Abstract
Mass cytometry (or CyTOF) is an atomic mass spectrometry-based single-cell immunoassay technology, which has provided an increasingly systematic and sophisticated view in basic biological and clinical studies. Using elemental reporters composed of stable heavy metal isotopes, more than 50 cellular parameters are measured simultaneously. However, this current multiplexing does not meet the theoretical capability of CyTOF instrumentation with 135 detectable channels, primarily due to the limitation of available chemistries for conjugating elemental mass tags to affinity reagents. To address this issue, we develop herein additional metallic mass tag based on bismuth-209 (209 Bi) for efficient conjugation to monoclonal antibody. This enables the use of an addtional channel m/z = 209 of CyTOF for single-cell immunoassays. Bismuth has nearly the same charge-to-radius ratio as lanthanide elements; thus, bismuth(III) cations (209Bi3+) could coordinate with DTPA chelators in the same geometry of O- and N-donor groups as that of lanthanide. In this report, the coordination chemistry of 209Bi3+ with DTPA chelators and Maxpar® X8 polymers were investigated in details. Accordingly, the protocols of conjugating antibody with bismuth mass tag were provided. A method based on UV-Vis absorbance at 280 nm of 209Bi3+-labeling DTPA complexes was developed to evaluate the stoichiometric ratio of 209Bi3+ cations to the conjugated antibody. Side-by-side single-cell analysis experiments with bismuth-and lanthanide-tagged antibodies were carried out to compare the analytical sensitivities. The measurement accuracy of bismuth-tagged antibody was validated within in vitro assay using primary human natural killer cells. Furthermore, bismuth-tagged antibodies were successfully employed in cell cycle measurements and high-dimensional phenotyping immunoassays.
Key terms: elemental mass tags, bismuth, labeling efficiency, antibody conjugation, mass cytometry, NK cell phenotyping, cell cycle measurement
Over the past several decades, the investigations of diverse features of individual cells have been heavily influenced by the use of fluorescence-based flow cytometry (1,2). During the same period, elemental mass spectrometry, especially inductively couple plasma mass spectrometry (ICP-MS), has become a well-established technique to determine elemental and isotopic compositions with ultrahigh sensitivity and widely dynamic range. ICP-MS has been mainly applied in the fields of materials science characterizing, environmental contamination monitoring and geological rock dating and so on (3,4). Recently, a hybrid instrument that couples flow cytometry with ICP-MS was developed (5,6) with a simultaneous multiple ion detector of time-of-fight to measure the single transient event as single cells. This new cytometric technology is known as mass cytometry or CyTOF for “cytometry by time-of-flight”. This combination of these two technologies combines their strengths by offering single-cell assays with measurements of over 50 simultaneously detected parameters. This advance significantly improves the analytical capabilities of single-cell cytometry and enables the high-dimensional investigation of basic biological and clinical research (7–9). Additional benefits in CyTOF technology have resulted in minimal spectral overlap between detection channels, absence of auto-fluorescence background and of natural contaminants, as well as promisingly enhanced sensitivity (10–15)
The detection reporters for CyTOF are referred as mass tags, which employ elements or stable (nonradioactive) isotopes of heavy metals or halogens. These constituents rarely exist in biological systems and are thus suitable as uniquely detectable tags. In the past decade, the elemental labeling strategies (16–18) have been increasingly developed to conjugate a variety of elements or stable isotopes to biological molecules such as proteins, antibodies, nucleic acids, and others affinity-based molecules. Generally, the strategies include nanoparticle-based and polymer-based approaches. For example, noble-metal particles with silver (19), gold (20), platinum (21), or chromium-containing quantum dots (22) have been conjugated to the antibodies against cellular surface CD markers or sequence-specific DNA oligonucleotide probes. Lanthanide-based biocompatible nanoparticles are also synthesized for the potential applications in CyTOF assays (23,24). However, nanoparticle tags have the “sticky” effects that result in high background signals caused by non-specific adsorption of cells, and the large stereoscopic volumes resulting in inefficient diffusion through cell membranes. Consequently, the biocompatible polymers containing chelators of 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA) or pentetic acid (DTPA) are widely used to trap heavy metal cations and to conjugate to monoclonal antibody (25,26). To date, stable isotopes of indium, yttrium, and lanthanides have been successfully chelated into Maxpar® X8 polymers and conjugated to antibodies for single-cell phenotype or function assays of CyTOF (27–29). There are also several organometallic and metallic molecules that have been developed to measure specific biological functions of single cells, such as iridium- or rhodium-based intercalators for cell identification (30), 5-iodo-2′-deoxyuridine (IdU) for cell cycle analysis (31), platinum-based cisplatin for cell viability (32) and antibody conjugation (33), organotelluriums for analysis of cell hypoxia (34,35), EDTA-chelating palladium for cell barcoding (36,37), and osmium and ruthenium tetroxide species (38) for cell barcoding and cell size assay (39). Despite these many advances to develop various mass tags for CyTOF assays, conjugation of monoclonal antibody with mass tags requires significant validation as the presence of these tagged heavy metallic cations can possibly alter the capability of antibody to recognize its antigen (40). Because of this, and limitations of labeling strategies for heavy metals, less than one-third of the available CyTOF channels, across the mass range from 75 to 209 Da, are currently utilized for measurement of protein expression in single cells.
Bismuth has been considered the element with the highest atomic mass that is stable on the earth. There is only one natural isotope, bismuth-209 (209Bi), with 83 protons and 126 neutrons. Previous studies showed that trivalent bismuth cations have strong coordinating affinity for DTPA and DOTA chelators. The bismuth radioactive isotope 213Bi has been investigated in radioimmune assays (41,42). Though Fluidigm has sold a few preconjugated antibodies with bismuth metal as commercial reagents of mass cytometry (43), to date, there have not been any public investigations and protocol reports for the development of bismuth-based mass tag of CyTOF. In this report, we developed new elemental labeling approaches for chelating stable bismuth isotope 209Bi3+into DTPA-containing polymers and for conjugating this mass tag to monoclonal antibody via site-specific coupling reactions between cysteine and maleimide-functionalized moieties. This method thus enables a new channel of m/z = 209 for CyTOF single-cell immunoassays. The stability factors and kinetics of chelating 209Bi3+ cations with DTPA molecules and Maxpar X8 polymers were investigated and characterized in details. In quantitative analysis, a BCA assay was developed to determine the conjugated antibody and a novel UV–Vis approach was established to evaluate bismuth labeling efficiency. Comparison of analytical sensitivity of bismuth- and lanthanide-tagged antibodies was performed in singe-cell immunoassays. The measurement accuracy of bismuth-tagged antibody was validated within in vitro assay using primary human natural killer cells as well.
Materials and Methods
Experimental Overview
The following methods are primarily for: 1) antibody conjugation procedures; 2) SDS-PAGE characterization; 3) BCA quantitative assay; 4) evaluation of bismuth labeling efficiency; 5) comparison of sensitivity and validation of measurement accuracy; and 6) single-cell applications of cell cycle measurement and phenotyping immunoassays.
Reagents
Elemental standard solutions were as follows: natural abundance of rare earth element mixture containing Sc, Y, La, Ce, Pr, Nd, Sm, Eu, Gd, Tb, Dy, Ho, Er, Tm, Yb, and Lu at 50 mg/L each in 2% nitric acid (cat. no. 67349-100ML; Sigma-Aldrich, St. Louis, MO); bismuth standard solution, at 1000 mg/L in 5% nitric acid (cat. no. 05719-100ML; Sigma-Aldrich); bismuth(III) nitrate pentahydrate, 99.999% (cat. no. 254150; Sigma-Aldrich); nitric acid, 70%, purified by re-distillation, ≥99.999% free from trace metals basis (cat. no. 225711; Sigma-Aldrich); pentetic acid, DTPA (cat. no. 1505506-100MG; Sigma-Aldrich); tris-(2-carboxyethyl) phosphine, hydrochloride, TCEP (cat. no. 77720; Thermo Fisher Scientific, Rockford, IL); Maxpar X8 polymer (cat. no. 201153B; Fluidigm, South San Francisco, CA), R-buffer for partially reducing antibody (cat. no. 2591404; Fluidigm), C-buffer for conjugating antibody (cat. no. 2931412; Fluidigm), and W-buffer for washing conjugated antibody (cat. no. 2721401; Fluidigm); and PBS-based antibody stabilizer (CAN-DOR Bioscience, Wangen, Germany). The following antibodies against human blood cell surface epitopes in low-sodium azide buffer without carrier proteins were obtained from BD Biosciences (San Jose, CA): anti-CD3 (UCHT1), anti-CD4 (RPA-T4), anti-CD7 (M-T701), anti-CD8 (RPA-T8), anti-CD11b (ICRF44), anti-CD19 (H1B19), anti-CD20 (2H7), anti-CD45 (HI30), and anti-CD56 (NCAM16.2). 164Dy-Cyclin B1 antibody and 166Er-pRb antibody for cell cycle measurement were obtained from Fluidigm. Cellular events were identified by iridium DNA intercalator (cat. no. 201192A; Fluidigm) with cell length range from 10 to 75 pushes. The viability of cells was measured with cisplatin (cat. no. P4394; Sigma-Aldrich). IdU (cat. no. I7125-25G; Sigma-Aldrich) was used to detect newly synthesized DNAs. The signal drift of CyTOF was normalized with EQ™ four-element calibration beads (cat. no. 201078; Fluidigm).
