Abstract
Radiology (imaging) and imaging-guided interventions, which provide multi-parametric morphologic and functional information, are playing an increasingly significant role in precision medicine. Radiologists are trained to understand the imaging phenotypes, transcribe those observations (phenotypes) to correlate with underlying diseases and to characterize the images. However, in order to understand and characterize the molecular phenotype (to obtain genomic information) of solid heterogeneous tumours, the advanced sequencing of those tissues using biopsy is required. Thus, radiologists image the tissues from various views and angles in order to have the complete image phenotypes, thereby acquiring a huge amount of data. Deriving meaningful details from all these radiological data becomes challenging and raises the big data issues. Therefore, interest in the application of radiomics has been growing in recent years as it has the potential to provide significant interpretive and predictive information for decision support. Radiomics is a combination of conventional computer-aided diagnosis, deep learning methods, and human skills, and thus can be used for quantitative characterization of tumour phenotypes. This paper discusses the overview of radiomics workflow, the results of various radiomics-based studies conducted using various radiological images such as computed tomography (CT), magnetic resonance imaging (MRI), and positron-emission tomography (PET), the challenges we are facing, and the potential contribution of radiomics towards precision medicine.
Keywords: Radiological imaging, Personalised medicine, Precision medicine, Quantitative imaging, Radiogenomics, Radiomics
1. Introduction
Precision medicine is gaining popularity for providing customised or personalised healthcare (Chen and Snyder, 2013). It employs diagnostic testing for the selection of appropriate and optimal therapies (treatments) based on the patient’s genetic context (Chen and Snyder, 2013). Thus, by practising precision medicine in healthcare, medical decisions and treatments can be tailored to suit every individual patient’s need. Diagnostic radiology and image-guided intervention are playing an increasingly significant role in precision medicine. For decades, radiology has been dealing with diagnosis and has provided anatomical abnormality information. With the advent of digital technology, quantitative imaging has been contributing to improved diagnosis and in addition, radiological imaging has become a recognised technology in the clinical assessment and confirmation of a disease (Cook et al., 2014; WHO, 2017). One of the significant contributions of radiology is in oncology, as cancer has become the number one source of mortality in recent years and there is an increasing proliferation of cancer cases in developing countries (Hawkins et al., 2014). Different radiological imaging modalities have the capability to detect different information that is present on the various types of tissue. However, extracting informative data from all radiological images is difficult in this era of ‘big data’. The latest progress in computational power and the exploitation of genomics have given rise to a recently developed field of research coined ‘Radiomics’.
Radiomics is a promising field of medical research that employs state-of-the-art machine learning techniques to extract imaging features from various modalities such as computed tomography (CT), magnetic resonance imaging (MRI), and positron-emission tomography (PET) to objectively and computably characterise tumour phenotypes. Radiomics was first formally introduced by Lambin et al. in 2012 (Lambin et al., 2012; Scrivener et al., 2016). It is the extraction and study of huge quantities of features from radiological images and these data are used to predict or decode concealed genetic and molecular traits for decision support (Kumar et al., 2012; Cook et al., 2014; Court et al., 2016; Gillies et al., 2016; Narang et al., 2016; Yip and Aerts, 2016; Sala et al., 2017). Radiomics features contain useful spatial and textural information on the greyscale patterns and the correlation between image pixels. Further, these features can be modelled into computer-aided systems that can supplement as an adjunct instrument for individualised diagnosis and treatment guidance (Parekh and Jacobs, 2016). Hence, information obtained from these radiological images can also be combined with additional ‘omics’ (genomics, proteomics, metabolomics, and transcriptomics) data for further analysis.
Radiomics has been extensively studied in oncology with a substantial contribution from the Quantitative Imaging Network (QIN) and National Cancer Institute (NCI) (Gillies et al., 2016). It has been researched and reported, notably in, breast cancer (Wu et al., 2016), glioblastoma (Narang et al., 2016), head and neck cancer (Wong et al., 2016), lung cancer (Scrivener et al., 2016), oesophageal cancer (van Rossum et al., 2016), prostate cancer (Stoyanova et al., 2016), and rectal cancer (Dinapoli et al., 2016). Furthermore, radiomics has been adopted in dermatology (Cho et al., 2015).
2. Workflow of radiomics
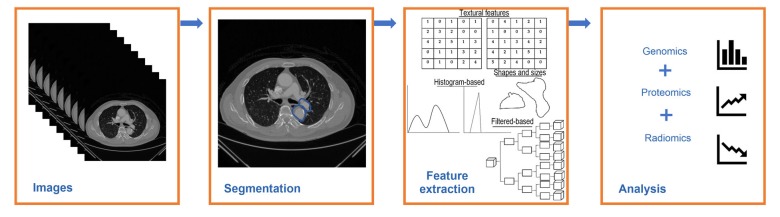
Fig. 1 shows a typical workflow of radiomics that consists of four main stages.
Fig. 1.
A typical workflow of radiomics
2.1. Images
The radiological images are acquired from different non-invasive imaging modalities such as CT, MRI, and PET (Castellino, 2005; Lambin et al., 2012; Aerts, 2016; Court et al., 2016; Yip and Aerts, 2016). These images are pre-processed to ensure that they are consistent and uniformity is maintained. Then, all images are progressively pooled together to form a large database. In addition, the image contrast is enhanced using a technique known as Contrast Limited Adaptive Histogram Equalization (CLAHE) and the noise and artefacts are removed in this pre-processing step (Pizer et al., 1987; Wang et al., 2005).
2.2. Segmentation
Segmentation is an essential step as highly distinctive features are obtained from the segmented area of the images and these features are used in the development of a model for automated screening of radiological images. The aim of segmentation is to simplify or to modify the image for more meaningful analyses (Aerts, 2016).
Once the images are pre-processed, segmentation of a region of interest (ROI) is performed. Initially, an ROI is selected as a part of an image that is of interest and where some machine learning algorithms have to be applied (https://www.mathworks. com/help/images/roi-processing-in-image-processing-toolbox.html). Segmentation of an ROI can be carried out manually, semi-automated, or fully automated. Comparative studies have been reported to increase the performance of fully automated, manual, or semi-automated segmentation of these images (Egger et al., 2013; Velazquez et al., 2013, 2015; Kumar et al., 2016; Narang et al., 2016). The 3D-Slicer is a free open source segmentation software for biomedical research that is widely adopted in the medical field (Velazquez et al., 2013).
Manual segmentation of the tumour volume is a normal clinical procedure in the planning process before patients receive radiotherapy (Eminowicz and McCormack, 2015). Manual delineation is easy but it is highly subjective and labour-intensive. In addition, manual segmentation is not recommended for radiomics evaluation as it requires a large amount of data and would be very time-consuming. Semi-automated segmentation and fully automated segmentation can be performed on radiological images with the help of software and segmentation algorithms. Furthermore, the present consensus is to implement computer-aided edge detection followed by manual adjustment (Gillies et al., 2016).
