Abstract
Transposable elements (TEs) are able to jump to new locations (transposition) in the genome, usually after replication. They constitute the so-called selfish or junk DNA and take over large proportions of some genomes. Due to their ability to move around they can change the DNA landscape of genomes and are therefore a rich source of innovation in genes and gene regulation. Surge of sequence data in the past years has significantly facilitated large scale comparative studies. Cephalochordates have been regarded as a useful proxy to ancestral chordate condition partially due to the comparatively slow evolutionary rate at morphological and genomic level. In this study, we used opsin gene family from three Branchiostoma species as a window into cephalochordate genome evolution. We compared opsin complements in terms of family size, gene structure and sequence allowing us to identify gene duplication and gene loss events. Furthermore, analysis of the opsin containing genomic loci showed that they are populated by TEs. In summary, we provide evidence of the way transposable elements may have contributed to the evolution of opsin gene family and to the shaping of cephalochordate genomes in general.
Introduction
Transposable elements (TEs) are complicated biological entities able to replicate and jump to new locations (transposition) in the genome. Rather simple models have been defined to study their dynamics1, while their classification is also problematic. The first TE-classification system2, distinguishes two classes of TEs, based on the transposition intermediate: RNA (class I or retrotransposons) and DNA (class II or DNA transposons), which follow a “copy-and-paste” and “cut-and-paste” mechanism, respectively. This system was later modified in order to include bacterial, non-autonomous TEs (such as the Miniature Inverted Repeat Transposable Elements - MITEs) and other types of TEs that couldn’t fall in any of these two categories. Curcio and Derbyshire3 categorized transposons according to the way they move, determined by their transposase proteins. A hierarchical classification system for eukaryotic TEs has been proposed by Wicker, et al.4, which takes into account not only the replication strategy but also the structure of the encoded proteins and of the non-coding domains, the presence and size of the target site duplication (TSD) and even some phylogenetic data.
It was long ago speculated that TEs can “control the time and type of activity of individual genes”5, or in other words they play key role in a variety of gene regulatory networks and lately there is accumulating information in favor of this theory (revised by Chuong et al.6 and Bourque7). This can be achieved either by the insertion of TEs in the proximity of genes and consequently the generation of new regulatory elements7 or the emergence of new regulatory proteins8. In fact, TEs occupy a large proportion of the regulatory control regions (revised by Feschotte8). On one hand, TEs alter gene expression (activate or inactivate genes); on the other hand, they promote inversions and deletions of chromosomal DNA, they can create new genes (or exons), or serve as illegitimate recombination hotspots. Consequently, they contribute to the shaping of the genome’s architecture, its evolution and the emergence of genetic innovations9–12. TE-associated chromosomal rearrangements can be driven by two mechanisms, in particular via homologous recombination13 or by an alternative transposition process14.
TEs are main components of eukaryote and prokaryote genomes and they are known to occupy large portions of vertebrate, invertebrate and plant genomes in particular15–19. Long-terminal repeat retrotransposons (LTRs) are the predominant order of TEs in plants20, whereas the Non-LTR TEs are the most commonly encountered in the human genome21 and Alu repetitive elements, in particular, are known to generate deletions, duplications and complex genomic rearrangements22.
The subphylum Cephalochordata, a.k.a. amphioxus or lancelets, have been regarded as a key animal group for understanding the origin of vertebrates, and a useful proxy to the ancestral chordate condition. This is in part due to the presumed slow evolutionary rate within the cephalochordate lineage both at the morphological and the genomic level. Cephalochordata are comprised of the three genera, namely Branchiostoma, Asymmetron and Epigonychtys23. It was recently found that Cephalochordata preserve a high TE diversity in comparison to modern vertebrates24. In fact, a comparative analysis of TEs in various genomes has revealed that they constitute 28% of B. floridae genome25. Amphioxus TEs belong to more than 30 superfamilies, which are highly heterogeneous as generally none of their members are drastically more abundant than others, and none of the TEs seems to have suffered any massive expansion26. The phylogenetic relationship within the extant amphioxus lineage was investigated27 providing divergence time estimates and suggesting a rather recent diversification within Branchiostoma genus, with divergence time similar e.g. to that between rodents belonging to Muridae family (mouse and rat)28. Whole genome comparative study of B. belcheri and B. floridae indicated high rate of proteome diversification24, which might however be explained at least in some cases by the gene prediction errors29.
In order to provide an insight into the possible role of TEs in cephalochordate genome evolution we focused on the opsin gene family, a member of the G-Protein Coupled Receptor (GPCR) gene superfamily. Opsins play crucial role in light detection in animals and their number differs significantly among species, with no apparent correlation to the overall level of body plan sophistication. Opsins classification, interfamily relationships and evolution of animal vision have been studied extensively30–39. Opsins can be roughly clustered into four major groups, namely the ciliary opsins expressed in ciliary photoreceptors (C-type), the rhabdomeric opsins expressed in rhabdomeric photoreceptors (R-type), the Group 4 opsins, and the Cnidarian opsins. Members of the three major groups were recently identified in the European lancelet40, whereas similar studies in the past were focused on the opsin complements of the Florida and Chinese lancelets41–43. By using manually curated and experimentally confirmed opsin complement of three Branchiostoma species, namely B. lanceolatum (Pallas 1774), B. floridae (Hubbs 1922) and B. belcheri (Gray 1847), we have identified gene duplication and loss events. Extrapolating from opsin gene family as an example, we try to address the question of how transposable elements may have been involved in the gene gain/losses and shaping of the Branchiostoma genus genome.
