PubMed] [Google Scholar]
- 26.Garaschuk O, Schneggenburger R, Schirra C, Tempia F, Konnerth A. Fractional Ca2+ currents through somatic and dendritic glutamate receptor channels of rat hippocampal CA1 pyramidal neurones. J Physiol. 1996;491(Pt 3):757–72. doi: 10.1113/jphysiol.1996.sp021255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobczyk A, Scheuss V, Svoboda K. NMDA receptor subunit-dependent [Ca2 + ] signaling in individual hippocampal dendritic spines. J Neurosci. 2005;25:6037–46. doi: 10.1523/JNEUROSCI.1221-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zahradníková A, Poláková E, Zahradník I, Zahradníková A. Kinetics of calcium spikes in rat cardiac myocytes: Kinetics of calcium spikes. The Journal of Physiology. 2007;578:677–691. doi: 10.1113/jphysiol.2006.117796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vauquelin G. Effects of target binding kinetics on in vivo drug efficacy: koff, kon and rebinding: Exploring drug rebinding in vivo. British Journal of Pharmacology. 2016;173:2319–2334. doi: 10.1111/bph.13504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swinney DC, Haubrich BA, Liefde IV, Vauquelin G. The Role of Binding Kinetics in GPCR Drug Discovery. Curr Top Med Chem. 2015;15:2504–22. doi: 10.2174/1568026615666150701113054. [DOI] [PubMed] [Google Scholar]
- 31.Weiss JN. The Hill equation revisited: uses and misuses. FASEB J. 1997;11:835–841. doi: 10.1096/fasebj.11.11.9285481. [DOI] [PubMed] [Google Scholar]
- 32.Ferrell JE. How responses get more switch-like as you move down a protein kinase cascade. Trends Biochem Sci. 1997;22:288–9. doi: 10.1016/S0968-0004(97)82217-7. [DOI] [PubMed] [Google Scholar]
- 33.Trunnell NB, Poon AC, Kim SY, Ferrell JE. Ultrasensitivity in the Regulation of Cdc25C by Cdk1. Mol Cell. 2011;41:263–74. doi: 10.1016/j.molcel.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldbeter A, Koshland DE. An amplified sensitivity arising from covalent modification in biological systems. Proc Natl Acad Sci USA. 1981;78:6840–4. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Antunes G, De Schutter E. A stochastic signaling network mediates the probabilistic induction of cerebellar long-term depression. J Neurosci. 2012;32:9288–300. doi: 10.1523/JNEUROSCI.5976-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomerening JR, Sontag ED, Ferrell JE. Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nature Cell Biology. 2003;5:346–351. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 37.De Koninck P, Schulman H. Sensitivity of CaM kinase II to the frequency of Ca2+ oscillations. Science. 1998;279:227–30. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 38.Olwin BB, Edelman AM, Krebs EG, Storm DR. Quantitation of energy coupling between Ca2+, calmodulin, skeletal muscle myosin light chain kinase, and kinase substrates. J Biol Chem. 1984;259:10949–55. [PubMed] [Google Scholar]
- 39.Yin N, Pei J, Lai L. A comprehensive analysis of the influence of drug binding kinetics on drug action at molecular and systems levels. Molecular BioSystems. 2013;9:1381. doi: 10.1039/c3mb25471b. [DOI] [PubMed] [Google Scholar]
- 40.Copeland RA, Pompliano DL, Meek TD. Drug–target residence time and its implications for lead optimization. Nature Reviews Drug Discovery. 2006;5:730–739. doi: 10.1038/nrd2082. [DOI] [PubMed] [Google Scholar]
- 41.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 42.Kiel C, Serrano L. Structural Data in Synthetic Biology Approaches for Studying General Design Principles of Cellular Signaling Networks. Structure. 2012;20:1806–1813. doi: 10.1016/j.str.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Faeder JR, Blinov ML, Hlavacek WS. Rule-based modeling of biochemical systems with BioNetGen. Methods Mol Biol. 2009;500:113–67. doi: 10.1007/978-1-59745-525-1_5. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P, et al. Single Turnover Autophosphorylation Cycle of the PKA RIIβ Holoenzyme. PLOS Biology. 2015;13:e1002192. doi: 10.1371/journal.pbio.1002192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayley PM, Findlay WA, Martin SR. Target recognition by calmodulin: dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–28. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou H-X, Bates PA. Modeling protein association mechanisms and kinetics. Current Opinion in Structural Biology. 2013;23:887–893. doi: 10.1016/j.sbi.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109:839–60. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faas GC, Raghavachari S, Lisman JE, Mody I. Calmodulin as a direct detector of Ca2+ signals. Nature Neuroscience. 2011;14:301–304. doi: 10.1038/nn.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintana AR, Wang D, Forbes JE, Waxham MN. Kinetics of calmodulin binding to calcineurin. Biochem Biophys Res Commun. 2005;334:674–80. doi: 10.1016/j.bbrc.2005.06.152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.