The various segmentation techniques namely active contour, level-set, region-based and graph-based methods have been used. The active contour model relies on knowledge of size, position, and structure of the ROI. Based on prior knowledge, the active contour can outline the ROI for segmentation (Kass et al., 1988). The level-set model retrieves the shapes and contours from medical images to two or three dimensions (Malladi et al., 1995). It is also a sort of active contour modelling (Malladi et al., 1995). The region-based technique, on the other hand, is a method based on grey level features. This technique relies on the principle of homogeneity where the ROI is determined by examining its neighbouring pixels relative to the initial seed point (Sonka et al., 2007). The graph-based approach is based on the selection of edges from the graph where every single pixel in the image corresponds to a node in the graph (Felzenszwalb and Huttenlocher, 2004). The segmentation criterion is then adjusted according to the variability of the pixels in the neighbourhood (Felzenszwalb and Huttenlocher, 2004). These segmentation techniques are commonly used in a computer-aided detection system.
Recently, deep learning-based approaches such as the convolutional neural network (CNN) have been used for medical image segmentation and demonstrated promising results (Ma et al., 2017a, 2017b).
2.3. Feature extraction
Feature extraction is the next step after an ROI is identified and segmented. Feature extraction is the selection of useful information to assist in the characterization of normal and abnormal radiological images. These features are comprised of intensity, shape, size, and texture of an ROI identified on the images.
The shape and size of the tumours are regularly used to define lesions which can be easily identified without computer assistance. On the other hand, the texture of a radiological image is defined by the way the grey levels are apportioned over the pixels in an image (van Rossum et al., 2016). These texture features are mathematically extracted using first-order, or second-order, and/or higher-order statistical methods (Davnall et al., 2012; Mitra and Shankar, 2015; van Rossum et al., 2016).
First-order statistics are characterised based on the distribution of values of individual voxels without concern for spatial relationships. They are generally histogram-based techniques which reduce an ROI to single value representation for entropy, kurtosis, maximum, mean, median, minimum, skewness, and uniformity of the intensities on the radiological image (Gillies et al., 2016). Second-order statistics are universally defined as textural features. They define statistical correlations between voxels with alike or unlike contrast value (Gillies et al., 2016). Grey level co-occurrence matrix (GLCM), grey level run-length matrix, and size-zone matrix are typically extracted from the images. Higher-order statistics set filter grids to obtain patterns on these images. Fractal analysis, Gabor transform, Laplacian transforms of Gaussian bandpass filters, laws kernel, and wavelets are commonly performed to extract textural information from the images (Yip and Aerts, 2016). Various textural features extracted from radiological images are summarised in Table 1.
Table 1.
Summaries of selected radiomics studies
| Author (year) | Objective of study | Radiomics features | Techniques used | Performance |
| Computed tomography | ||||
| Hunter et al. (2013) | Identify radiomics features that are consistent across multiple diagnostic tools | Geometry, intensity histogram, absolute gradient image, textural features | Hierarchical clustering, dice similaritycoefficient, Jaccard index, CCC | Feature reproducibility is depended on the machine and images |
| Velazquez et al. (2013) | Evaluate the clinical applicability of a semiautomatic segmentation method | 3D-Slicer algorithm | It is noted that 3D-Slicer illustrated high agreement, smaller uncertainty areas, and lower volume differences as compared to manual slice delineation | |
| Aerts et al. (2014) | Assess the correlation of radiomics features with clinical data and the predictive capability of radiomics features | Intensity histogram, shape, textural features, wavelet features | Friedman test, Wilcoxon test, Kaplan-Meier analysis | It is concluded that radiomics features possess prognostic capability |
| Balagurunathan et al. (2014b) | Assess the reproducibility of quantitative imaging features | Intensity histogram, textural features, Laws’ kernels features | Test-retest CCC, Kaplan-Meier plot | Texture features are statistically significant |
| Balagurunathan et al. (2014a) | Assess the reproducibility of image features | Morphological features, first-order features, wavelets feature | CCC, dynamic range, redundancy reduction, weighted Kappa index, optical threshold linear discriminant analysis, ROC curve | It is concluded that most features show high reproducibility with semi-automated segmentation |
| Fried et al. (2014) | Investigate if pre-treatment textural features can enhance the predictive capability in stage III non-small cell lung cancer | Intensity histogram, textural features | Kaplan-Meier curves, leave-one-out cross-validation, CCC | Pre-treatment textural features have better predictive values |
| Parmar et al. (2014) | Evaluate the performance of using 3D-Slicer | Intra-class correlation coefficient | Radiomics features obtained from 3D-Slicer had notably higher reproducibility than those from manual delineation | |
| Hawkins et al. (2014) | Propose an automated prognostic model | Intensity histogram, textural features, geometric features | Decision tree, leave-one-out cross-validation | Accuracy=77.50% |
| Coroller et al. (2015) | Prognostic capability to identify distant metastasis for lung adenocarcinoma patients | Intensity histogram, shape and texture | Laplacian of Gaussian filter, wavelet filter, minimum relevance feature selection | Radiomics features possess strong prognostic capability |
| Parmar et al. (2015a) | Compare radiomics features across different cancer types | Intensity histogram, shape, textural and wavelet features | Consensus clustering, ROC analysis, concordance index, logistic regression, Jaccard index | Radiomics features possess clustering and prognostic characteristics |
| Parmar et al. (2015b) | Assess the different feature selection and classification methods in terms of their performance and prognostic capability | Intensity histogram, textural features | Various feature selection and classification techniques implemented | It is concluded that conditional infomax feature extraction, minimum redundancy maximum relevance, and mutual information feature selection produced the highest stability and predictive capability |
| Cunliffe et al. (2015) | Investigate the correlation of radiation dose and radiomics features and the capability of radiomics features in the analysis of radiation pneumonitis | Textural features, fractal features | Laws’ filters, linear regression model, ANOVA, ROC curve | Radiomics is a promising methodology that can diagnose patients with radiation pneumonitis |
| Mackin et al. (2015) | Study the variability between different CT scanners | Textural features | General Electric, Philips, Siemens, and Toshiba scanners, feature noise introduced | It is verified that the variability in features corresponded in the variability seen in the radiomics features obtained from the images |
| Mattonen et al. (2016) | Physician assessment versus radiomic assessment in the detection of local cancer recurrence for lung cancer | First-order statistics features, textural features | PRTools 5.0 (Delft Pattern Recognition Research, Delft) was utilised for feature selection and classification, leave-one-out cross-validation, SVM classifier | Radiomics can detect early changes associated with local recurrence |
| Mattonen et al. (2015) | Analysis of features for prognostic capability of lung cancer recurrence | Textural features | PRTools 5.0 (Delft Pattern Recognition Research, Delft) was utilised for classification, Wilcoxon signed rank test | High prognostic accuracy using GLCM |