Materials and Methods
Gene Prediction, alignments, synteny and phylogenetic analysis
We analyzed both available Branchiostoma floridae genome assemblies, i.e. v1.0 through JGI, where two haplotypes are present (http://genome.jgi.doe.gov/Brafl1/Brafl1.home.html) and v2.0 through NCBI (http://www.ncbi.nlm.nih.gov/assembly/GCF_000003815.1/), from which most of the allelic scaffolds have been eliminated and is therefore a non-redundant mosaic of v1.0. All previously annotated opsin genes41 were validated through BLAST, Genscan44 and SpliceView45 analyses. In order to detect putative opsin homologs that were not previously reported, we conducted extensive keyword and BLAST searches. Newly identified opsin containing genomic loci were subjected to Genscan and SpliceView for de novo gene prediction. In the case of discrepancies between database gene models and our in silico analysis, PCR amplification of the “suspicious” regions was performed, followed by cloning and sequencing (see paragraph “Cloning and Sequencing of Opsin Gene Fragments/Transcripts”). Additionally, we thoroughly queried the B. belcheri HapV2(v7h2) and the v18h27.r3_ref_genome assemblies, available at the Chinese Lancelet (Amphioxus) Genome Sequencing project webpage (http://genome.bucm.edu.cn/lancelet/), applying both keyword and BLAST searches. In order to investigate the phylogenetic relationships of previously annotated and newly identified amphioxus opsins and thus establish orthology of opsin genes, a Maximum Likelihood tree was constructed according to Pantzartzi et al.40. The same dataset was used and it was enriched with B. floridae and B. belcheri sequences (Supplementary File 1 and Table 1). For each opsin gene, orthologs from the three Branchiostoma species were aligned using ClustalO46 and visualized using BoxShade. In the case of orthologs absent from one or two species, we used Circoletto47, in order to investigate synteny conservation and visualize sequence similarity among syntenic scaffolds from the Branchiostoma species. E-value for the BLAST run was set to e−20.
Transposable Elements Analysis
Genomic scaffolds containing opsins and those expected to contain opsin genes based on synteny analyses were screened for repetitive elements using Censor48 in the RepBase database49. NCBI Accession numbers for B. floridae scaffolds used are NW_003101565 (Bf_scaffold6), NW_003101418 (Bf_scaffold_187), NW_003101537 (Bf_scaffold_36), NW_003101507 (Bf_scaffold_98) and NW_003101409 (Bf_scaffold_196). The genomic regions used were: Bf_scaffold_6: 305,868–729,662 or 305,868–547140 (Comparison of Narrow Regions, CNR); Bf_scaffold_187: 4,135,366–4,628,754 or 4,135,366–4,378,895 (CNR); Bl_Sc0000005: 5,300,000–7,300,000 or 6,885,201–7,300,000 (CNR); Bb_scaffold48: 1–2,523,832 or 1,200,000–2,523,832 (CNR); Bf_scaffold_36: 4,567,754–4,488,902, Bf_scaffold_98: 4,107,000–4,213,900, Bl_Sc0000154: 143,384–219,100, Bl_Sc0000040: 850,000–1,050,000, Bb_Sc0000263: 1–200,000; Bb_scaffold123: 447,402–528,601; Bf_scaffold_196: 2,792,247–2,817,466, Bl_Sc0000011: 2,118,981–2,146,160; Bb_Sc0000116: 763,100–794,099.
Animal Collection
B. floridae adults were collected in Old Tampa Bay (Florida, USA, no permission required for amphioxus collection). Housing of animals and in vivo experiments in the present study were performed in accordance with guidelines established by the Institute of Molecular Genetics and in compliance with national guidelines (ID#12135/2010–17210). All animal works were also conducted according to the National Institute of Health standards as underlined by the “Guide for Care and Use of Laboratory Animals’’. Gametes were obtained and embryos raised, as previously described50. Staging of all collected embryos was performed according to Hirakow and Kajita51, specimens from late neurula (N3), larvae (L1–L3) and adult stage were collected and frozen in RNAlater® Stabilization Solution (ThermoFisher Scientific), under light conditions.
RNA Isolation/cDNA Preparation
Total RNA was isolated from B. floridae embryos stored in RNAlater® Stabilization Solution using the Trizol reagent (Ambion). To avoid genomic DNA contamination, isolated RNA was treated with DNaseI and purified on RNeasy Mini Kit (Qiagen) column. Random-primed cDNA was prepared from 250ng of RNA in a 20 μl reaction using SuperScript VILO cDNA Synthesis kit (Invitrogen).
Cloning and Sequencing of Opsin Gene Fragments/Transcripts
For validation of the in silico predicted gene models, cloning and sequencing of opsin gene fragments and complete transcripts from B. floridae was performed, according to Pantzartzi et al.40. Primers used are included in Supplementary Table 2.
qRT-PCR
Primers used are provided in Supplementary Table 2. Experiments and analysis of results were performed according to Pantzartzi et al.40. TBP was used as the housekeeping gene.
Results
Identification, classification and genome organization of opsin genes in the Branchiostoma genus
We initially performed a thorough comparative analysis of the opsin gene repertoires of three cephalochordate species. We used the recently reported genes from B. lanceolatum40 together with previously reported genes from B. floridae and B. belcheri41–43 many of which had to be re-predicted and some were de novo identified in the current study (Supplementary Table 1). Final transcripts and encoded proteins for newly characterized and modified opsins from B. floridae and B. belcheri as well as details on gene organization and genomic location are provided in Supplementary File 1. Orthology of identified genes was validated by synteny and phylogenetic analysis (Supplementary Fig. 1). The alignments of orthologs for each opsin gene from the three Branchiostoma species are provided in Supplementary File 1. Orthologs have the same number of exons; the sole exceptions are op7 and op20. Orthologous exons have almost identical size, however, pronounced changes are observed in the size of the last exon. Furthermore, there is a great similarity among orthologs in terms of sequence, with the C-terminus being the most variable. Evidently, opsin genes are spread over 16 genomic regions (scaffolds) in B. floridae and 14 in B. belcheri (Supplementary Fig. 2). Phylogenetic analysis (Supplementary Fig. 1) in combination with the arrangement of opsin genes in the genomes of the three species (Supplementary Fig. 2) supports the fact that the majority of opsin genes are represented by an ortholog in all three species (Table 1). This is not the case for op6, op12b, op13b, and op17b, which seem to be the result of a gene duplication.