Conjugation of IgG Antibody with Bismuth Mass Tag
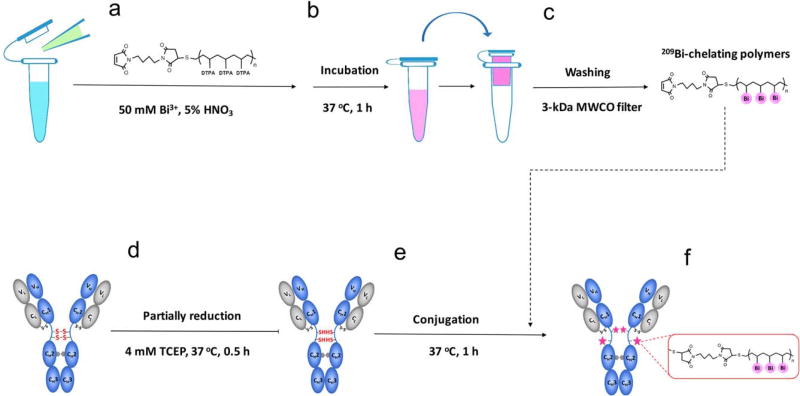
The 209Bi3+ solution was prepared by dissolving approximately 25 mg of bismuth(III) nitrate pentahydrate in an appropriate volume of 5% HNO3 to obtain 50 mM 209Bi3+ solution. Figure 1 shows the protocol for conjugating bismuth mass tag to antibody in the following six main steps: 1) Retrieve one tube 200 µg of Maxpar X8 polymers (for conjugating 100 µg of IgG antibody), and re-suspend the polymers in 95 µl of 5% HNO3, then add 5 µl of previously prepared 50 mM 209Bi3+ solution; 2) Incubate the mixture at 37°C for 1 h. Mix the solution approximately every 20 min by pipetting up and down using filter tips; 3) Transfer the incubation into a 3-kDa MWCO filter column, and wash away free 209Bi3+ cations twice by spinning the filter column at 14,000g for 30 min at room temperature. Importantly, the first wash is in 300 µl of ddH2O and the second wash is in 400 µl of C-buffer. Because C-buffer at pH 7.0–7.4 will cause the precipitation of the concentrated 209Bi3+ solution, the first wash using water is necessary; 4) Partially reduce IgG antibody in a 50-kDa MWCO filter 0.5-ml column using 4 mM TCEP in R-buffer, and incubate at 37°C for 30 min. Note, this antibody reduction step should be carried out when the second wash of step 3 starts, to ensure that bismuth-chelating polymers and the reduced antibody will be ready for the next step of conjugation at the same time; 5) Resuspend 209Bi-chelating polymers in 200 µl of C-buffer and transfer into the partially reduced antibody, incubate the mixture at 37°C for 1 h; and 6) Wash 209Bi-tagged antibody four times in W-buffer by spinning the filter column at 12,000g for 10 min at room temperature. Finally, collect 209Bi-tagged antibody by spinning the inverted filter in 50 µl of W-buffer twice.
Figure 1.
Strategy for conjugating bismuth mass tag to IgG antibody. (a) 209Bi3+ solution is mixed with Maxpar® X8 polymers in 5% HNO3 acid buffer. (b) The mixture is incubated at 37°C for 1 h. (c) The solution is transferred into a 3-kDa MWCO filter column, and free 209Bi3+ cations are washed away. (d) The disulfide bonds within the Fc region of antibody are partially reduced using 4 mM TCEP. (e) The 209Bi-chelating polymers are conjugated to the reduced antibody. (f) A 50-kDa MWCO filter column is used to wash away unconjugated polymers, and 209Bi-tagged antibody is collected.
SDS-PAGE Gel Characterization of Bismuth-Tagged Antibody
To characterize the conjugated antibody, SDS-PAGE gel electrophoresis (NuPAGE™ 4–12% Bis-Tris gels, cat. no. NP0322BOX; Thermo Fisher Scientific) was carried out under denaturing conditions. A 2.5-µl aliquot (about 250 ng) of purified CD56 antibody, 209Bi-tagged CD56 antibody or 176Yb-tagged CD56 antibody were mixed with 2.5 µl of sample buffer, 1.0 µl of reducing agent, and 4.0 µl of deionized water. After incubation at 70°C for 10 min, samples and protein standards were loaded into gel wells and the gels were run at 180 V for 30–40 min and stained with Coomassie blue. The stained bands of purified CD56 antibody showing at ~27 and ~53 kDa were used as the controls for the light and heavy chains of IgG antibody. 176Yb-tagged CD56 antibody was analyzed as a comparable counterpart to 209Bi-tagged CD56 antibody.
BCA Protein Assay of Bismuth-Tagged Antibody
Bismuth mass tag has the UV–Vis absorbance in the range from 250 to 300 nm, which interferes with protein absorption at 280 nm mostly used for the quantification. Therefore, a Nanodrop spectrophotometry approach based-on bicinchoninic acid (BCA) assay was employed to determine the concentration of 209Bi-tagged antibody. The procedures are as following steps. First, to prepare working solution, 1,000 µl of BCA reagent A (cat. no. 23225; Thermo Fisher Scientific) was combined with 20 µl of BCA reagent B at the ratio of 50:1 (Reagent A:B), and then mixed well by vortexing. Second, to make IgG antibody standards, 1.0 mg of lyophilized IgG control antibody (cat. no. MAB004; R&D Systems, Minneapolis, MN) was dissolved in 1.0 ml of PBS to obtain a 1,000 µg/ml IgG antibody standard. A series of diluted IgG antibody standards were then prepared at the concentrations of 100, 200, 500, and 750 µg/ml. Next, 2.0 µl of each IgG antibody standard and 2.0 µl of 209Bi-tagged anti-body were pipetted into pre-labeled PCR tubes and then 30 µl of prepared working solution was added into each PCR tube. After mixing by pipetting up and down, the samples were incubated at 37°C for 30 min and cooled down to room temperature. The absorbance at 562 nm (A562) generated from each IgG antibody standard and 209Bi-tagged antibody were measured within 10 min. Then, A562 of each IgG antibody standard was plotted vs. its concentration to create a calibration curve for quantitative analysis. Finally, the concentration of 209Bi-tagged antibody was determined as described in the “Results” section.
Evaluation of Bismuth Labeling Efficiency Using UV–Vis Spectrophotometry
The coordinative complex of Bi3+cation with DTPA molecule (Bi@DTPA) has a unique UV–Vis absorption peak in the range from 250 to 300 nm, which is not present in either Bi3+ or DTPA. Therefore, UV–Vis absorbance at 280 nm of Bi@DTPA was measured to evaluate bismuth labeling efficiency. Standard solutions of Bi@DTPA were prepared by dissolving approximately 20 mg of DTPA solid powder in an appropriate volume of PBS to obtain a 50.0 mM DTPA solution. Then, an equal volume of 50.0 mM of 209Bi3+ solution was added. After mixing by pipetting up and down, the solution was incubated at room temperature for 1 h. Then, 1.0 mM Bi@DTPA solution was obtained by dilution in PBS and the value of pH was adjusted to between 7.0 and 7.5 with 2.0 M of NaOH solution. Next, a series of dilutions of Bi@DTPA were prepared at the concentrations of 0.1, 0.2, 0.5, and 0.75 mM in PBS. A280 of each Bi@DTPA standard was plotted vs. its concentration to create a calibration curve for quantitative analysis. Finally, the stoichiometric ratio of 209Bi3+ cations to the conjugated antibody was determined as described in the “Results” section.
Preparation of Cell Samples
Human peripheral blood was obtained from the Stanford University Blood Center for anonymous healthy human donors. Collection procedure followed a Stanford University Institutional Review Board-approved protocol. Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll–Hypaque density gradient centrifugation. The fixation and permeabilization conditions are a standard for our laboratory, as previously described (6). PBMCs were either used immediately for immunostaining or frozen in fetal bovine serum (FBS) with 10% DMSO in liquid nitrogen for long-term storage. Cryopreserved cells were thawed and washed in RPMI-1640 supplemented with 10% FBS (Sigma-Aldrich) and 1% penicillin–streptavidin (Corning Cellgro, Manassas, VA), and L-glutamine (Corning Cellgro). Cell sample preparation was performed using Ba2+-free PBS and cell staining media (CSM).