| Wang et al. (2016) | Initiate and evaluate a normalised set of features obtained from CT images and their correlations with overall survival | Lexicon of BI-RADS, Fleischner Society as a guide to establish features | Weighted Kappa index, Kaplan-Meier analysis, CART classifier, PCA | It was reported that textural features are important in the characterisation of lung adenocarcinomas |
| Fave et al. (2015) | Validate the usefulness of predictive models using radiomics features extracted from CT images | Textural features | CCC analysis | Certain radiomics features are resistant to poor-quality CBCT images and noises |
| Huynh et al. (2016) | Analyse stereotactic body radiation therapy with lung cancer | Textural, shape, and statistical features | PCA, factor analysis, Wilcoxon rank-sum test | Radiomics features possess the potential to be predictive |
| Szigeti et al. (2016) | Propose an innovative technique to diagnose lung diseases | Nonlinear features | Radiomics-based fractal dimension analysis, Kruskal-Wallis test, Mann-Whitney post hoc test, Gaussian curves | It is reported that the proposed method is promising and can be implemented in clinical practices |
| Coroller et al. (2016) | Investigate if pre-treatment radiomics features have the prognostic capability in non-small cell lung cancer | Textural features | Wilcoxon-test, Kaplan-Meier analysis, logistic regression, ROC curve | Radiomics features are predictive |
| Zhao et al. (2016) | Investigate the reproducibility of radiomics features | Intensity histogram, morphological features, textural features | Gabor energy, wavelets, Laplacian of Gaussian, model-based feature of fractal dimension, CCC analysis | It is noted that radiomics features are reproducible over different algorithms |
| Yang et al. (2016) | Analyse quantitative imaging features in lung tumours | Intensity histogram, textural features, geometric shape | CCC analysis, Spearman’s correlation, PCA, edge-preserving smoothing filter | Texture features could be utilised for predictive analysis in the future |
| Song et al. (2016) | Investigate if tumour heterogeneity of non-small cell lung cancer can be predicted with radiomics | Textural features | Statistical analysis | Radiomics has prognostic capability for clinical aided diagnosis |
| Wu et al. (2016) | Identify the classifiers for radiomics in lung cancer histology | Intensity histogram, shape features, textural features | Twenty-four feature selection methods, random forest classifier, naïve Bayes classifier, K-NN classifier, ROC curve | Naïve Bayes classifier performed the best; the area under ROC curve: accuracy=0.72 |
| Fave et al. (2016) | Evaluate how different image pre-processing techniques may impact the predictive outcome in univariate analysis | Intensity histogram, textural features | Harrell’s concordance index (c-index), Benjamini-Hochberg procedure | Pre-processing of CT images has an impact on the volume dependence of a feature |
| He et al. (2016) | Examine the effects of contrast-enhancement, reconstruction slice thickness, and convolution kernel on the diagnosis performance of radiomics | Textural features | Laplacian of Gaussian spatial band-pass filter, statistical analysis, Mann-Whitney U-test | It is noted that contrast-enhancement, reconstruction slice thickness, and convolution kernel affect the performance of radiomics |
| Emaminejad | Develop a new quantitative image feature analysis scheme and | Textural features | Naïve Bayesian network-based classifier, leave-one-case-out cross-validation, synthetic minority oversampling technique | Accuracy=80.00% |
| Liang et al. (2016) | Review the prognostic ability of radiomics features for the staging of colorectal cancer | Textural features | Logistic regression model, Mann-Whitney U-test, ROC curve | The area under ROC curve: accuracy=0.792; sensitivity=0.611; specificity=0.680 |
| Huang et al. (2016) | Justify the prognostic ability of lymph node metastasis in patients using radiomics nomogram | Textural features | Statistical analysis, multivariable logistic regression analysis | Radiomics nomogram is proved to be useful in clinical settings |
| Kumar et al. (2016) | Analyse an automatic liver segmentation | Statistical parameter-based approach | It is proven that the proposed method has prognostic capability | |
|
Magnetic resonance imaging | ||||
| Egger et al. (2013) | Assess the advantages of 3D-Slicer over manual segmentation | Dice similarity coefficient, Hausdorff distance | It is reported that 3D-Slicer is more time-efficient and is statistically comparable to manual segmentation | |
| Zhou et al. (2014) | Determine the differentiating ROIs within the tumour for clinical practices | Textural features | K-NN classifier | K-NN classifier: accuracy=93.75% |
| Coquery et al. (2014) | Assess the possibility if histologic properties can be extracted based on an automated analysis | Gaussian mixture modelling, t-tests, linear discriminant analysis | It is reported that the integration of spatial selection and cluster analysis can be implemented to form information obtained from the multiparametric MRI | |
| Chaddad et al. (2015) | Validate the effectiveness of features extracted from GLCM in glioblastoma phenotypes | Textural features | Nearest neighbour classifier | Accuracy=75.58%; sensitivity=63.95%; specificity=90.69% |
| Chaudhury | Validate a new method in the study of heterogeneity in breast cancer | Textural kinetic features | CCC-based random subspace method, naïve Bayes classifier, decision tree classifier, SVM classifier, Kappa statistic | It was concluded that textural kinetic features were more prognostic than features obtained from the whole tumour |
| Upadhaya et al. (2015a) | Study on multimodal MRI in glioblastoma multiforme | Intensity histogram, textural features | SVM classifier | Accuracy=90.00%; sensitivity=85.00%; specificity=95.00% |
| Upadhaya et al. (2015b) | Propose a workflow for a predictive model based on textural features | Intensity histogram, textural features | SVM classifier | T1 pre-contrast: accuracy=60.00%; T1 post-contrast: accuracy=82.50%;T2: accuracy=72.50%;FLAIR: accuracy=75.00%; T1 pre-contrast/T1 post-contrast: accuracy=90.00% |
| Lee et al. (2015) | Identify the correlation of radiomics features with EGFR-driven tumours | Spatial diversity features | Spatial diversity analysis, ROC curve, | Characterising EGFR-driven tumours, the area under ROC curve: accuracy=0.790 |
| Depeursinge et al. (2015) | Assess the significance of intensity and textural features | Nonlinear features, textural features | Riesz wavelet, concordance index, SVM classifier, Cox-LASSO predictive model | C-index=0.81±0.02. It is concluded that accuracy is better when features are extracted based on solid components of a tumour instead of the entire tumour |
| Khalvati et al. (2015) | The automatic detection system of prostate cancer using texture features | Statistical features, textural features, Gabor features | Gabor filter, SVM classifier | Proposed technique surpasses conventional model |
| Velazquez et al. (2015) | Assess the predictive value of glioblastoma automatically segmented and compare it with manual segmentation | VASARI features | The brain tumour image analysis software was utilised, statistical analysis, Spearman rank correlation, CCC analysis, decision tree classifier | It is reported that the automatically segmented datasets have potential in medical imaging research |
| Guo et al. (2015) | Determine the efficiency of the prognostic outcome when combining genomic and radiomics features | Morphological texture, kinetic curve assessment, enhancement-variance kinetic features | Quantitative radiomics analysis, logistic regression, Benjamini-Hochberg procedure, cross-validation with ROC curve | Radiomics features have prognostic capability in determining pathological stage |
| Chung et al. (2015) | Present an innovative automated prostate detection algorithm | Intensity histogram, textural features | Radiomics-driven conditional random field framework, Kirsch edge detection, Gabor filters, SVM classifier | Accuracy=91.17%; sensitivity=71.47%; specificity=91.93% |