Table 1.
Opsin repertoire in the Branchiostoma genus.
| B. lanceolatum | B. floridae | B. belcheri | ||
|---|---|---|---|---|
| C-type opsins | op1 | + | + | + |
| op2 | + | + | + | |
| op3 | + | + | + | |
| op4 | + | + | + | |
| op5 | + | + | + | |
| Neuropsins | op6 | − | + | − |
| op7 | + | + | + | |
| op8 | + | + | + | |
| Go opsins | op9 | + | + | + |
| op10 | + | + | + | |
| op11 | + | + | + | |
| op12a | + | + | + | |
| op12b | − | − | + | |
| op13a | + | + | + | |
| op13b | + | − | − | |
| Peropsins | op14 | + | + | + |
| Melanopsins | op15 | + | + | + |
| Amphiop6 | op16 | + | + | + |
| op17a | + | + | − | |
| op17b | + | − | − | |
| op18 | + | + | + | |
| op19 | + | + | + | |
| op20 | + | + | + | |
| op21 | + | + | + | |
| TOTAL | 21+1pseudo | 21 | 20 | |
Note: Numbering of genes is based on Pantzartzi et al.40.
We further analyzed the opsin expression pattern across different developmental stages (Supplementary Fig. 3) of B. floridae. Onset of several opsin genes expression starts at L1 stage, in which frontal eye and lamellar body (ciliary photoreceptive organs) start to develop. In agreement with B. lanceolatum40, the majority of the B. floridae opsins show most predominant expression in L2/3 stages, where all of the known amphioxus photoreceptor organs are differentiated. Nevertheless, differences are observed between the two species in regard to the onset of expression of op13a. Interestingly, op6, a gene detected only in B. floridae, follows a distinct pattern in regard to the other two neuropsins (i.e. op7 and op8), for which expression patterns are the same for both B. floridae and B. lanceolatum.
Transposable elements and opsin genes in the Branchiostoma genus
Differences have been noted among the three Branchiostoma species in regard both to the structure and the number of opsin genes (Table 1 and Supplementary File 1). Since transposable elements (TEs) have been vastly implicated in gene structure alteration as well as gene duplications and losses, we scanned scaffolds containing altered genes against RepBase to locate TEs populating these regions; for opsin orthologs that are absent from one or two Branchiostoma species (Table 1), we found the syntenic scaffolds and also scanned them against RepBase.
The beginning of forth exon of Bl_op2 is occupied by small repeated sequences, a fact that leads to elongation of the third cytoplasmic loop (Supplementary File 1). Noticeably, the fifth intron of Bl_op8 highly resembles a satellite locus from Salmo salar (SAT-11_SSa in RepBase). In fact, the beginning of the last exon is one of the repeat units. It is also worth mentioning that the last exon of Bl_op16 is longer in size than the respective exons from the B. floridae and B. belcheri orthologs due to palindromic repeats at its end (Supplementary File 1). Bl_op16 is flanked by a truncated and a complete copy of the DNA transposon Ginger2-1 and the non-autonomous DNA transposon Harbinger-N11 (data not shown).
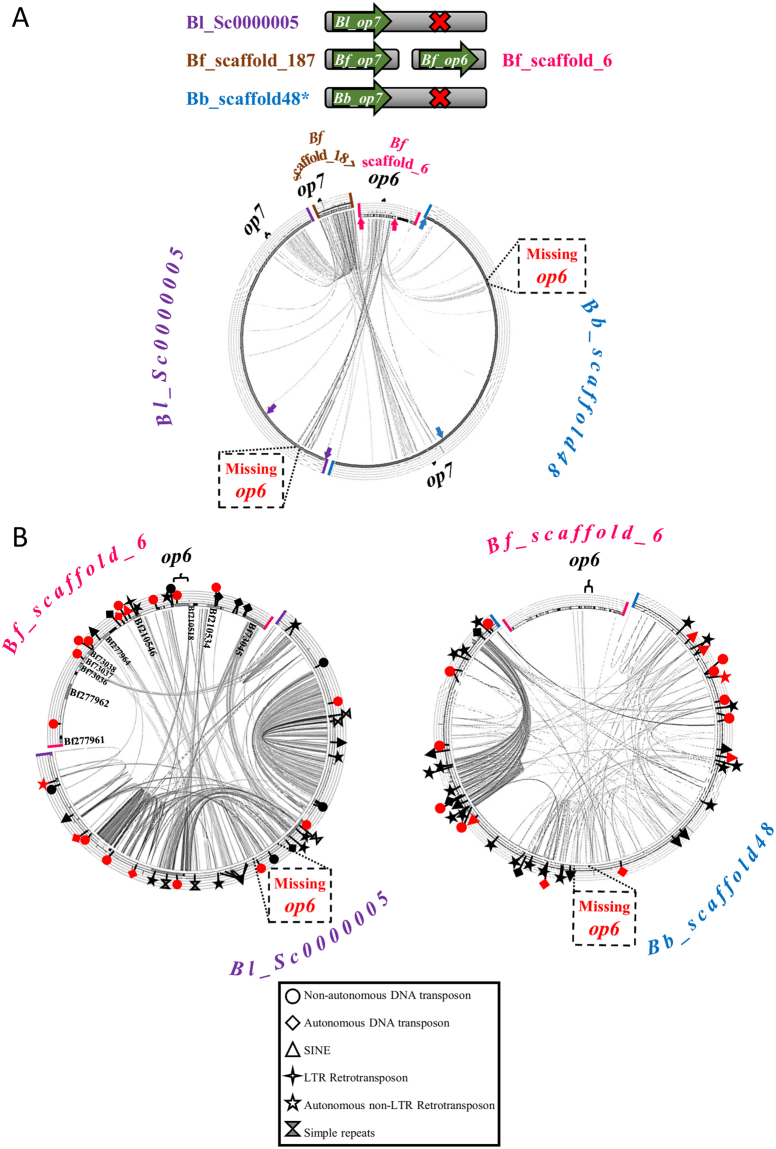
Comparison of the syntenic scaffolds related to op6 is portrayed in Fig. 1. High similarity is observed among the genomic regions containing op7 in B. floridae, B. lanceolatum and B. belcheri (Fig. 1A). Similarity is also observed between the genomic regions flanking op6 in B. floridae and B. lanceolatum Sc0000005 and B. belcheri scaffold48, however, there are no traces of op6 in the other two species. Some of the immediately flanking genes of Bf_op6 have their orthologs in B. lanceolatum (only one seems to be eliminated, namely Bf210534), but are duplicated in the latter, with more striking example that of Bf73045 (Fig. 1B). Duplication of other genomic fragments in the region where Bl_op6 was supposed to be is also evident. Numerous families of transposable elements and simple repeated sequences of varying size (2–65 bp) have been detected within and in the proximity of the duplicated genes and genomic fragments in Bl_Sc0000005 (see Supplementary Fig. 4A for names of TEs). A similar case of duplicated genomic fragments populated by transposable elements is also observed in B. belcheri. What is even more appealing is the number and type of transposable elements within Bf_op6 and Bf_op7 genes and in their vicinity (Supplementary Fig. 4B). No other conservation at genomic level is observed between B. floridae scaffolds 6 and 187, apart from the opsin genes and various transposable elements, as shown in Supplementary Fig. 4B.