Mass Cytometry Data Acquisition
Mass cytometry measurements were performed on CyTOF version 1, CyTOF version 2, and CyTOF Helios instruments (Fluidigm). In order to quantify 209Bi3+ cations of bismuth-tagged antibody, CyTOF was performed in solution-mode analysis as well, which is using the same procedure as the measurement of elemental tuning solutions. In solution-mode CyTOF assays, bismuth standards and bismuth-tagged antibody were diluted in 2% HNO3. Pulse counts and dual counts of mass signals were acquired for 2–5 min. In single-cell assays, cell suspensions were diluted in ddH2O to a density of 0.5–1 × 106 cells/ml and passed through a cell strainer cap with 35-µm pores (BD Biosciences) immediately before acquisition. The solution-mode data were collected in CSV files and analyzed in Excel. Single-cell data were collected in FCS files. Manually cell gating, density plots and histograms as well as SPADE illustrations and representations were performed using Cytobank (Cytobank, Inc., Mountain View, CA)
Results
The general strategy for conjugating bismuth mass tag to IgG antibody is shown in Figure 1. Preparatory analysis for potential use of bismuth is discussed herein first and then proceeding to the conjugation chemistries as outlined below. We then include testing and comparison of bismuth as mass tag as compared to other isotope mass tags used in CyTOF.
Mass Spectrum Profile and Quantification of the Dynamic Range of Bismuth
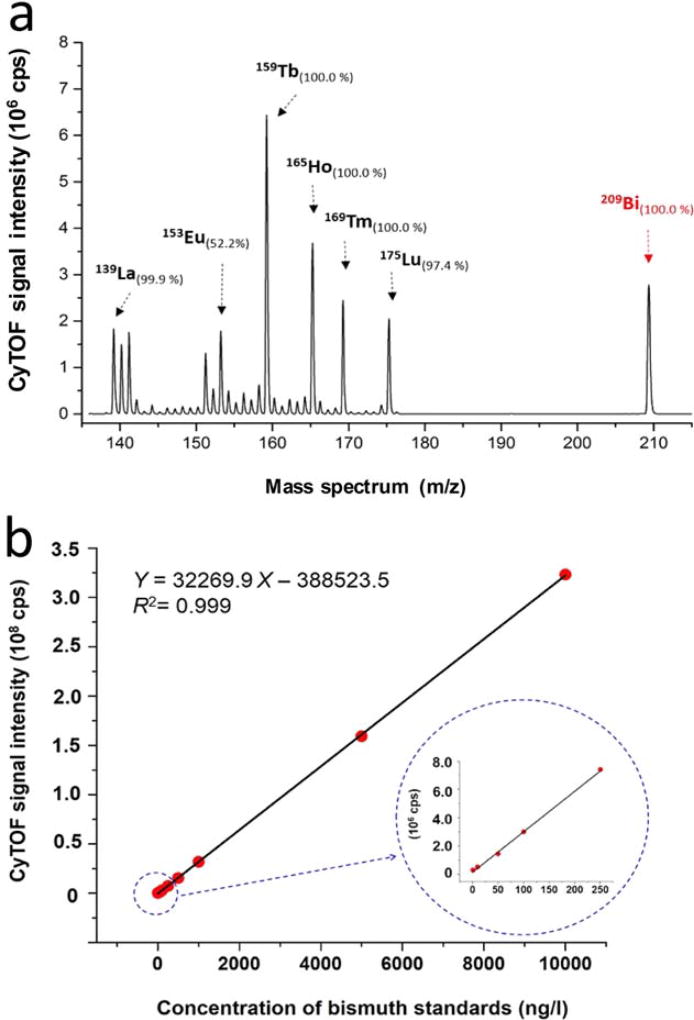
To employ bismuth as a mass tag of CyTOF, the bismuth mass spectrum and quantification of the dynamic range were investigated with the solution-mode measurements of CyTOF. In the mass spectra of bismuth and lanthanide elements at concentrations of 100 ng/l, bismuth mass peak showing at m/z of 209 is >32-Da (mass of O2) far away from the mass region of the lanthanides, as shown in Figure 2a. It indicates that there are no any oxide polyatomic ions from the lanthanides that could result in mass interference with bismuth ions. In CyTOF, the ion transmission efficiency is optimized to obtain the highest sensitivity in the central of the mass range of the lanthanides within an asymmetric bell curve (28,44). Additionally, due to the space-charge effects of ICP-MS, heavier bismuth ions are preferentially transmitted over lighter lanthanide ions. As the above two factors, as shown in Figure 2a, the sensitivity of bismuth is lower than those of 159Tb (100%) and 165Ho (100%) but higher than those of 139La (99.9%) and 175Lu (97.4%). Within the dynamic range of quantitative measurement of bismuth, there is a four-magnitude linear regression between CyTOF signal intensities and bismuth concentrations ranging from 1 to 10,000 ng/l, as shown in Figure 2b.
Figure 2.
Mass spectrum and quantitatively dynamic range of bismuth via CyTOF. (a) CyTOF mass spectra of 13 natural abundance lanthanide elements and 100% abundance bismuth at concentrations of 100 ng/l in 2% HNO3. (b) The CyTOF signal intensities are plotted vs. concentrations of bismuth standards. The dynamic range of quantification of bismuth is linear over four orders of magnitude ranging from 1 to 10,000 ng/l. All measurements were carried out in solution-mode CyTOF analysis
Coordination Chemistry of Bismuth(III) Cations with DTPA-Chelators and -Polymers
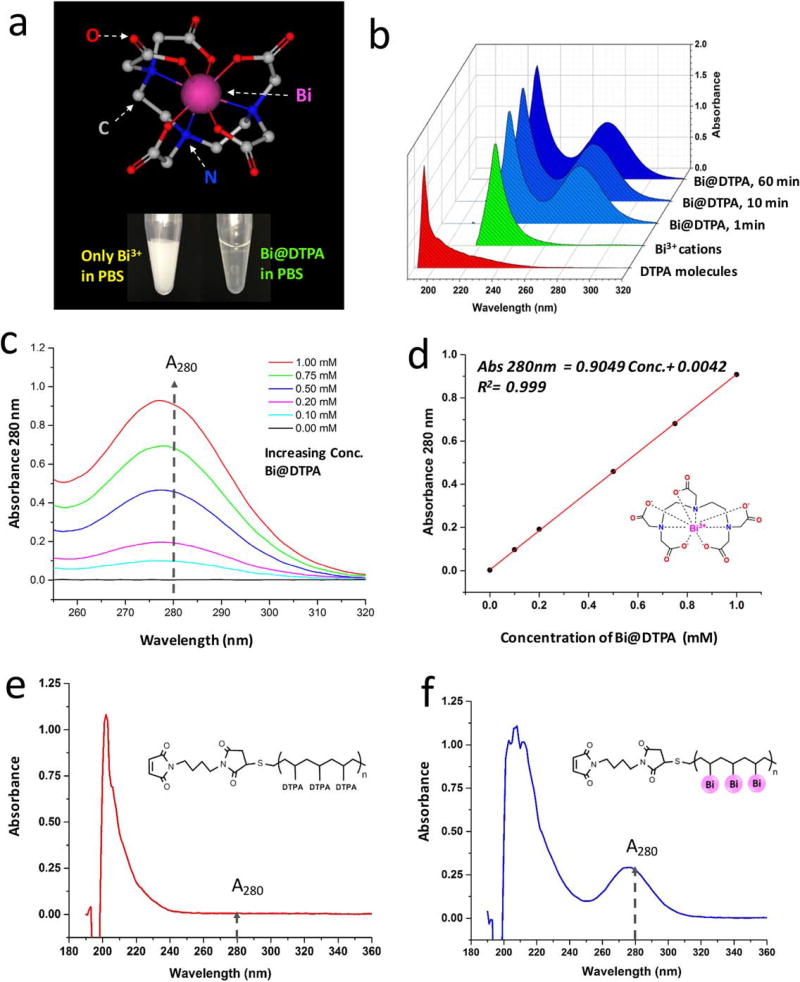
The eight-coordination Shannon ionic radius of Bi3+ is 1.17 Å, and that of La3+ is comparably 1.16 Å. Thus, these two trivalent cations have very close charge-to-radius ratios (2.564 and 2.586) (45). Consequently, Bi3+ cations are able to coordinate with DTPA chelators in the same geometry with O- and N-donor groups as that of lanthanum, as shown in the top of Figure 3a. However, Bi3+ cations can only be stable in strongly acidic solutions, otherwise Bi3+ cations are susceptible to hydrolysis in the formation of basic salts even at pH values of 1–2. In our experiments, Bi3+ cations have been investigated to be stable in 2–5% nitric acid at concentrations ranging from 1 ng/l to 10,000 mg/l for at least 2 years. Noticeably, after the complexation with DTPA, Bi@DTPA can be stable in neutral solutions or slightly basic mediums for a long term. As shown in the bottom of Figure 3a, 1.0 mM of Bi3+ cations precipitated in 100 mM pH 7.2 PBS, and in contrast, the precipitation was not observed within 1.0 mM of Bi@DTPA in the same buffer.
Figure 3.
Coordinative chemistry of bismuth(III) cations with DTPA-chelators and -polymers. (a) Top, eight-coordination structural presentation of Bi3+ cations chelating with DTPA molecules (Bi@DTPA). Bottom, comparison of stability of Bi3+ cations and Bi@DTPA in 100 mM, pH 7.4, PBS. (b) UV–Vis spectra of 1.0 mM of DTPA, Bi3+, and Bi@DTPA chelating for 1, 10, and 60 min. (c) UV–Vis spectra of Bi@DTPA at concentrations from 0.10 to 1.00 mM, with the increasing absorbance at 280 nm. (d) Plots of A280 vs. the concentrations of Bi@DTPA, and a quantitatively linear regression. (e) UV–Vis spectrum of Maxpar® X8 polymer before the complexation of Bi3+ cations. (f) UV–Vis Spectrum of Maxpar X8 polymer after the complexation of Bi3+ cations.