| van den Burg et al. (2016) | Analysis of labyrinth | Intensity histogram | Fourier DCT filter, edge detection, gradient orientation filter, entropy filter, Laplacian filter, ridge filter, discrete wavelet transform, image saliency filter, clustering components, morphological components, and binarize colour tone mapping | There was significant statistical difference between normal and patients |
| Grossmann et al. (2016) | Investigate the correlation of radiomics features and molecular pathways | Volumetric features | Gene expression, pathway analysis | It is noted that radiomics features contain highly significant features |
|
Positron emission tomography | ||||
| Nair et al. (2012) | Investigate radiogenomics with PET | Textural features | Student’s t-test, chi-squared test, Fisher’s exact test, PCA, SUV, Kaplan-Meier curves | It is noted that there is a correlation between gene signatures and features extracted with ʏ18һF-FDG |
| Leijenaar et al. (2013) | Carry out a stability analysis of radiomics features in non-small cell lung carcinoma | Intensity histogram, textural features | SUV discretisation, ICC, non-parametric ANOVA | Radiomics features possess prognostic capability |
| Leijenaar et al. (2015) | Determine the importance of a standardised procedure in tumour texture analysis | Textural features | SUV discretisation, Kruskal-Wallis one-way ANOVA | Experiments showed the importance of standardised procedure in tumour texture analysis |
| Ypsilantis et al. (2015) | Determine if machine learning techniques can predict cancer’s metabolic profile and compare the performance of machine learning algorithms with 3S-convolutional neural network | Textural features | Convolutional neural networks, SUV, PCA, SVM classifier, logistic regression | Sensitivity=80.70%; specificity=81.60%. Convolutional neural networks possess better prognostic capability of response to therapy |
| Tixier et al. (2015) | Compare the results of the visual and prognostic assessment in non-small cell lung cancer | Textural features | SUV, t-test, Spearman rank coefficient, Kaplan-Meier method, Cox regression model | The assessments of both visual and prognostic using textural features are not contradictory to each other |
| Nyflot et al. (2015) | Study the influence of stochastic outcome on textural features | Intensity histogram, textural features | Power analysis | It is concluded that protocols should be implemented to ensure that textural features extracted are standardised |
| Grootjans et al. (2016) | Investigate the effect of respiratory gating and noises in PET images | Textural features | Fuzzy locally adaptive Bayesian segmentation algorithm, ranking tests | It is reported that textural features in this work are resilient in the existence of respiratory motion |
| Lian et al. (2016) | Propose a novel methodology for the prognostic tool in PET imaging | Textural features, SUV-based features | SUV, Dempster-Shafer theory for feature selection, ADASYN, K-NN classifier | Proposed methodology showed promising results |
|
Positron emission tomography/computed tomography | ||||
| Cheebsumon et al. (2012) | Comparing PET-and CT-based methods to pathology | SUV, statistical analysis | Diameter from PET-based delineation has reported a more similar result with pathology as compared to CT-based delineation | |
| Yoon et al. (2015) | Discover the predictors of tumours for lung adenocarcinoma | Morphological features, histogram-based, regional, and local features | Chi-squared test, Student’s t-test, SUV, ten-fold cross-validation method | This methodology has potential to be utilised in clinical practice |
| Oliver et al. (2015) | Investigate the difference of image features between RG and 3D images | Sphericity, spherical disproportion, entropy, sum entropy, textural features | SUV, long axis calculation and rotation analysis | It is reported that features obtained from 3D and RG images are different |
| van Velden et al. (2016) | Evaluate the impact of reconstruction techniques and the delineation of radiomics features in non-small cell lung cancer | Fractal features, textural features | SUV, statistical analysis, Wilcoxon signed-rank test | It is noted that the performance of radiomics features is better in delineation than that of applied reconstruction technique |
| Bailly et al. (2016) | Assess the robustness of textural features | Textural features | SUV, coefficient of variation, one-way ANOVA, Tukey HSD test | SUV-based metrics (energy, entropy, RP, and ZP) are found to be robust features |
| Desseroit et al. (2016) | Initiate a nomogram by blending clinical and imaging features | Textural features | SUV, Kaplan-Meier method, log-rank test | Textural features can be used to create a nomogram but need to be further validated |
|
Positron emission tomography/magnetic resonance imaging | ||||
| Vallières et al. (2015) | Develop a joint FDG-PET/CT and MRI texture-based model for early diagnosis of lung metastasis risk | Textural features | SUV, discrete wavelet transform, wavelet band-pass filtering, logistic regression, multivariable analysis, ROC curve | The area under ROC curve: accuracy=98.4%; sensitivity=95.5%; specificity=92.6% |
| Antunes et al. (2016) | Investigate the capability of radiomics features in FLT-PET and MRI | Filtered-based, entropy and textural features from PET images | Sobel and Kirsch edge filters, Gaussian low-pass filter, SUV, Bhattacharyya distance | Radiomics has potential to be an effective tool for categorising treatment response in PET/MRI |
|
Ultrasonography | ||||
| Acharya et al. (2016b) | Propose a novel algorithm to accurately classify non-alcoholic fatty liver disease | DCT | Radon transform, fuzzy sugeno, fatty liver disease index | Accuracy=100.00%; sensitivity=100.00%; specificity=100.00% |
| Acharya et al. (2016c) | Develop an automated thyroid screening system | Entropies | Gabor transform, statistical analysis, C4.5 decision tree | Accuracy=94.30% |
| Acharya et al. (2016a) | Assess the reliability and robustness of an automated fatty liver disease and cirrhosis diagnosis system | Higher-order spectra and entropies | Probabilistic neural network, liver disease index | Accuracy=97.33%;sensitivity=96.00%;specificity=100.00% |
| Acharya et al. (2017) | Evaluate the performance of a new ultrasonography procedure | Second-order statistics | Quadratic discriminant analysis, shear wave breast cancer risk index | Accuracy=93.59%; sensitivity=90.41%; specificity=96.39% |
| Raghavendra et al. (2017) | Develop a computer-aided diagnosis system to automatically differentiate the different stages of thyroid cancer | Textural features | SVM, thyroid clinical risk index | Accuracy=97.52%; sensitivity=90.32%; specificity=98.57% |
| Ma et al. (2017a) | Initiate and assess an automated thyroid nodules detection system | Convolutional neural network | The area under ROC curve: accuracy=98.51% | |
| Ma et al. (2017b) | Propose a hybrid approach to automatically classify benign and malignant thyroid nodules | Convolutional neural network | Accuracy=83.02%; sensitivity=82.41%; specificity=84.96% | |
CCC: concordance correlation coefficient; ROC: receiver operating characteristic; ANOVA: analysis of variance; SVM: support vector machine; GLCM: grey level co-occurrence matrix; CT: computed tomography; BI-RADS: breast imaging reporting and data system; CART: classification and regression tree; PCA: principal component analysis; CBCT: cone beam computed tomography; K-NN: K-nearest neighbour; ROI: region of interest; MRI: magnetic resonance imaging; FLAIR: fluid-attenuated inversion recovery; EGFR: epidermal growth factor receptor; LASSO: least absolute shrinkage and selection operator; VASARI: Visually Accessible Rembrandt Images; DCT: discrete cosine transform; SUV: standardised uptake value; ICC: intraclass correlation; PET: positron-emission tomography; ADASYN: adaptive synthetic; HSD: honest significant difference; FDG: fludeoxyglucose; FLT: fluorothymidine
From Table 1, it is observed that textural features are persistently used in the study of radiomics regardless of the radiological modalities (CT, MRI, or PET). Furthermore, Fig. 2 shows a summary of the various feature extraction techniques implemented based on the literature review (Table 1) and it can be noted that textural-based methods are commonly used.