Figure 1.
Comparison of genomic loci containing or lacking op6 and op7. (A) Comparison of the op6 containing Bf_scaffold_6 and the op7 containing Bf_scaffold_187 with the Bl_Sc0000005 and the Bb_scaffold48 that apparently contain only op7 and lack op6. (B) Comparison of more narrow regions of Bf_scaffold_6 (delimited by arrows in (A)), with Bl_Sc0000005 (left) and Bb_scaffold48 (right). A high degree of duplicated regions was observed for B. lanceolatum, with the most striking example that of Bf73045 (left). Duplicated regions were also observed for B. belcheri (right). Red and black (complete and partial copies based on the RepBase database) symbols mark the position of simple tandem repeats and various families of Transposable Elements (TEs) (see key legend for explanation and Supplementary Fig. 4A for TE names). For the sake of clarity, predicted B. floridae gene models are listed only in the internal part of the Bf_scaffold_6 in (B). Ribbons connecting syntenic scaffolds under comparison denote similarity at genomic level.
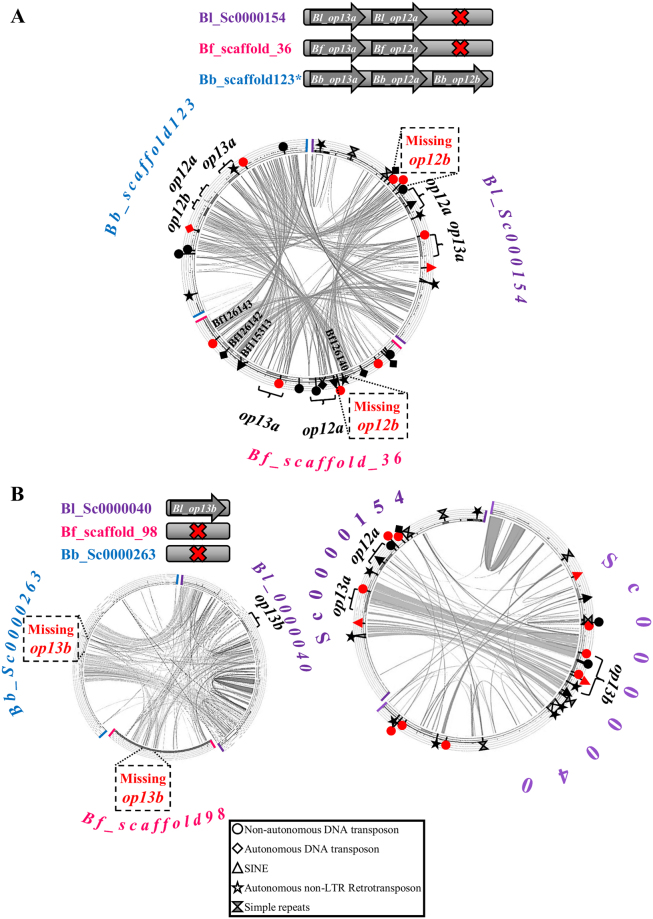
Differences are observed among the three species in regard to op12 and op13 copies (Table 1, Fig. 2 and Supplementary Fig. 2). In general, these genes exhibit high sequence similarity and contain the same number of exons. Size of the exons is almost identical, with a strikingly smaller last exon in Bb_op12b (Supplementary File 1). Comparison of scaffolds bearing op12a, op12b and op13a from the three species (Fig. 2A) shows that there is high conservation in opsin genes as well as in their flanking regions. However, no significant similarity exists in the intergenic regions of op12a and op13a. Interestingly, opsin genes in B. belcheri are flanked by complete copies of DNA transposons (Supplementary Fig. 4C). The absence of op13b ortholog from B. floridae and B. belcheri is evident from the comparison of syntenic scaffolds (Fig. 2B). On the other hand, scaffolds containing the B. lanceolatum op13a and op13b paralogs (Fig. 2C, Supplementary Fig. 4C) show a high degree of similarity only in the genic regions and their immediate neighborhood which does not extend further in the region of Bl_op12a. The region of similarity is bordered by simple repeats as well as complete or partial copies of TEs.
Figure 2.