Furthermore, to study reaction kinetics of chelating Bi3+ with DTPA, UV–Vis absorbance of reaction precursors of 1.0 mM of Bi3+ and DTPA and the product of Bi@DTPA were measured in a time course experiment. As shown in Figure 3b, the absorption of DTPA molecules is mainly from 190 to 250 nm and that of Bi3+cations from 220 to 250 nm. After their complexation, a new absorption peak of Bi@DTPA ranging from 250 to 300 nm was distinguishably observed. The intensities of absorbance at 280 nm (A280) of Bi@DTPA were measured at three time points of 1, 10, and 60 min. Their values were 0.79, 0.80, and 0.80, respectively. This indicate the coordinative complexation of Bi@DTPA has achieved kinetic stability in <10 min at room temperature. In Figure 3c, the spectra of Bi@DTPA at concentrations of 0.1, 0.2, 0.5, 0.75, and 1.0 mM were profiled with the increase of absorbance at 280 nm. Furthermore, there is a quantitatively linear regression between the concentrations of Bi@DTPA and their A280 in Figure 3d. The calibration curve is presented in the following equation:
| (1) |
where A280 (Bi@DTPA) is absorbance at 280 nm of the complex of Bi@DTPA; [Bi@DTPA] is the concentration of Bi@DTPA (mM); 0.9049 mM−1 is the slope of the equation; 0.0042 is the absorbance of the blank. The coefficients of the equation are dependent on the individual spectrophotometer and measurement parameters.
Additionally, the coordinative complexation of Bi3+ cations with Maxpar X8 polymers containing multiple DTPA molecules was investigated. In Figure 3e, it shows the UV–Vis spectrum of Maxpar X8 polymers before complexing with Bi3+ cations, in which 200 µg of Maxpar X8 polymers were directly dissolved in 200 µl of 100 mM pH 7.2 PBS. On the other side, Figure 3f shows the UV–Vis spectrum of Maxpar X8 polymer after complexing with Bi3+ cations (Bi@Maxpar X8). To prepare the complex of Bi@Maxpar X8, 200 µg polymers were dissolved in 195 µl of 5% HNO3 and 5 µl of 50 mM Bi3+ solution were added. The mixture was incubated for 1 h, washed twice in deionized water, and resuspended in 200 µl of 100 mM pH 7.2 PBS. Comparing the difference of UV–Vis absorbance between Maxpar X8 and Bi@Maxpar® X8, obviously a new absorption peak ranging from 250 to 300 nm was observed because of the existence of Bi3+ cations. The property of this new absorption spectrum of Bi@Maxpar X8 is consistent with that of Bi@DTPA.
Characterization of Bismuth-Tagged Antibody by UV–Vis Spectrum and SDS-PAGE
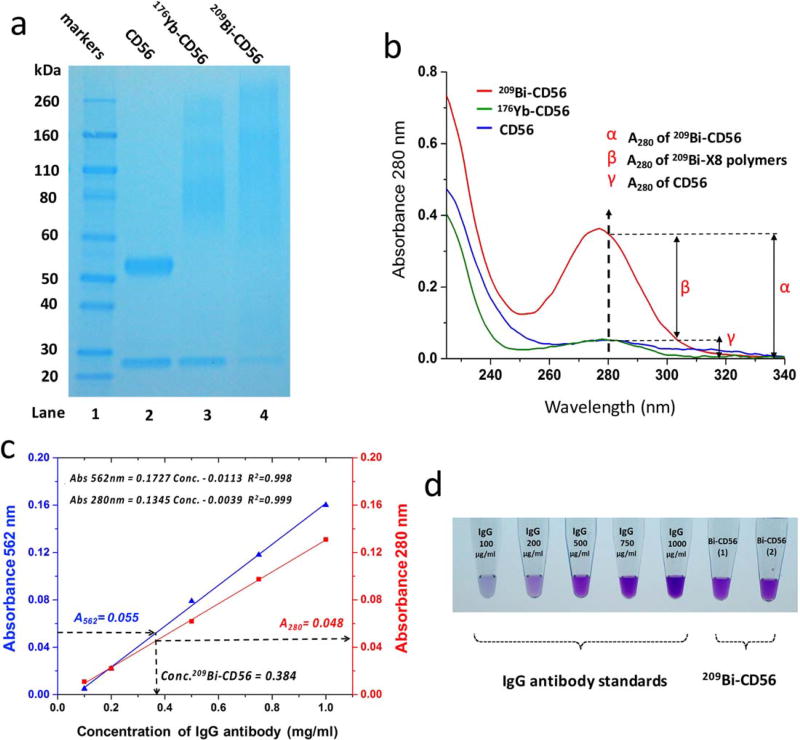
CD56 antibody was conjugated to bismuth mass tag as the procedures illustrated in Figure 1. Then, 209Bi-tagged CD56 antibody (209Bi-CD56) was characterized by UV-Vis spectrophotometry and SDS-PAGE. As shown in lane 2 of Figure 4a, unconjugated anti-CD56 migrated as two bands: the band at ~27 kDa corresponds to light chains, and the other band at ~53 kDa to heavy chains. Comparably, in lane 4 of 209 Bi-CD56, the band corresponding to the light chains was present as well, but differently the heavy chains ran as a smear ranging from 60 kDa to over 260 kDa. According to the fact that inter-chain disulfide bonds located in the hinge region between two heavy chains are readily exposed to the reducing solvent, these disulfide bonds tend to be reduced under mild conditions (46). The SDS-PAGE result indicates the successful conjugation of bismuth mass tag to CD56 antibody, because maleimide-functionalized Bi@Maxpar X8 polymers were covalently coupled to the reduced cysteine sulfhydryl groups of the fragment crystallizable (Fc) region in heavy chains of CD56 antibody. Due to the distribution of molecular weight of heterogeneous polymers, bismuth labeling efficiency, and the ratio of conjugated polymers to antibody, the band of Bi-tagged heavy chains appeared as a distribution of molecular weights. In lane 3, the comparable pattern of bands of 176Yb-CD56 was observed with that of 209Bi-CD56. The polymer attachment to antibody was dependent on Michael Addition of thiol and maleimide groups, therefore the processes of antibody coupling with Bi@Maxpar X8 and Ln@Maxpar®X8 were identical as expected. To demonstrate the consistent electrophoretic separations of other lanthanide-tagged antibodies and bismuth-tagged antibodies, additional SDS-PAGE results are shown in Supporting Information Figure S1.
Figure 4.
Characterization and quantification of bismuth-tagged antibody. (a) SDS-PAGE image of conjugated and unconjugated CD56 antibodies. Lane 1: protein markers; Lane 2: unconjugated CD56 antibody (CD56); Lane 3: 176Yb-tagged CD56 antibody (176Yb-CD56); Lane 4: 209Bi-tagged CD56 antibody (209Bi-CD56). (b) Comparison of UV–Vis spectra of CD56, 176Yb-CD56, and 209Bi-CD56 at 0.40 mg/ml. Their A280 were 0.051 ± 0.01, 0.051 ± 0.02, and 0.362 ± 0.02, respectively. (c) Calibration curves for BCA assay and A280 protein quantification: blue line obtained by plotting A562 vs. IgG standard concentration, and red line by A280 vs. IgG standard concentration. (d) BCA colorimetric images of a series of IgG antibody standards and duplicate samples of 209Bi-CD56.
UV–Vis spectra of CD56, 176Yb-CD56, and 209Bi-CD56 at the same concentration of 0.40 mg/ml were compared. As shown in Figure 4b, CD56 and 176Yb-CD56 had the identical absorbance of A280 within experimental error. However, A280 of 209Bi-CD56 was considerably higher than them. This phenomenon results from the UV–Vis absorption property of Bi@Maxpar X8, as explained in Figure 3f. In the quantitative illustration, the total absorbance A280 of 209 Bi-CD56, denoted as α, was 0.362; the partial contribution of A280 resulting from unconjugated anti-CD56, denoted as γ, was 0.051. The difference of α and γ, denoted as β, was 0.311, which is the partial contribution of A280 resulting from Bi@Maxpar X8. Combining the value of β and Equation (1), the concentration of tagged bismuth cations could be quantitatively estimated.
Quantification of Conjugated Antibody and Evaluation of Bismuth Labeling Efficiency
The conventional approach of protein quantification using A280 cannot be used to determine the concentration of 209Bi-tagged antibody, because of the interference caused by the UV–Vis absorbance of Bi@Maxpar X8 in the range from 250 to 300 nm. To address this issue, a method based on BCA assay using a Nanodrop spectrophotometer was developed to measure 209 Bi-tagged antibody. The quantification is based on UV–Vis absorbance at 562 nm (A562) of 209 Bi-tagged anti-body stained with BCA colorimetric dyes. IgG antibody standards and duplicated samples of 209Bi-CD56 were stained with BCA dyes as the procedures described in the “Materials and Methods” section. As shown in the colorimetric images of Figure 4d, with gradually increasing the concentrations of IgG antibody standards, the violet color resulting from the absorption of was enhanced. A calibration curve was generated by plotting A562 vs. the concentration of antibody in Figure 4c. There is a linear regression for the quantification of 209Bi-tagged antibody, as shown in the following equation:
| (2) |
where A562 (Bi-Ab) is the absorbance at 562 nm of 209 Bi-tagged antibody; [Bi-Ab] is the concentration of 209Bi-tagged antibody (mg/ml); 0.1727 ml/mg is the slope of the equation; and −0.0113 is the absorbance of the blank. The coefficients of the equation are dependent on the individual spectrophotometer and measurement parameters.