Fig. 2.
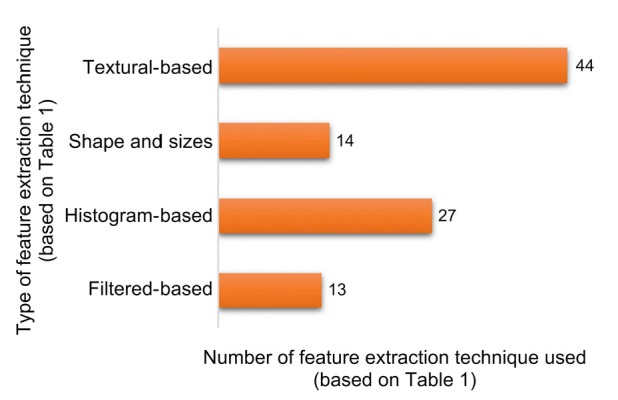
Types of feature extraction methods used
Generally, the feature extraction process produces a huge number of features; among them few features are redundant. Therefore, feature reduction or selection is performed to identify useful features from the pool of features extracted. This step significantly boosts the performance of the classifier by selecting only the highly significant features and eliminating redundant features. In addition, statistical tests, namely analysis of variance (ANOVA), Benjamini-Hochberg procedure, chi-squared test, Cox regression model, Friedman test, Kaplan-Meier analysis, Kruskal-Wallis test, Mann-Whitney test, Student’s t-test, Tukey honest significant difference (HSD) test, and Wilcoxon test, are performed to select highly significant features for optimal performance.
These statistical tests are used based on different conditions (SPSS Tutorials 2017; https://www.spss-tutorials.com). Hence, there are different types of statistical tests to implement based on the conditions of the dataset. ANOVA is used for a three-class or more problem with the assumption that the input data are normally distributed. Benjamini-Hochberg procedure is used in a three-class or more problem and when the data are independent of each other. Chi-squared test is a distribution-free test and can be used only when the two-class problem data are independent of each other. Cox regression model works on a normally distributed two-class problem data. Friedman test can be used in a three-class or more problem that does not assume that the data are normally distributed. Also, the Kaplan-Meier analysis can be used in a three-class or more problem in abnormally distributed data. Kruskal-Wallis test is used when there is a three-class or more problem and the data are assumed to be skewed; Mann-Whitney test, also known as the Wilcoxon rank sum test, is used when the data are independent and assumed to be non-normally distributed. Wilcoxon signed rank test, on the other hand, is used when the two-class problem data are dependent and assumed to be non-normally distributed. Student’s t-test is used only when the two-class problem data are assumed to be normally distributed. The Tukey HSD test is used when the three-class or more problem dataset is assumed to be normally distributed.
2.4. Analysis
In the final stage, the selected highly significant features are fed into classifiers for further analysis and assessment. These extracted radiomics features are then examined together with clinical data obtained from other ‘omics’ studies in order to analyse the mutuality of clinical features (information) and radiomics features (Narang et al., 2016). The combination of data is known as multilevel data. Classifier models are machine learning algorithms for making accurate predictions based on the training dataset and on the features extracted from radiological images.
Machine learning uses algorithms to analyse data, learn the data, and make a prediction with the data. The machine is ‘trained’ with a huge amount of data using algorithms to extract information from the data and to analyse them. Moreover, with the advancement and improvement of machine learning, a subset of machine learning known as deep learning is established. Deep learning is inspired by the anatomy and function of the brain and it involves neural networks with at least three or more hidden layers (Chen et al., 2014). The difference between deep learning and conventional machine learning is in the segmentation and feature extraction processes. Deep learning does not require any algorithm to select ROI or to extract significant features for classification. This technique can learn to focus on the right features with least guidance; hence, the potential of deep learning in high-throughput medicine is straightforward (Angermueller et al., 2016). However, deep learning is relatively new and has tremendous potential waiting to be explored. Nevertheless, Ma et al. (2017a, 2017b) reported the potential by implementing deep learning (CNN) in their work.
From the literature review (Table 1), it can be seen that the different classifiers, namely bagging (Parmar et al., 2015b), Bayesian (Parmar et al., 2015b), boosting (Parmar et al., 2015b), classification and regression tree (CART) (Wang et al., 2015), decision tree (Hawkins et al., 2014; Chaudhury, 2015; Parmar et al., 2015b; Velazquez et al., 2015), discriminant analysis (Balagurunathan et al., 2014b; Coquery et al., 2014; Parmar et al., 2015b), generalised linear (Parmar et al., 2015b), K-nearest neighbour (K-NN) (Zhou et al., 2014; Chaddad et al., 2015; Parmar et al., 2015b; Lian et al., 2016; Wu et al., 2016), logistic regression (Guo et al., 2015; Parmar et al., 2015a; Vallières et al., 2015; Ypsilantis et al., 2015; Coroller et al., 2016; Huang et al., 2016; Liang et al., 2016), naïve Bayes (Chaudhury, 2015; Emaminejad et al., 2016; Wu et al., 2016), neural network (Parmar et al., 2015b), random forest (Parmar et al., 2015b; Wu et al., 2016), and support vector machine (SVM) (Zhou et al., 2014; Chaudhury, 2015; Chung et al., 2015; Depeursinge et al., 2015; Khalvati et al., 2015; Parmar et al., 2015b; Upadhaya et al., 2015a, 2015b; Ypsilantis et al., 2015; Mattonen et al., 2016), are used. Fig. 3 depicts a summary of the types and times of the classifiers used in various studies.
Fig. 3.
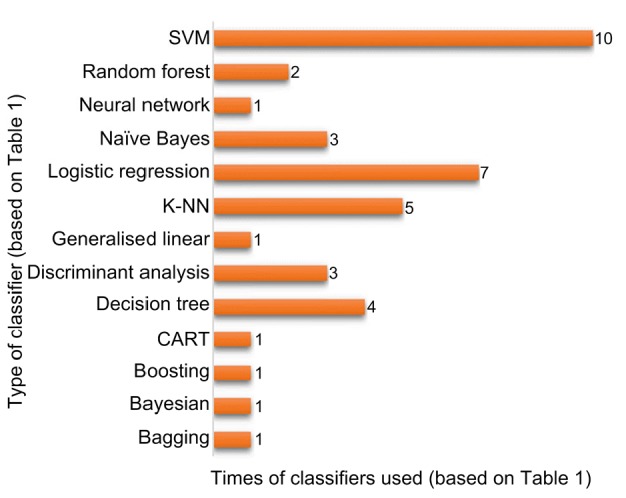
Types of classifiers used
SVM: support vector machine; K-NN: K-nearest neighbour; CART: classification and regression tree
3. Discussion
In this review, the results of radiomics and various radiomics-based studies conducted using various radiological images such as CT, MRI, PET, and ultrasonography are discussed. The findings of those studies are summarised in Table 1.