Comparison of genomic loci containing or lacking op12a, op12b, op13a and op13b opsins. (A) Comparison of Bl_Sc0000154 with Bf_scaffold_36 and Bb_scaffold_23 (B) Comparison of B. lanceolatum scaffold containing the op13b gene with the syntenic scaffolds from B. floridae (Bf_scaffold_98) and B. belcheri (Bb_Sc0000263). (C) Comparison of B. lanceolatum scaffolds bearing opsins op13a (Sc0000154) and op13b (Sc0000040). Red and black (complete and partial copies based on the RepBase database) symbols mark the position of simple tandem repeats and various families of transposable elements (TEs) (see key legend for explanation and Supplementary Fig. 4B and C for TE names). Predicted B. floridae gene models are listed in the internal part of the scaffolds.
Another example of putative gene duplication and loss event is that of op17a and op17b (Fig. 3). Using the neighboring genes of Bf_op17a we detected the syntenic scaffold in B. belcheri. Comparison of the three scaffolds shows conservation in the flanking regions but no traces of a Bb_op17a gene. Instead, in the region where Bb_op17a is expected to be, there are copies of retrotransposons52 (Supplementary Fig. 4D). Bl_op17a and Bl_op17b genes are as well flanked by autonomous and non-autonomous transposons.
Figure 3.
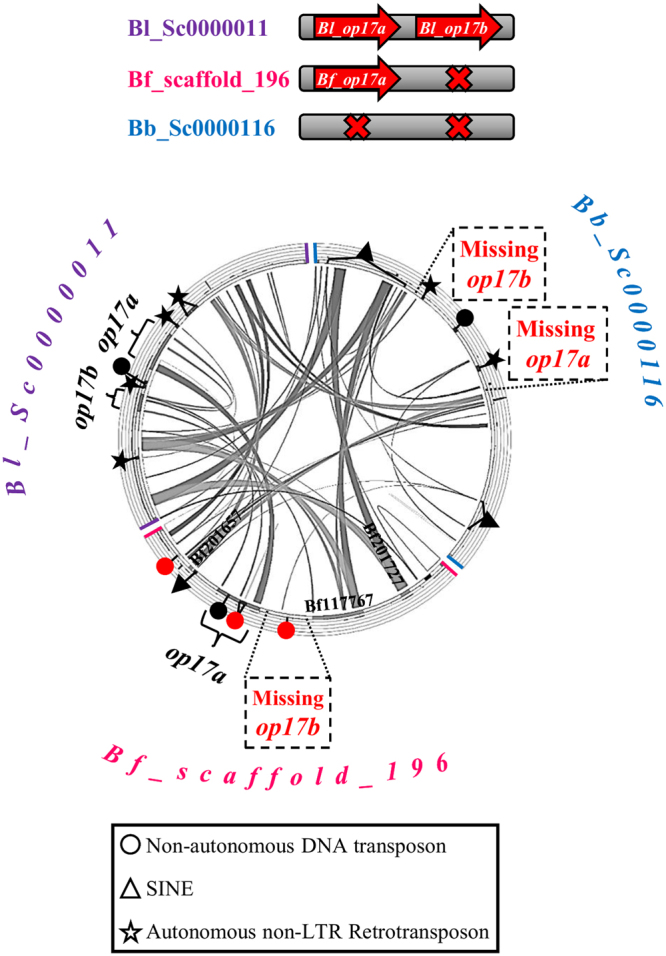
Comparison of genomic loci containing or lacking op17a and op17b opsins. Comparison of the op17a and op17b containing scaffold from B. lanceolatum with the op17a-containing B. floridae scaffold and the syntenic scaffold from B. belcheri that obviously lacks both op17a and op17b. A clear conservation of the genomic regions is observed. Red and black (complete and partial copies based on the RepBase database) symbols mark the position of various families of transposable elements (TE) (see key legend for explanation and Supplementary Fig. 4D for TE names). Predicted B. floridae gene models are listed in the internal part of the scaffolds.
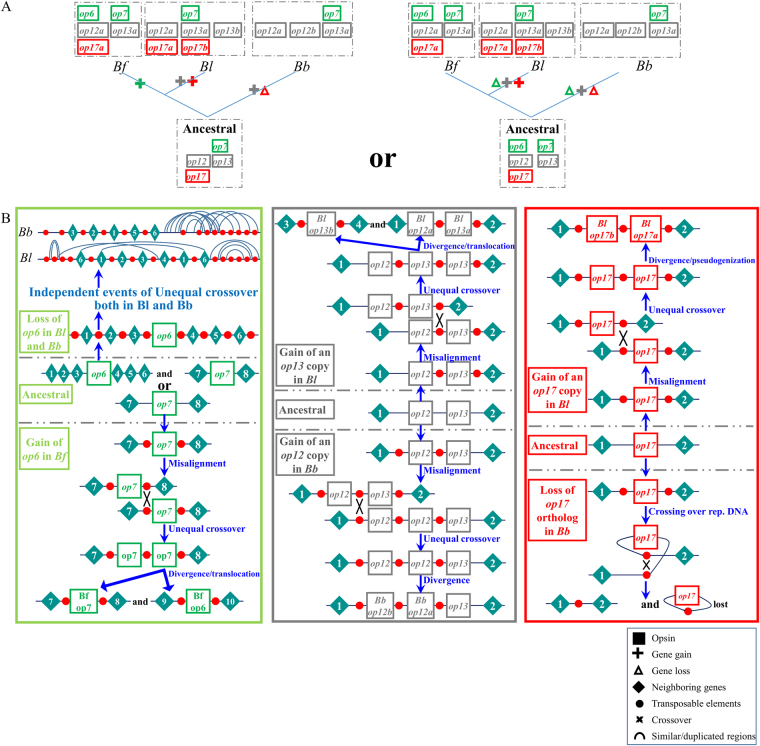
To summarize our previous findings, we could say that independent events of gene duplications and losses occurred during the evolution of Branchiostoma opsins (Fig. 4A). Taking into account the higher similarity between B. lanceolatum and B. belcheri regions, the almost identical structure of Bf_op6 and Bf_op7 and the presence of common transposable elements within and outside these two genes, we could conclude that op6 is the result of a duplication event in B. floridae, after its split from B. lanceolatum. However, we cannot rule out the possibility that op6 existed in the common ancestor of the Branchiostoma species and it was eliminated in the lineages of B. lanceolatum and B. belcheri. We could also conclude that Bb_op12a and Bl_op13a were independently duplicated in B. belcheri and B. lanceolatum. Finally, we assume that op17a was lost in B. belcheri and op17b is the result of a gene duplication only in B. lanceolatum (Fig. 4A). Figure 4B outlines what the ancestral state could have been for each of the duplicated/lost genes and the putative mechanisms through which gene gains and losses took place. Complete and partial copies of TEs identified in the vicinity of opsin genes probably served as illegitimate spots for recombination, leading to misalignment, unequal crossover and hence duplication of an opsin gene, as in the case of op12 and op13, or caused crossing over of the same chromosome, leading to the deletion of op17 in B. belcheri.