To determine the partial contribution A280 of IgG anti-body, there is a calibration curve generated by plotting A280 vs. the concentration of IgG antibody standard, as shown in the following equation:
| (3) |
where A280 (Ab) is the absorbance at 280 nm of IgG antibody; [Ab] is the concentration of IgG antibody, mg/ml; 0.1345 ml/mg is the slope of the equation; and −0.0039 is the absorbance of the blank. The coefficients of the equation are dependent on the individual spectrophotometer and measurement parameters.
In the determination of an unknown sample of 209Bi-CD56, as shown in Figure 4c, the measurement of A562 (Bi-CD56) was 0.055 ± 0.002. Using Equations (2) and (3), the concentration of 209Bi-CD56 was determined as 0.384 ± 0.012 mg/ml and the partial A280 resulting from CD56 anti-body was 0.048 ± 0.002. The measurement of A280 (Bi-CD56) was 0.348 ± 0.003; thus, the partial A280 resulting from Bi@Maxpar X8 was 0.300 ± 0.003. Using Equation (1), the concentration of Bi3+ cations was determined as 0.327 ± 0.018 mM. Finally, the stoichiometric ratio of Bi3+ cations to 209Bi-CD56 was 128 ± 10. The results were consistent with the measurements of solution-mode CyTOF analysis within experimental error, as shown in Supporting Information Figure S2. Additionally, to evaluate the quantification accuracy of BCA assay of mass-tagged antibodies, 176Yb-tagged CD56 antibody was quantified by using both BCA assay and conventional protein assay of A280, and these results were in agreement within experimental error, as shown in Supporting Information Figure S3. In conclusion, taking the advantages of BCA assay and the absorbance of A280 of bismuth mass tag, the concentration of 209Bi-tagged antibody and bismuth labeling efficiency can be determined.
Comparison of Analytical Sensitivity of Bismuth and Lanthanide Mass Tags
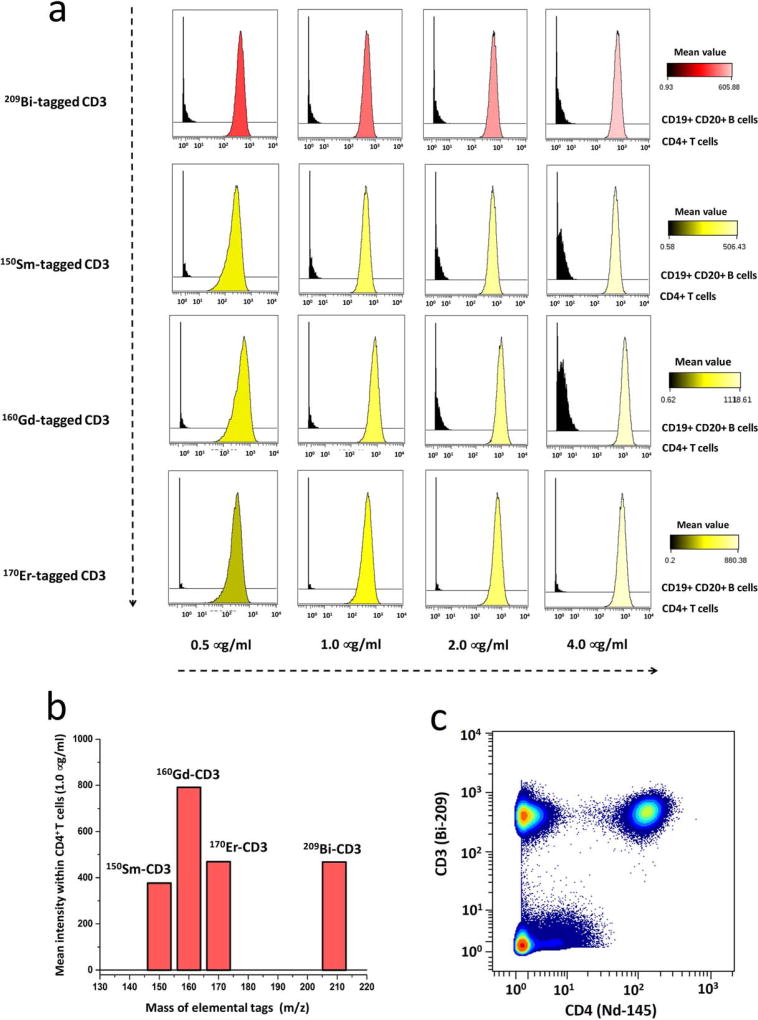
The analytical sensitivity of bismuth mass tag in single-cell CyTOF assays was compared with conventional lanthanide mass tags. CD3 antibody was therefore conjugated with the mass tags consisting of 209Bi, 150Sm, 160Gd, or 170Er. The anti-body titration experiments were performed with human PBMCs, which were stained with 89Y-CD45, 145Nd-CD4, 146Nd-CD8, 142Nd-CD19, and 147Sm-CD20 antibodies as well as each mass-tagged CD3 antibody above. The gating hierarchy was manually clustered to identify CD4+ T cell subsets and CD19+ CD20+ B-cell subsets, which are used as positive and negative controls of the expression of CD3 proteins. Bismuth- and lanthanide-tagged CD3 antibodies were titrated at the concentrations of 0.5, 1.0, 2.0, and 4.0 µg/ml, as shown in Figure 5a. At the optimal concentration of 1.0 µg/ml, the mean intensity value of each conjugated CD3 antibody within CD4+ T cells were 376, 790, 469, and 468, indicating the sensitivity order of 160Gd > 170Er ≈ 209Bi > 150Sm, as shown in Figure 5b. Therefore, 209Bi mass tag has relatively high analytical sensitivity in CyTOF assays, which is comparable with 170Er lanthanide counterpart. In the biaxial scatter plots of Figure 5c, 209Bi-tagged CD3 antibody indicated the obvious distinction between CD3-positive and -negative cell subsets within human PBMCs.
Figure 5.
Comparison of analytical sensitivity of bismuth- and lanthanide-tagged CD3 antibodies. (a) CyTOF titration histograms of 209Bi-, 150Sm-, 160Gd-, or 170Er-tagged CD3 antibody. The stained cells are human PBMCs, with positive control of CD4+ T cells and negative control of CD19+ CD20+ B cells. The concentrations of conjugated CD3 antibodies were 0.5, 1.0, 2.0, and 4.0 µg/ml. (b) Bar graphs of mean intensity of each conjugated CD3 antibody at the concentration of 1.0 µg/ml in the subset of CD4+ T cells. (c) Biaxial scatter plots of 209Bi-tagged CD3 antibody vs. 145Nd-tagged CD4 antibody within human PBMCs at the optimal concentration of 1.0 µg/ml.
Validation of Measurement Accuracy of Bismuth Mass Tag
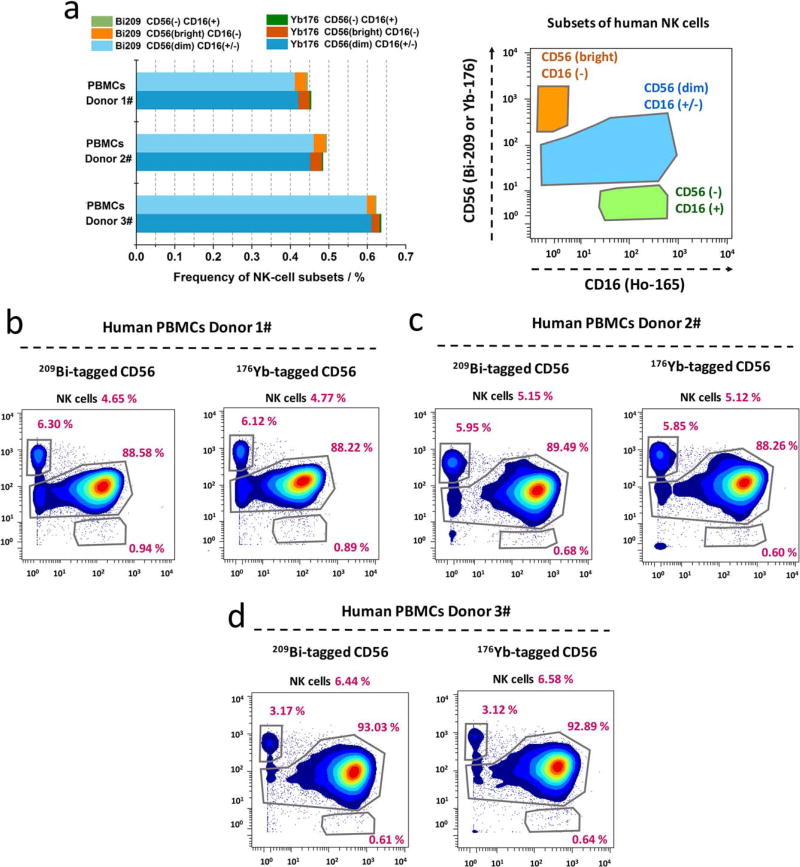
Human natural killer (NK) cells can be subdivided into several different cell populations based on the relative expression of surface proteins of CD16 and CD56. To evaluate the measurement accuracy of bismuth-tagged antibody in immunophenotyping assays, the percentages of NK cell subsets within human PBMCs from three healthy donors were determined using either 209 Bi- or 176Yb-tagged CD56 antibody. NK cells were gated based on CD3− CD20− CD7+ HLADR− from PBMCs, as shown in Supporting Information Figure S4. Three NK-cell subsets are defined on the basis of the relative expression of the marker CD56 and CD16: 1) CD56brightCD16−, 2) CD56dimCD16±, and 3) CD56−CD16+, as shown in Figure 6a. Highly comparable percentage of each NK cell subset was achieved by using either 209Bi- or 176Yb-tagged CD56 antibody for each human PBMCs donor as illustrated by biaxial scatter plots in Figures 6b–d. As a result, a linear regression of these two sets of data was obtained with Pearson correlation, r = 0.999; P < 0.00001, two-tailed t test, in Supporting Information Figure S5. Therefore, it indicates bismuth-tagged antibody provides the accurate quantification for single-cell immunophenotyping measurements. That proves there is not any cross-talking issue, when bismuth- and lanthanide-tagged antibodies are mixed and simultaneously employed in the CyTOF processing procedures.