Cho et al. (2015) experimented on an automated screening of melanoma and noted that radiomics improved the overall efficacy of characterising benign from malignant melanoma. They achieved an accuracy of 82.4% using 367 dermal radiomics features with the random forest classifier. In the oesophageal cancer review paper by van Rossum et al. (2016), they reported that textural radiomics features proved to be correlated to the underlying physiologic processes as textural features are valuable characteristics used to extract subtle information from ROIs (Haralick et al., 1973).
Some researchers have studied the reproducibility and resilience of radiomics features in CT images and concluded that radiomics features can be reproducible (Hunter et al., 2013; Balagurunathan et al., 2014a, 2014b; Fried et al., 2014; Zhao et al., 2016). Hunter et al. (2013) utilised two different CT scanners obtained from 59 non-small cell lung cancer patients in their experiment to single out radiomics features that are highly significant, non-redundant, and reproducible across the scanners. They found that average derived image features showed superior inter-machine reproducibility as compared to end-exhale and breath-hold images, as well as cine 4D-CT compared to helical 3D-CT. Mackin et al. (2015) also evaluated the repeatability of radiomics features in images acquired from different scanners and found that there was inter-scanner variability in the mean CT numbers and noise for the 17 CT scanners. In addition, Oliver et al. (2015) reported a difference between the radiomics features extracted from a static CT image and a respiratory-gated CT image, thus exhibiting the effect of respiratory motion on radiomics features. Hence, we can comment that radiomics features reproducibility and redundancy are dependent on the CT scanners and the type of CT image, as well as the standardisation of image acquisition and reconstruction.
To date, there is still limited study on radiomics features using MRI and its reliability. One of the diseases that have been studied is glioblastoma multiforme (GBM), a highly malignant neurologic tumour in the brain exhibiting complex intra-tumoural and inter-patient heterogeneity. Hence, because of the difficulty in obtaining representative biopsies, the underlying radiomics features extracted by processing radiological imaging data are hypothesized to provide detailed phenotypic information (Lambin et al., 2012). Narang et al. (2016) found two main approaches used to develop radiomics features in GBM. Firstly, the standardised semantic features scored by radiologists are used. The features include tumour location, lesion size, proportion enhancing, proportion necrosis, proportion oedema, and definition of enhancing margin. Secondly, fully computational derived features using imaging and statistical techniques are extracted from the images. Semantic features are known to be highly correlated with clinical outcomes, as shown in a study by Lacroix et al. (2001) on 416 patients with GBM. They found that necrosis and tumour enhancement were a strong prognostic indicator. A Visually Accessible Rembrandt Images (VASARI) feature set comprising 30 semantic features was used to standardise radiological assessment of GBM (https://wiki. cancerimagingarchive.net/display/Public/VASARI+Research+Project). The neuroradiology domain experts agreed that these distinct VASARI features could quantify the phenotype comprehensively and robustly, and these measurements and assessments can be reproducible to provide clinically meaningful and biologically relevant information (Gutman et al., 2013). These VASARI features can be extracted using automated phenotype quantification, i.e. radiomics.
PET is progressively being used for analysis, cancer staging, and therapy response assessments in tumours (Boellaard, 2009; Kato and Nakajima, 2012; Rahim et al., 2014). In addition, radiomics features extracted from PET images have been used to broaden clinical parameters such as the standardised uptake value (SUV) used to aid visual interpretation (Thie, 2004) in clinical practices (Chicklore et al., 2013; Tixier et al., 2015). Most PET studies included SUVs in their work (Cheebsumon et al., 2012; Nair et al., 2012; Leijenaar et al., 2013, 2015; Oliver et al., 2015; Tixier et al., 2015; Vallières et al., 2015; Yoon et al., 2015; Ypsilantis et al., 2015; Antunes et al., 2016; Bailly et al., 2016; Desseroit et al., 2016; Grootjans et al., 2016; Lian et al., 2016; van Velden et al., 2016). Leijenaar et al. (2013) reported a high test-retest stability of approximately 71% using radiomics features in the analysis of non-small cell lung carcinoma (NSCLC) from PET scans. Moreover, the performance of analysis using radiomics features outperformed inter-observer’s diagnosis. Furthermore, Tixier et al. (2015) compared inter-observer’s analysis with the analysis made with radiomics for NSCLC, and concluded that a computer-aided diagnosis system using radiomics has a higher prognostic capability and can reduce subjective diagnosis. Tixier et al. (2012) in another study on oesophageal cancer found that for some textural features such as entropy, dissimilarity, homogeneity, size variability, and intensity of homogenous tumour areas (regional characterization) had reproducibility similar to or better than that of simple SUV measurements. However, several studies found that PET radiomics features are susceptible to reconstruction parameters and the type of image acquisition. Galavis et al. (2010) in their cohort study of 20 patients with various solid tumours found that 40 out of 50 features tested demonstrated large variations (>30%) when the number of iterations, grid size, reconstruction algorithm, and/or post-reconstruction filter were changed. Their findings were further supported by van Velden et al. (2016) and Yan et al. (2015). Respiratory motion artefact is also a well-known concern in any imaging modality. Grootjans et al. (2016) and Oliver et al. (2015) studied lung cancer with PET and reported that respiratory motion has a considerable effect on the quantification of tumour heterogeneity. From these recent studies, we can conclude that PET radiomics features can be reproducible; however, interpretation of the images and results must be done carefully bearing in mind that there are limitations.
The combination of CT and PET imaging is an established diagnostic imaging tool in oncology (Tixier et al., 2015; Lu and Chen, 2016). PET-CT has an advantage over just PET or CT scan only as PET-CT provides additional information on a tumour (Lu et al., 2015). Also, a combination of MRI and PET-CT has been reported (Vallières et al., 2015; Antunes et al., 2016). Vallières et al. (2015) proposed a collaborative study with PET and MRI to analyse lung metastasis risk in soft-tissue sarcomas. It was concluded that the radiomics features extracted from the fusion of PET-MRI possess additional prognostic characteristics and could reveal the underlying details from the ROI. It is also noted that there are very few radiomics-based studies using ultrasound modality mainly due to the subjectivity and operator-dependent nature of this modality.
As mentioned here, there are very few studies using ultrasound modality mainly due to the subjective and operator-dependent nature of this modality. However, there have been several attempts with very good results (Acharya et al., 2016a, 2016b, 2016c, 2017). It is anticipated that more studies will be embarked on this area. Acharya et al. (2016b) proposed a novel technique to distinguish fatty liver disease using discrete cosine transform and radon transform. They have achieved 100% accuracy, sensitivity, and specificity with the fuzzy sugeno classifier. Also, they developed a fatty liver disease index using a single number to differentiate normal from the fatty liver disease class. Moreover, they extended their work to a three-class problem to differentiate normal, fatty liver disease, and cirrhosis (Acharya et al., 2016a). They also formulated a fatty liver disease index to mathematically distinguish the three classes. Further, Acharya et al. (2016c) explored the possibility of thyroid lesion classification obtained from ultrasound images. They implemented Gabor transform on the images and then extracted entropy features. They attained a performance rate of 94.30% with C4.5 decision tree classifier. In their later work (Acharya et al., 2017), they also studied the classification of breast lesion in shear wave ultrasound. In their work, they devised a shear wave breast cancer risk index to distinguish benign and malignant breast lesions. They reported a performance of 93.59% accuracy using a quadratic discriminant analysis classifier.