Figure 4.
Reconstruction of the evolutionary history of opsin family in the Branchiostoma genus. (A) Schematic representation of gene gains and losses in the lineages of B. lanceolatum, B. floridae and B. belcheri. (B) op6 was either lost independently in the lineages of B. lanceolatum and B. belcheri or duplicated in B. floridae, due to misalignment and unequal cross-over events, where Transposable Elements (TEs) were used as illegitimate recombination hotspots. Likewise, Bb_op12b and Bl_op13b were duplicated independently only in the genomes of B. belcheri and B. lanceolatum, respectively. Finally, Bl_op17b was duplicated in the genome of B. lanceolatum and later was rendered non-functional, whereas recombination over transposable elements eliminated Bl_op17a from the B. belcheri genome.
Discussion
Cephalochordates are often used as a proxy to the ancestral chordates. This is in large part due to the presumed slow evolutionary rate of their genomes. In this study we used the Branchiostoma opsin gene family as an example of how TEs can shape cephalochordate genomes, by deleting or creating new genes, by altering the number and size of exons or influencing their expression patterns. We further reconstructed the evolutionary history of opsin family in the Branchiostoma genus, via comparison of primary sequence, structure and expression patterns of opsin genes from three cephalochordate species.
The species-specific duplicates Bl_op13a and Bl_op13b differ in their spatial (tissue-specific) but overlap in their temporal expression patterns and are already detected at an earlier stage than B. floridae (Pantzartzi et al.40 and Supplementary Fig. 3). The first one is indicative of subfunctionalization, where the two genes seem to have optimized for specific tasks in tissues with different type of photoreceptor cells (ciliary and rhabdomeric), while the latter implies that Bl_op13a underwent neofunctionalization, due to which expression is triggered at an earlier stage. The relatively large size of the Go group and the retention in the genome of the duplicated opsins (Bb_op12b and Bl_op13b) could be an indication of fine tuning between these opsins in order for specific photoreception-related tasks to be fulfilled. Similarly, retention of Bf_op6 and Bf_op7 in the genome of B. floridae could be attributed to subfunctionalization, since changes are noted in their temporal expression pattern (Supplementary Fig. 3B).
The role of transposable elements (TEs) in shaping the genome and promoting evolution has been the focus of many studies, and what was formerly characterized as “junk” or “selfish DNA” is gaining more and more value and functional importance53. TEs may act in the same or completely different way, depending on selection forces. This is nicely exemplified by the ParaHox loci in Ciona, amphioxus and vertebrates54,55. ParaHox cluster in Ciona has lost the tight organization present in chordates and this degeneration could be attributed to the invasion of TEs in the locus, specifically of MITEs55. On the other hand, even though the amphioxus ParaHox cluster was found to be a hotspot for TE insertion, selection constraints probably inhibit this disruptive elements from influencing the ParaHox locus54. Another example of how TEs may influence the gene structure is that of PRHOXNB gene, for which the gain of an intron was reported, in which the miniature inverted-repeat transposable element (MITE) LanceletTn-2 was detected56.
An increase in the number of opsin gene has been previously reported for various species, owing either to local gene duplications57 or whole genome duplications58. In some cases, the number or structure of opsin genes seems to be shaped under the influence of TEs59–61. The presence of an incomplete Alu element upstream the human middle wavelength sensitive (MW) opsin gene may imply that Alu elements have been involved in the initial gene duplication responsible for the MW and long-wavelength sensitive (LW) genes in the Old World primates and the high frequency of gene loss and gene duplication within the opsin gene array60. It is suggested that unequal crossover is the mechanism through which this duplication occurred60. In the swordtail fish, Xiphophorus helleri, one of the four LW copies was found to be the result of a retrotransposition event59. On the other hand, the loss of function of the Takifugu rubripes RH2-2 gene is reported to follow a transposon-induced deletion that truncated the N-terminal of the protein61.
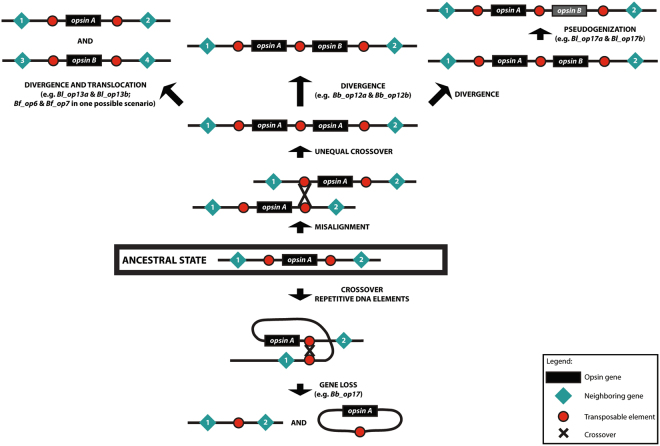
We have provided information about how TEs might have led to gene duplications and losses in the Branchiostoma opsin family, or alterations in the number and size of exons. In fact, the Branchiostoma opsin family could serve as an example of how TEs can play an important role in the shaping of a gene family and of the genome per se, through gene gain and loss events due to unequal cross-over or moving of genes between different loci in the genome (Fig. 5). Moreover, TEs may also lead to neofunctionalization of duplicate genes, which typically occurs by the acquisition of new regulatory elements. Overrepresentation of transcription factor binding sites is evident for TEs residing in promoter regions of not only human genes11, but of amphioxus as well62. Retention of Branchiostoma gene copies in the genome and differences in their spatiotemporal expression pattern, together with the presence of different types of TEs, could also imply that TEs were not implicated only in the birth or death of opsin genes but in their control as well.