Figure 6.
NK cell subsets quantified using either bismuth- or lanthanide-tagged CD56 antibodies. (a) Comparable frequencies of NK-cell subsets determined using either 209Bi- or 176Yb-tagged CD56 antibodies in human PBMCs from three healthy donors. NK cells are gated as Supporting Information S4, and three NK-cell subsets are gated as CD56brightCD16−, CD56dimCD16+/−, and CD56−CD16+. (b–d) Comparable frequencies of NK-cell subsets in each donor, depicted with biaxial scatter plots. (Pearson correlation, r = 0.999, P < 0.00001; Supporting Information Figure S5).
Single-Cell Biological Applications with Bismuth-Tagged Antibody
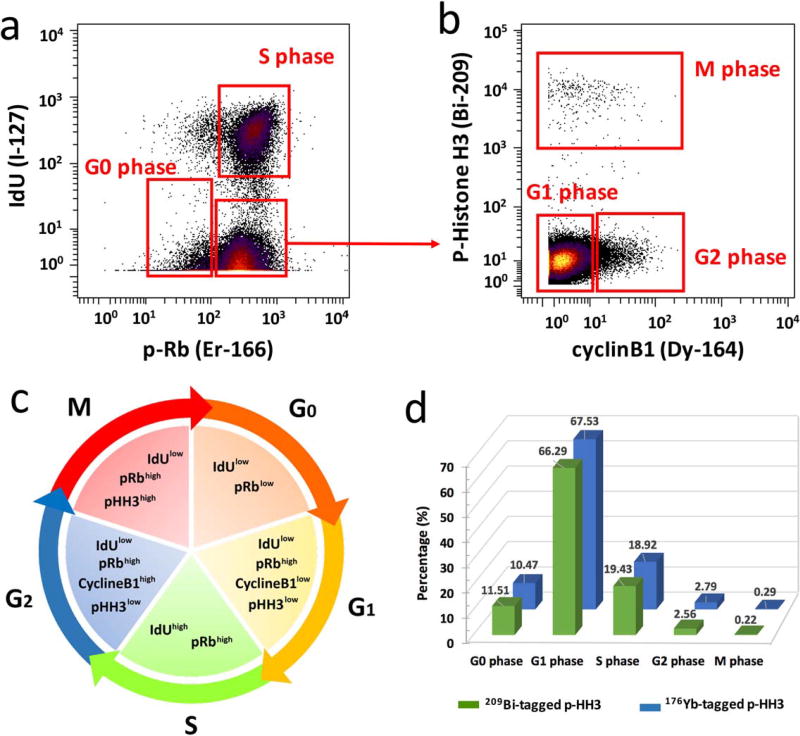
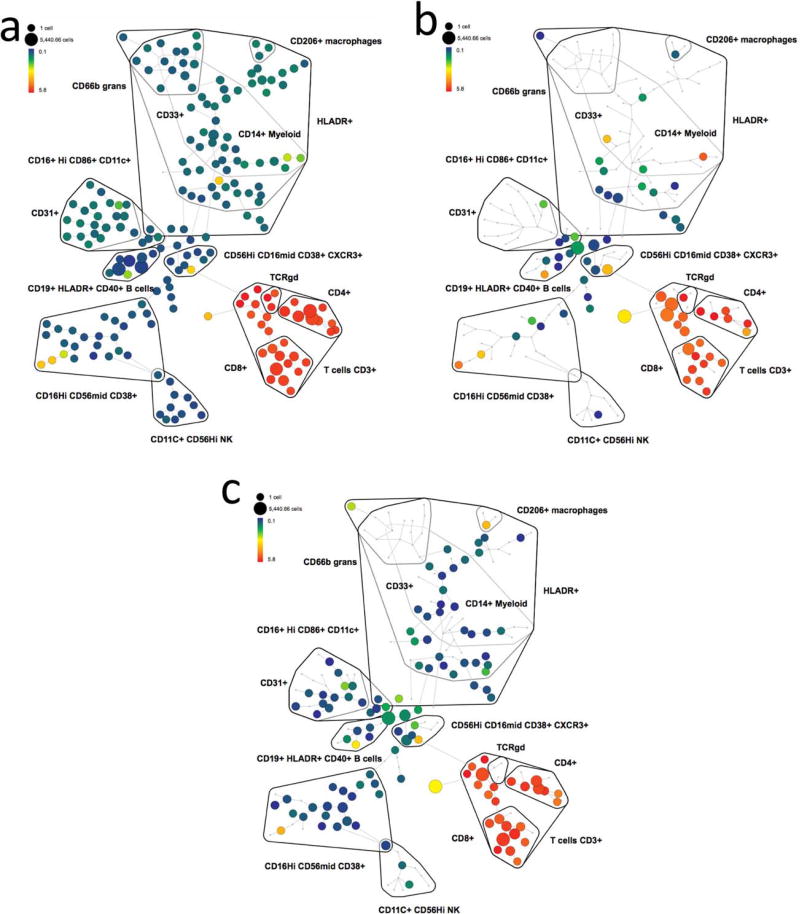
Using the presented conjugation methods, bismuth mass tag was conjugated to the antibody against intracellular marker of phosphorylated histone H3 (S28) as well. Then, the measurement of cell cycle phases of G0, G1, S, G2, and M was carried out using 209Bi-tagged histone H3, and lanthanide conjugated antibodies against cyclin B1, cyclin A, and phosphorylated retinoblastoma (p-Rb) protein (Ser 807/811), as well as IdU. Mesodermal progenitors derived from human embryonic stem cells were stained and analyzed as the procedures described in our previous studies (31). As shown in Figure 7a, the IdUlowp-Rblow population was defined as G0 phase cells, and the IdUhighp-Rbhigh as S phase cells. The IdUlowp-Rbhigh population contained G1, G2, and M phase cells. As shown in Figure 7b, the population with high expression of p-Histone H3 (pHH3) measured by bismuth-tagged antibody was the cells in all stages of M phase. The p-HH3low cycline-B1low population was identified as G1 phase cells and the p-HH3lowcyclineB1high as G2 phase cells. Figure 7c illustrates the summary of gating strategy for the cell phases. Consequentially, the analytical results of the percentage of each phase of the cell cycle using 209Bi-tagged pHH3 was demonstrated in Figure 7d, which were comparable with the measurements using the lanthanide counterpart of 176Yb-tagged pHH3 (Supporting Information Figure S6). Furthermore, in order to demonstrate the applicability of bismuth-tagged antibody for comprehensive high-dimensional immunoassays of CyTOF, Figure 8 depicts the SPADE analysis of 209Bi-tagged CD3 antibody on the immunophenotypic assays of human PBMCs and dissociated cells from human skin and kidney tissues. The panel of antibodies used for staining cell samples is listed in Supporting Information Table S1. Live single cells were manually gated using biaxial scatter plots as shown in Supporting Information Figure S7. Then, SPADE clustering was performed on the following markers: HLA-DR, CD4, CD8, CD206, CD66b, CD11c, CD33, CD19, CD16, CD163, CD14, CD3, and CD56, with the default configuration settings of 10% of down-sampling and 200 target nodes.
Figure 7.
Cell cycle measurements using 209Bi-tagged p-HH3 antibody. The analysis of mesodermal progenitors derived from human embryonic stem cells on day 6 of differentiation. (a) G0 and S phase populations are identified by biaxial scatter plots of IdU vs. p-Rb(Ser807/811). (b) G1, G2, and M phase populations are defined by the expression of phospho-histone H3(Ser28) and cyclin B1 among non-G0/S phase populations. (c) Gating strategy for each phase of the cell cycle. (d) Comparable percentages of cell phases determined using either 209Bi- or 176Yb-tagged phospho-histone H3 antibody.