Raghavendra et al. (2017) explored the fusion of different texture features and extracted various entropies for the discrimination of benign and malignant thyroid lesions. Similarly, they constructed a thyroid clinical risk index to distinguish the two classes using numerical values. Also, Ma et al. (2017a, 2017b) proposed CNN-based automatic thyroid nodule detection system. These high performances confirm the application of CNN in developing robust computer-aided diagnosis systems using medical images.
4. Radiogenomics
However, utilising radiomics alone is insufficient to qualify as or achieve the goal for precision medicine. More information is needed to consider individual variability in genes for more accurate diagnosis and treatment. Nevertheless, studies have demonstrated that there is a correlation between genomics, proteomics, and radiomics (Segal et al., 2007; Zinn et al., 2011; Aerts et al., 2014; Sala et al., 2017). Furthermore, genomics and proteomics have brought overwhelming progress in medicine (Wanichthanarak et al., 2015; Aerts, 2016). They belong to the ‘omics’ data which have facilitated the state-of-the-art analysis of diverse organismal and molecular processes (Horgan and Kenny, 2011; Wanichthanarak et al., 2015). Thus, the term radiogenomics was coined as it highlights the synergistic combination of genomics and radiomics. Radiogenomics signifies the broadening of clinical imaging into genomic and molecular imaging (Kuo and Jamshidi, 2014) as it focuses on the relationship between features obtained from radiological images and biomolecular markers.
Yamamoto et al. (2014) performed a radiogenomic analysis in non-small cell lung cancer and achieved an accuracy of 78.8%, a sensitivity of 83.3%, and specificity of 77.9%. They concluded that the molecular phenotype and CT imaging, when combined, can distinguish tumours from non-tumours. Moreover, Yamamoto et al. (2015) conducted another radiogenomic study on breast cancer to investigate the correlations between MRI phenotype, quantitative imaging, and RNA expression. Their experiment showed the potential of radiogenomics in identifying early metastasis.
Although radiogenomics is still in its infancy, radiogenomic analysis does possess the potential to reveal a predictive radiomic signature and underlying gene expression patterns. Therefore, for radiogenomics to be formally introduced to clinical settings, radiomics must be firmly established first.
5. Challenges
Radiomics is a new multi-disciplinary field and hence, new challenges are inevitable (Kumar et al., 2012). Firstly, there is no standardised procedure in image acquisition (Leijenaar et al., 2015; Scrivener et al., 2016). Different protocols have been implemented by different centres for acquisition and analysis of radiological images. The variation of protocol parameters includes, among others, resolution, field of view, and slice thickness of the images. Thus, studies using these heterogeneous radiological images will not produce accurate comparisons across different sets of radiological images.
Secondly, it can be seen from Table 1 that there are numerous techniques and algorithms being used. However, there is no standard methodology being developed and therefore, it is unclear which is the best way forward (Dinapoli et al., 2016). Furthermore, there is a lack of international protocols and standards for validating results (Sala et al., 2017). It is important to validate the re-usability and suitability of a radiomic model to establish rules in this new discipline for a reliable and consistent prognostic tool to be implemented in a clinical setting (Coquery et al., 2014; van Rossum et al., 2016).
Thirdly, sharing of radiological images publicly has always been a problem in clinical research (Nelson, 2009; Gillies et al., 2016). It is beneficial to have shared databases for maximum information to be extracted from radiological images (Gillies et al., 2016). Having an integrated shared database with radiological images and the extracted features will ensure standardisation in radiomic algorithms (Nyflot et al., 2015; Gillies et al., 2016; Wong et al., 2016). Moreover, using the same set of data across various radiomic algorithms will ensure consistency when it comes to inter-comparison and validation of results.
6. Future directions
An important achievement is to correlate ‘omics’ data with radiomics features extracted from radiological images and integrate them to create a more efficient and robust prognostic model to aid clinicians in practising personalised medicine. The next step is to implement radiomics as an adjunct tool in clinical settings. However, the challenges discussed here must be addressed first before radiomics could be employed in routine clinical settings. Currently, radiomics is at best fragmented with multiple players. Thus, it is imperative to form an international consortium to bring all researchers together for collaboration.
Another focus will be to incorporate deep learning to radiomics as deep learning is currently not being extensively deployed. The deep learning technique reduces the need for feature engineering and thus, it is much more time-efficient and boosts accuracy.
7 Conclusions
The increasing use of biomarkers in cancer diagnosis has paved the way for personalised medicine; however, the accuracy and predictive diagnosis and treatment alternatives are further improved by using imaging biomarkers derived from radiomics. In this review, the potentials of radiomics, the current workflow of radiomics, and employment of radiomics are highlighted. Thus, by introducing radiomics in the clinical setting, significant features can be extracted from the radiological images for subsequent analysis of image phenotypes in order to aid clinical decision-making. However, we need to overcome the challenges before radiomics can be successfully introduced into clinical settings. Further development in radiogenomics will bring us closer towards precision medicine.