Figure 5.
Suggested mechanisms for the evolution of a gene family, under the effect of Transposable Elements, as exemplified by the opsin family in the Branchiostoma genus.
Electronic supplementary material
Acknowledgements
This work was supported by the Czech Science Foundation (GACR, 17-15374S); Ministry of Education, Youth and Sports (LO1220 CZ-OPENSCREEN, LQ1604 NPU II, BIOCEV- CZ.1.05/1.1.00/02.0109) and the Institute of Molecular Genetics institutional support (RVO 6878050). This work was supported by the Branchiostoma lanceolatum genome consortium that provided access to the B. lanceolatum genome sequence.
Author Contributions
C.N.P., J.P. and Z.K. conceived and designed the experiments; C.N.P. performed in silico analysis, J.P. performed expression analysis experiments; C.N.P., J.P. and Z.K. wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20683-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lankenau, D. & Volff, J. N. Transposons and the Dynamic Genome. Springer-Verlag Berlin Heidelberg, (2009).
- 2.Finnegan DJ. Eukaryotic transposable elements and genome evolution. Trends Genet. 1989;5:103–107. doi: 10.1016/0168-9525(89)90039-5. [DOI] [PubMed] [Google Scholar]
- 3.Curcio MJ, Derbyshire KM. The outs and ins of transposition: from mu to kangaroo. Nat. Rev. Mol. Cell Biol. 2003;4:865–877. doi: 10.1038/nrm1241. [DOI] [PubMed] [Google Scholar]
- 4.Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 5.McClintock B. Controlling elements and the gene. Cold Spring Harb. Symp. Quant. Biol. 1956;21:197–216. doi: 10.1101/SQB.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 6.Chuong, E. B., Elde, N. C. & Feschotte, C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. advance online publication; 10.1038/nrg.2016.139 (2016). [DOI] [PMC free article] [PubMed]
- 7.Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr. Opin. Genet. Dev. 2009;19:607–612. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Feschotte C. Transposable elements and the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feschotte C. The contribution of transposable elements to the evolution of regulatory networks. Nat. Rev. Genet. 2008;9:397–405. doi: 10.1038/nrg2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McVean G. What drives recombination hotspots to repeat DNA in humans? Phil. Trans. R. Soc. B. 2010;365:1213–1218. doi: 10.1098/rstb.2009.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thornburg BG, Gotea V, Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 12.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arguello, J. R., Fan, C., Wang, W. & Long, M. Origination of chimeric genes through DNA-level recombination. Karger (2007). [DOI] [PubMed]
- 14.Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/S0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 15.Canapa A, Barucca M, Biscotti MA, Forconi M, Olmo E. Transposons, genome size, and evolutionary insights in animals. Cytogenet. Genome Res. 2015;147:217–239. doi: 10.1159/000444429. [DOI] [PubMed] [Google Scholar]
- 16.Joly-Lopez Z, Bureau TE. Diversity and evolution of transposable elements in Arabidopsis. Chromosome Res. 2014;22:203–216. doi: 10.1007/s10577-014-9418-8. [DOI] [PubMed] [Google Scholar]
- 17.Kaminker, J. S. et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3, 10.1186/gb-2002-3-12-research0084 (2002). [DOI] [PMC free article] [PubMed]
- 18.Mahillon J, Chandler M. Insertion Sequences. Microbiol. Mol. Biol. Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 20.Baucom RS, et al. Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 2009;5:e1000732. doi: 10.1371/journal.pgen.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu S, et al. Alu-mediated diverse and complex pathogenic copy-number variants within human chromosome 17 at p13.3. Hum. Mol. Genet. 2015;24:4061–4077. doi: 10.1093/hmg/ddv146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertrand S, Escriva H. Evolutionary crossroads in developmental biology: amphioxus. Development. 2011;138:4819–4830. doi: 10.1242/dev.066720. [DOI] [PubMed] [Google Scholar]
- 24.Huang S, et al. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 2014;5:5896. doi: 10.1038/ncomms6896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalopin D, Naville M, Plard F, Galiana D, Volff JN. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol. Evol. 2015;7:567–580. doi: 10.1093/gbe/evv005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canestro C, Albalat R. Transposon diversity is higher in amphioxus than in vertebrates: functional and evolutionary inferences. Briefings in functional genomics. 2012;11:131–141. doi: 10.1093/bfgp/els010. [DOI] [PubMed] [Google Scholar]
- 27.Igawa T, et al. Evolutionary history of the extant amphioxus lineage with shallow-branching diversification. Sci. Rep. 2017;7:1157. doi: 10.1038/s41598-017-00786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim EB, et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banyai L, Patthy L. Putative extremely high rate of proteome innovation in lancelets might be explained by high rate of gene prediction errors. Sci. Rep. 2016;6:30700. doi: 10.1038/srep30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porter ML, et al. Shedding new light on opsin evolution. Proceedings. Biological sciences / The Royal Society. 2012;279:3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liegertova M, et al. Cubozoan genome illuminates functional diversification of opsins and photoreceptor evolution. Scientific reports. 2015;5:11885. doi: 10.1038/srep11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terakita A. The opsins. Genome biology. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shichida Y, Matsuyama T. Evolution of opsins and phototransduction. Phil Trans R Soc B. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuda R, Hamilton SC, McInerney JO, Pisani D. Metazoan opsin evolution reveals a simple route to animal vision. Proc. Natl. Acad. Sci. USA. 2012;109:18868–18872. doi: 10.1073/pnas.1204609109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plachetzki DC, Degnan BM, Oakley TH. The origins of novel protein interactions during animal opsin evolution. PLoS One. 2007;2:e1054. doi: 10.1371/journal.pone.0001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peirson SN, Halford S, Foster RG. The evolution of irradiance detection: melanopsin and the non-visual opsins. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2009;364:2849–2865. doi: 10.1098/rstb.2009.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Aniello S, et al. Opsin evolution in the Ambulacraria. Marine Genomics. 2015;24(Part 2):177–183. doi: 10.1016/j.margen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 38.Ramirez MD, et al. The last common ancestor of most bilaterian animals possessed at least 9 opsins. Genome Biol Evol. 2016;8:3640–3652. doi: 10.1093/gbe/evw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suga H, Schmid V, Gehring WJ. Evolution and functional diversity of jellyfish opsins. Curr. Biol. 2008;18:51–55. doi: 10.1016/j.cub.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 40.Pantzartzi, C. N., Pergner, J., Kozmikova, I. & Kozmik, Z. The opsin repertoire of the European lancelet: a window into light detection in a basal chordate. Int. J. Dev. Biol.61, 763–772, 10.1387/ijdb.170139zk (2017). [DOI] [PubMed]
- 41.Holland LZ, et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome research. 2008;18:1100–1111. doi: 10.1101/gr.073676.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koyanagi M, Kubokawa K, Tsukamoto H, Shichida Y, Terakita A. Cephalochordate melanopsin: evolutionary linkage between invertebrate visual cells and vertebrate photosensitive retinal ganglion cells. Curr. Biol. 2005;15:1065–1069. doi: 10.1016/j.cub.2005.04.063. [DOI] [PubMed] [Google Scholar]
- 43.Koyanagi M, Terakita A, Kubokawa K, Shichida Y. Amphioxus homologs of Go-coupled rhodopsin and peropsin having 11-cis- and all-trans-retinals as their chromophores. FEBS Lett. 2002;531:525–528. doi: 10.1016/S0014-5793(02)03616-5. [DOI] [PubMed] [Google Scholar]
- 44.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 45.Rogozin IB, Milanesi L. Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol. 1997;45:50–59. doi: 10.1007/PL00006200. [DOI] [PubMed] [Google Scholar]
- 46.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darzentas N. Circoletto: visualizing sequence similarity with Circos. Bioinformatics. 2010;26:2620–2621. doi: 10.1093/bioinformatics/btq484. [DOI] [PubMed] [Google Scholar]
- 48.Kohany O, Gentles AJ, Hankus L, Jurka J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics. 2006;7:474. doi: 10.1186/1471-2105-7-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holland LZ, Yu JK. Cephalochordate (amphioxus) embryos: procurement, culture, and basic methods. Methods in cell biology. 2004;74:195–215. doi: 10.1016/S0091-679X(04)74009-1. [DOI] [PubMed] [Google Scholar]
- 51.Hirakow R, Kajita N. Electron microscopic study of the development of amphioxus, Branchiostoma belcheri tsingtauense: the neurula and larva. J. Anat. 1994;69:1–13. [PubMed] [Google Scholar]
- 52.Permanyer J, Albalat R, Gonzalez-Duarte R. Getting closer to a pre-vertebrate genome: the non-LTR retrotransposons of Branchiostoma floridae. International journal of biological sciences. 2006;2:48–53. doi: 10.7150/ijbs.2.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum. Mol. Genet. 2007;16(R2):R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- 54.Osborne PW, Ferrier DE. Chordate Hox and ParaHox gene clusters differ dramatically in their repetitive element content. Mol. Biol. Evol. 2010;27:217–220. doi: 10.1093/molbev/msp235. [DOI] [PubMed] [Google Scholar]
- 55.Ferrier DE, Holland PW. Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol Phylogenet Evol. 2002;24:412–417. doi: 10.1016/S1055-7903(02)00204-X. [DOI] [PubMed] [Google Scholar]
- 56.Xing F, et al. Characterization of amphioxus GDF8/11 gene, an archetype of vertebrate MSTN and GDF11. Dev. Genes Evol. 2007;217:549–554. doi: 10.1007/s00427-007-0162-3. [DOI] [PubMed] [Google Scholar]
- 57.Matsumoto Y, Fukamachi S, Mitani H, Kawamura S. Functional characterization of visual opsin repertoire in Medaka (Oryzias latipes) Gene. 2006;371:268–278. doi: 10.1016/j.gene.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Porath-Krause AJ, et al. Structural differences and differential expression among rhabdomeric opsins reveal functional change after gene duplication in the bay scallop, Argopecten irradians (Pectinidae) BMC Evol. Biol. 2016;16:250. doi: 10.1186/s12862-016-0823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watson CT, Lubieniecki KP, Loew E, Davidson WS, Breden F. Genomic organization of duplicated short wave-sensitive and long wave-sensitive opsin genes in the green swordtail, Xiphophorus helleri. BMC Evol. Biol. 2010;10:87–87. doi: 10.1186/1471-2148-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dulai KS, von Dornum M, Mollon JD, Hunt DM. The evolution of trichromatic color vision by opsin gene duplication in New World and Old World primates. Genome research. 1999;9:629–638. [PubMed] [Google Scholar]
- 61.Neafsey DE, Hartl DL. Convergent loss of an anciently duplicated, functionally divergent RH2 opsin gene in the fugu and Tetraodon pufferfish lineages. Gene. 2005;350:161–171. doi: 10.1016/j.gene.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 62.Holland LZ, Short S. Gene duplication, co-option and recruitment during the origin of the vertebrate brain from the invertebrate chordate brain. Brain Behav. Evol. 2008;72:91–105. doi: 10.1159/000151470. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.