Figure 8.
SPADE plots of analysis of cell samples using 209Bi-tagged CD3 antibody. The minimum spanning tree was constructed using SPADE analysis based on 13 cell surface markers. Each node of the SPADE tree is colored for the median value of CD3 expression from low (blue) to high (red). Three SPADE analyses were performed within the cell samples of (a) human PBMCs, (b) human skin tissue, and (c) human kidney tissue.
Discussion
Mass cytometry has enabled deeply high-dimensional and systematic single cell analysis beyond the capabilities of conventional fluorescence-based flow cytometry. The advances of elemental labeling reagents including metal-chelating polymers (25,26), metal-embedding normalization beads (47), and cell identification mass tags (30) have facilitated the analysis of heterogeneous immune cells in the ways that were previously not possible. In order to extend parameterization capabilities of mass cytometry technology, we employed the posttransition metal of bismuth as a mass tag for CyTOF immunoassays. The protocols for conjugation of bismuth mass tag to antibody, and characterization and quantification of antibody conjugates were optimized as detailed above. This work enabled an additional channel m/z = 209 of CyTOF to measure phenotypic and functional protein expressions in single cells from normal and diseased states. Using the presented protocols, bismuth mass tag was also conjugated to mouse MHC class II antibody, which was employed in the systemic immune assays for effective cancer immunotherapy (48).
Bismuth has nearly the same charge-to-radius ratio as that of lanthanide element and is therefore capable of complexing with DTPA chelator. However, the coordinative conditions of Bi3+ cations required for ligands are significantly different from those of Ln3+ cations. As a result, Ln3+ solutions are able to be stable in ammonium acetate buffer pH 5.5–6.0; however, Bi3+ solutions will precipitate in the same butter. The optimal acidic buffer for Bi3+ cations is 2.0–5.0% nitric acid. Notably, after complexation with DTPA chelators, both lanthanide and bismuth complexes (Ln@DTPA and Bi@DTPA) are stable in neutral solutions or a slightly basic buffer such as PBS pH 7.4, which is a good media for the coupling reactions of maleimide and thiol groups.
Another distinct feature of bismuth mass tag is the unique UV–Vis absorption spectrum in the range from 250 to 300 nm, which is resulting from molecular electronic transitions of Bi@DTPA complex from the ground to the excited energy states. The effluence of this property of bismuth mass tag is that the conventional approach for antibody quantification using A280 is not suitable to bismuth-tagged antibody due to the overlap absorption spectra. To address this issue, a modified BCA protein assay based on the measurement of UV–Vis absorbance at A562 was developed as a new quantification method. Furthermore, by taking the advantage of the unique UV–Vis absorbance of Bi@DTPA, a novel and convenient approach was developed to calculate the concentration of labeled Bi3+ cations and further to evaluate bismuth labeling efficiency without requiring the conventional method based on ICP-MS.
While bismuth element has different ion transition efficiency and ionization potential than lanthanide elements, the CyTOF sensitivity of bismuth is at the relatively high level compared to that of the series of lanthanide elements from 139La to 176Yb, which is comparable to the sensitivity of 170Er. Bismuth-tagged antibody provides the accuracy measurements of cell surface and intracellular markers, which give the evidence that there is not cross-talking between bismuth- and lanthanide-chelating Maxpar X8 polymers in the processes of simultaneously cell staining and CyTOF analysis. Additionally, bismuth has a single 100%-abundance isotope, which is easily accessible, and does not have polyatomic ion mass interference to lanthanides. Therefore, bismuth element is an excellent mas tag to increase the capacity of multiplexing of immunoassays in mass cytometry.
Supplementary Material
Acknowledgments
We thank Gina Jager for administrative support and Astraea Jager for mass cytometry quality control and instrument maintenance. We thank Gregory K. Behbehani, Sean C. Bendall, Matthew H. Spitzer, Christina Loh, and Elena W. Y. Hsieh for helpful discussions.
Grant sponsor: US National Institutes of Health, Grant numbers: U19 AI057229; 1U19AI100627; R01CA184968; R33 CA183654; R33 CA183692
Grant sponsor: US National Heart, Lung, and Blood Institute, Grant number: N01-HV-00242
Grant sponsor: US Department of Defense, Grant number: OC110674, 11491122
Grant sponsor: Bill & Melinda Gates Foundation, Grant number: OPP1113682
Grant sponsor: Food and Drug Administration, Grant number: HHSF223201210194C
Footnotes
Author Contributions
GH investigated bismuth complexation chemistry and developed bismuth mass tag and antibody conjugation and quantification protocols, performed the experiments and wrote the manuscript. SC performed cell cycle experiments and provided the advice for NK-cell experiments. VGM performed ovarian cancer experiments. ERZ provided advice and assistance with the experiments. WJF helped design the experiments and edited the manuscript. GPN helped design the experiments and edited the manuscript.
Competing Financial Interests
GPN had a personal financial interest in Fluidigm, the manufacturer of the mass cytometer used in this study, for the duration of this project.
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Robinson JP, Roederer M. Flow cytometry strikes gold. Science. 2015;350:739–740. doi: 10.1126/science.aad6770. 6262. [DOI] [PubMed] [Google Scholar]
- 2.Chattopadhyay PK, Roederer M. Cytometry: today’s technology and tomorrow’s horizons. Methods. 2012;57(3):251–258. doi: 10.1016/j.ymeth.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houk RS, Fassel VA, Flesch GD, Svec HJ, Gray AL, Taylor CE. Inductively coupled argon plasma as an ion-source for mass-spectrometric determination of trace-elements. Analyt Chem. 1980;52(14):2283–2289. [Google Scholar]
- 4.Thomas R. Practical Guide to ICP-MS: A Tutorial for Beginners. 2. Boca Raton: Crc Press-Taylor & Francis Group; 2008. pp. 1–339. [Google Scholar]
- 5.Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, Pavlov S, Vorobiev S, Dick JE, Tanner SD, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Analyt Chem. 2009;81(16):6813–6822. doi: 10.1021/ac901049w. [DOI] [PubMed] [Google Scholar]
- 6.Bendall SC, Simonds EF, Qiu P, Amir E-aD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner SD, Baranov VI, Ornatsky OI, Bandura DR, George TC. An introduction to mass cytometry: fundamentals and applications. Cancer Immunol Immunother. 2013;62(5):955–965. doi: 10.1007/s00262-013-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. A deep profiler’s guide to cytometry. Trends Immunol. 2012;33(7):323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaudillière B, Ganio EA, Tingle M, Lancero HL, Fragiadakis GK, Baca QJ, Aghaeepour N, Wong RJ, Quaintance C, El-Sayed YY, et al. Implementing mass cytometry at the bedside to study the immunological basis of human diseases: distinctive immune features in patients with a history of term or preterm birth. Cytometry Part A. 2015;87A(9):817–829. doi: 10.1002/cyto.a.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjornson ZB, Nolan GP, Fantl WJ. Single-cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol. 2013;25(4):484–494. doi: 10.1016/j.coi.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nicholas KJ, Greenplate AR, Flaherty DK, Matlock BK, Juan JS, Smith RM, Irish JM, Kalams SA. Multiparameter analysis of stimulated human peripheral blood mononuclear cells: a comparison of mass and fluorescence cytometry. Cytometry Part A. 2016;89A(3):271–280. doi: 10.1002/cyto.a.22799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Abbasi F, Ornatsky O, Cole KD, Misakian M, Gaigalas AK, He H-J, Marti GE, Tanner S, Stebbings R, et al. Human CD4(+) lymphocytes for antigen quantification: Characterization using conventional flow cytometry and mass cytometry. Cytometry Part A. 2012;81A(7):567–575. doi: 10.1002/cyto.a.22060. [DOI] [PubMed] [Google Scholar]
- 14.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, Pe’er D, Nolan GP, Bendall SC. Normalization of mass cytometry data with bead standards. Cytometry Part A. 2013;83A(5):483. doi: 10.1002/cyto.a.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai L, Ong R, Li J, Albani S. A CD45-based barcoding approach to multiplex mass-cytometry (CyTOF) Cytometry Part A. 2015;87A(4):369–374. doi: 10.1002/cyto.a.22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baranov VI, Quinn Z, Bandura DR, Tanner SD. A sensitive and quantitative element-tagged immunoassay with ICPMS detection. Analyt Chem. 2002;74(7):1629–1636. doi: 10.1021/ac0110350. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Zhang Z, Yu B, Shi J, Zhang X. Application of the biological conjugate between antibody and colloid Au nanoparticles as analyte to inductively coupled plasma mass spectrometry. Analyt Chem. 2002;74(1):96–99. doi: 10.1021/ac0103468. [DOI] [PubMed] [Google Scholar]
- 18.Han G, Zhang S, Xing Z, Zhang X. Absolute and relative quantification of multiplex DNA assays based on an elemental labeling strategy. Angew Chem Int Ed. 2013;52(5):1466–1471. doi: 10.1002/anie.201206903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz AR, Stanislawiak S, Baumgart S, Grützkau A, Mei HE. Silver nanoparticles for the detection of cell surface antigens in mass cytometry. Cytometry Part A. 2017;91A(1):25–33. doi: 10.1002/cyto.a.22904. [DOI] [PubMed] [Google Scholar]
- 20.Han G, Xing Z, Dong Y, Zhang S, Zhang X. One-step homogeneous DNA assay with single-nanoparticle detection. Angew Chem Int Ed. 2011;50(15):3462–3465. doi: 10.1002/anie.201006838. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Han G, Xing Z, Zhang S, Zhang X. Multiplex DNA assay based on nanoparticle probes by single particle inductively coupled plasma mass spectrometry. Analyt Chem. 2014;86(7):3541–3547. doi: 10.1021/ac404245z. [DOI] [PubMed] [Google Scholar]
- 22.Montoro Bustos AR, Garcia-Cortes M, González-Iglesias H, Ruiz Encinar J, Costa-Fernández JM, Coca-Prados M, Sanz-Medel A. Sensitive targeted multiple protein quantification based on elemental detection of quantum dots. Analyt Chim Acta. 2015;879:77–84. doi: 10.1016/j.aca.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Tong L, Lu E, Pichaandi J, Zhao G, Winnik MA. Synthesis of uniform NaLnF(4) (Ln: Sm to Ho) nanoparticles for mass cytometry. J Phys Chem C. 2016;120(11):6269–6280. [Google Scholar]
- 24.Lin W, Hou Y, Lu Y, Abdelrahman AI, Cao P, Zhao G, Tong L, Qian J, Baranov V, Nitz M, et al. A high-sensitivity lanthanide nanoparticle reporter for mass cytometry: tests on microgels as a proxy for cells. Langmuir. 2014;30(11):3142–3153. doi: 10.1021/la403627p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Polymer-based elemental tags for sensitive bioassays. Angew Chem Int Ed. 2007;46(32):6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majonis D, Herrera I, Ornatsky O, Schulze M, Lou X, Soleimani M, Nitz M, Winnik MA. Synthesis of a functional metal-chelating polymer and steps toward quantitative mass cytometry bioassays. Analyt Chem. 2010;82(21):8961–8969. doi: 10.1021/ac101901x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinsteuber K, Corleis B, Rashidi N, Nchinda N, Lisanti A, Cho JL, Medoff BD, Kwon D, Walker BD. Standardization and quality control for high-dimensional mass cytometry studies of human samples. Cytometry Part A. 2016;89A(10):903–913. doi: 10.1002/cyto.a.22935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi C, Au-Yeung A, Fuh F, Ramirez-Montagut T, Bolen C, Mathews W, O’Gorman WE. Mass cytometry panel optimization through the designed distribution of signal interference. Cytometry Part A. 2017;91A(1):39–47. doi: 10.1002/cyto.a.22977. [DOI] [PubMed] [Google Scholar]
- 29.Sumatoh HR, Teng KWW, Cheng Y, Newell EW. Optimization of mass cytometry sample cryopreservation after staining. Cytometry Part A. 2017;91A(1):48–61. doi: 10.1002/cyto.a.23014. [DOI] [PubMed] [Google Scholar]
- 30.Ornatsky OI, Lou X, Nitz M, Sheldrick WS, Baranov VI, Bandura DR, Tanner SD. Study of cell antigens and intracellular DNA by identification of element-containing labels and metallointercalators using inductively coupled plasma mass spectrometry. Analyt Chem. 2008;80(7):2539–2547. doi: 10.1021/ac702128m. [DOI] [PubMed] [Google Scholar]
- 31.Behbehani GK, Bendall SC, Clutter MR, Fantl WJ, Nolan GP. Single-cell mass cytometry adapted to measurements of the cell cycle. Cytometry Part A. 2012;81A(7):552–566. doi: 10.1002/cyto.a.22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP, Bodenmiller B. A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry Part A. 2012;81A(6):467–475. doi: 10.1002/cyto.a.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mei HE, Leipold MD, Maecker HT. Platinum-conjugated antibodies for application in mass cytometry. Cytometry Part A. 2016;89A(3):292–300. doi: 10.1002/cyto.a.22778. [DOI] [PubMed] [Google Scholar]
- 34.Edgar LJ, Vellanki RN, Halupa A, Hedley D, Wouters BG, Nitz M. Identification of hypoxic cells using an organotellurium tag compatible with mass cytometry. Angew Chem Int Ed. 2014;53(43):11473–11477. doi: 10.1002/anie.201405233. [DOI] [PubMed] [Google Scholar]
- 35.Edgar LJ, Vellanki RN, McKee TD, Hedley D, Wouters BG, Nitz M. Isotopologous organotellurium probes reveal dynamic hypoxia in vivo with cellular resolution. Angew Chem Int Ed. 2016;55(42):13159–13163. doi: 10.1002/anie.201607483. [DOI] [PubMed] [Google Scholar]
- 36.Zunder ER, Finck R, Behbehani GK, Amir ED, Krishnaswamy S, Gonzalez VD, Lorang CG, Bjornson Z, Spitzer MH, Bodenmiller B, et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc. 2015;10(2):316–333. doi: 10.1038/nprot.2015.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mei HE, Leipold MD, Schulz AR, Chester C, Maecker HT. Barcoding of live human peripheral blood mononuclear cells for multiplexed mass cytometry. J Immunol. 2015;194(4):2022–2031. doi: 10.4049/jimmunol.1402661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Catena R, Özcan A, Zivanovic N, Bodenmiller B. Enhanced multiplexing in mass cytometry using osmium and ruthenium tetroxide species. Cytometry Part A. 2016;89A(5):491–497. doi: 10.1002/cyto.a.22848. [DOI] [PubMed] [Google Scholar]
- 39.Stern AD, Rahman AH, Birtwistle MR. Cell size assays for mass cytometry. Cytometry Part A. 2017;91A(1):14–24. doi: 10.1002/cyto.a.23000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majonis D, Ornatsky O, Kinach R, Winnik MA. Curious results with palladium- and platinum-carrying polymers in mass cytometry bioassays and an unexpected application as a dead cell stain. Biomacromolecules. 2011;12(11):3997–4010. doi: 10.1021/bm201011t. [DOI] [PubMed] [Google Scholar]
- 41.Behr TM, Béhé M, Stabin MG, Wehrmann E, Apostolidis C, Molinet R, Strutz F, Fayyazi A, Wieland E, Gratz S, et al. High-linear energy transfer (LET) alpha versus low-LET beta emitters in radio immunotherapy of solid tumors: Therapeutic efficacy and dose-limiting toxicity of Bi-213-versus Y-90-labeled CO17-1A Fab ’ fragments in a human colonic cancer model. Cancer Res. 1999;59(11):2635–2643. [PubMed] [Google Scholar]
- 42.Wild D, Frischknecht M, Zhang H, Morgenstern A, Bruchertseifer F, Boisclair J, Provencher-Bolliger A, Reubi J-C, Maecke HR. Alpha-versus beta-particle radiopeptide therapy in a human prostate cancer model (Bi-213-DOTA-PESIN and Bi-213-AMBA versus Lu-177-DOTA-PESIN) Cancer Res. 2011;71(3):1009–1018. doi: 10.1158/0008-5472.CAN-10-1186. [DOI] [PubMed] [Google Scholar]
- 43.Fluidigm Corporation Maxpar Reagents. http://maxpar.fluidigm.com.
- 44.Tricot S, Meyrand M, Sammicheli C, Elhmouzi-Younes J, Corneau A, Bertholet S, Malissen M, Le Grand R, Nuti S, Luche H, et al. Evaluating the efficiency of isotope transmission for improved panel design and a comparison of the detection sensitivities of mass cytometer instruments. Cytometry Part A. 2015;87A(4):357–368. doi: 10.1002/cyto.a.22648. [DOI] [PubMed] [Google Scholar]
- 45.Casely IJ, Ziller JW, Mincher BJ, Evans WJ. Bismuth coordination chemistry with allyl, alkoxide, aryloxide, and tetraphenylborate ligands and the {2,6-(Me2NCH2)(2)C6H3 (2)Bi}(+) cation. Inorg Chem. 2011;50(4):1513–1520. doi: 10.1021/ic102119y. [DOI] [PubMed] [Google Scholar]
- 46.Liu HC, May K. Disulfide bond structures of IgG molecules: structural variations, chemical modifications and possible impacts to stability and biological function. Mabs. 2012;4(1):17–23. doi: 10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdelrahman A, Dai S, Thickett SC, Ornatsky O, Bandura D, Baranov V, Winnik MA. Lanthanide-containing polymer microspheres by multiple-stage dispersion polymerization for highly multiplexed bioassays. J Am Chem Soc. 2009;131(42):15276–15283. doi: 10.1021/ja9052009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spitzer MH, Carmi Y, Reticker-Flynn NE, Kwek SS, Madhireddy D, Martins MM, Gherardini PF, Prestwood TR, Chabon J, Bendall SC, et al. Systemic Immunity Is Required for Effective Cancer Immunotherapy. Cell. 2017;168(3):487–502. doi: 10.1016/j.cell.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.