Footnotes
Compliance with ethics guidelines: U. Rajendra ACHARYA, Yuki HAGIWARA, Vidya K. SUDARSHAN, Wai Yee CHAN, and Kwan Hoong NG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Acharya UR, Raghavendra U, Fujita H, et al. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput Biol Med. 2016;79:250–258. doi: 10.1016/j.compbiomed.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Acharya UR, Fujita H, Sudarshan VK, et al. An integrated index for identification of fatty liver disease using radon transform and discrete cosine transform features in ultrasound images. Inform Fusion. 2016;31:43–53. doi: 10.1016/j.inffus.2015.12.007. [DOI] [Google Scholar]
- 3.Acharya UR, Chowriappa P, Fujita H, et al. Thyroid lesion classification in 242 patient population using Gabor transform features from high resolution ultrasound images. Knowl-Based Syst. 2016;107:235–245. doi: 10.1016/j.knosys.2016.06.010. [DOI] [Google Scholar]
- 4.Acharya UR, Ng WL, Rahmat K, et al. Data mining framework for breast lesion classification in shear wave ultrasound: a hybrid feature paradigm. Biomed Signal Proces. 2017;33:400–410. doi: 10.1016/j.bspc.2016.11.004. [DOI] [Google Scholar]
- 5.Aerts HJWL. The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol. 2016;2:1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 6.Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun, 5:4006. 2014 doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angermueller C, Pärnamaa T, Parts L, et al. Deep learning for computational biology. Mol Syst Biol, 12: 878. 2016 doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antunes J, Viswanath S, Rusu M, et al. Radiomics analysis on FLT-PET/MRI for characterisation of early treatment response in renal cell carcinoma: a proof of concept study. Transl Oncol. 2016;9(2):155–162. doi: 10.1016/j.tranon.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailly C, Bodet-Milin C, Couespel S, et al. Revisiting the robustness of PET-based textural features in the context of multi-centric trials. PLoS ONE, 11:7. 2016 doi: 10.1371/journal.pone.0159984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balagurunathan Y, Gu YH, Wang H, et al. Reproducibility and prognosis of quantitative features extracted from CT images. Transl Oncol. 2014;7(1):72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balagurunathan Y, Kumar V, Gu YH, et al. Test-retest reproducibility analysis of lung CT image features. J Digit Imaging. 2014;27(6):805–823. doi: 10.1007/s10278-014-9716-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boellaard R. Standards for PET image acquisition and quantitative data analysis. J Nuclear Med, 50:11S-20S. 2009 doi: 10.2967/jnumed.108.057182. [DOI] [PubMed] [Google Scholar]
- 13.Castellino RA. Computer-aided detection (CAD): an overview. Cancer Imaging. 2005;5:17–19. doi: 10.1102/1470-7330.2005.0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaddad A, Zinn PO, Colen RR. Radiomics texture feature extraction for characterising GBM phenotypes using GLCM. IEEE 12th International Symposium on Biomedical Imaging (ISBI). New York, USA.2015. [Google Scholar]
- 15.Chaudhury B. The Use of Textural Kinetic Habitats to Mine Diagnostic Information from DCE MR Images of Breast Tumours. Fowler Avenue, Tampa, USA: University of South Florida,; 2015. PhD Theses. [Google Scholar]
- 16.Cheebsumon P, Boelaard R, de Ruysscher D, et al. Assessment of tumour size in PET/CT lung cancer studies: PET-and CT-based methods compared to pathology. EJNMMI Res. 2012;2(1):56. doi: 10.1186/2191-219X-2-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2013;5(1):73–82. doi: 10.1002/wsbm.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YS, Lin ZH, Zhao X, et al. Deep learning-based classification of hyperspectral data. IEEE J-STARS. 2014;7(6):2094–2107. doi: 10.1109/JSTARS.2014.2329330. [DOI] [Google Scholar]
- 19.Chicklore S, Goh V, Siddique M, et al. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133–140. doi: 10.1007/s00259-012-2247-0. [DOI] [PubMed] [Google Scholar]
- 20.Cho DS, Clausi DA, Wong A. Dermal radiomics for melanoma screening. Vision Lett. 2015;1(1):23. doi: 10.15353/vsnl.v1i1.58. [DOI] [Google Scholar]
- 21.Chung AG, Khalvati F, Shafiee MJ, et al. Prostate cancer detection via a quantitative radiomics-driven conditional random field framework. IEEE Access. 2015;3:2531–2541. doi: 10.1109/ACCESS.2015.2502220. [DOI] [Google Scholar]
- 22.Cook GJR, Siddique M, Taylor BP, et al. Radiomics in PET: principles and applications. Clin Transl Imaging. 2014;2(3):269–276. doi: 10.1007/s40336-014-0064-0. [DOI] [Google Scholar]
- 23.Coquery N, Francois O, Lemasson B, et al. Microvascular MRI and unsupervised clustering yields histology-resembling images in two rat models of glioma. J Cerebr Blood Met. 2014;34(8):1354–1362. doi: 10.1038/jcbfm.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coroller TP, Grossmann P, Hou Y, et al. CT-based radiomic signature predicts metastasis in lung adenocarcinoma. J Eur Soc Therapeut Radiol Oncol. 2015;114(3):345–350. doi: 10.1016/j.radonc.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coroller TP, Agrawal V, Narayan V, et al. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother Oncol. 2016;119(3):480–486. doi: 10.1016/j.radonc.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Court LE, Fave X, Mackin D, et al. Computational resources for radiomics. Transl Cancer Res. 2016;5(4):340–348. doi: 10.21037/tcr.2016.06.17. [DOI] [Google Scholar]
- 27.Cunliffe A, Armato III SG, Castillo R, et al. Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development. Int J Radiat Oncol. 2015;91(5):1048–1056. doi: 10.1016/j.ijrobp.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davnall F, Yip CSP, Ljungqvist G, et al. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3(6):573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Depeursinge A, Yanagawa M, Leung AN, et al. Predicting adenocarcinoma recurrence using computational texture models of nodule components in lung CT. Med Phys. 2015;42(4):2054–2063. doi: 10.1118/1.4916088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desseroit MC, Visvikis D, Tixier F, et al. Development of a nomogram combining clinical staging with 18F-FDG PET/CT image features in non-small-cell lung cancer stage I‒III. Eur J Nucl Med Mol Imaging. 2016;43(8):1477–1485. doi: 10.1007/s00259-016-3325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dinapoli N, Casa C, Barbaro B, et al. Radiomics for rectal cancer. Transl Cancer Res. 2016;5(4):424–431. doi: 10.21037/tlcr.2016.08.01. [DOI] [Google Scholar]
- 32.Egger J, Kapur T, Fedorov A, et al. GBM volumetry using the 3D Slicer medical image computing platform. Sci Rep. 2013;3:1364. doi: 10.1038/srep01364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emaminejad N, Qian W, Guan YB, et al. Fusion of quantitative image and genomic biomarkers to improve prognosis assessment of early stage lung cancer patients. IEEE Trans Biomed Eng. 2016;63(5):1034–1043. doi: 10.1109/TBME.2015.2477688. [DOI] [PubMed] [Google Scholar]
- 34.Eminowicz G, McCormack M. Variability of clinical target volume delineation for definitive radiotherapy in cervix cancer. Radiother Oncol. 2015;117(3):542–547. doi: 10.1016/j.radonc.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 35.Fave X, Mackin D, Yang JZ, et al. Can radiomics features be reproducibly measured from CBCT images for patients with non-small cell lung cancer? Med Phys. 2015;42(12):6784–6797. doi: 10.1118/1.4934826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fave X, Zhang LF, Yang JZ, et al. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl Cancer Res. 2016;5(4):349–363. doi: 10.21037/tcr.2016.07.11. [DOI] [Google Scholar]
- 37.Felzenszwalb PF, Huttenlocher DP. Efficient graph-based image segmentation. Int J Comput Vision. 2004;59(2):167–181. doi: 10.1023/B:VISI.0000022288.19776.77. [DOI] [Google Scholar]
- 38.Fried DV, Tucker SL, Zhou SH, et al. Prognostic value and reproducibility of pretreatment CT texture features in stage III non-small cell lung cancer. Int J Radiat Oncol Biol Phys. 2014;90(4):834–842. doi: 10.1016/j.ijrobp.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galavis PE, Hollensen C, Jallow N, et al. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol. 2010;49(7):1012–1016. doi: 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grootjans W, Tixier F, van der Vos CS, et al. The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med. 2016;57(11):1692–1698. doi: 10.2967/jnumed.116.173112. [DOI] [PubMed] [Google Scholar]
- 42.Grossmann P, Gutman DA, Dunn Jr WD, et al. Imaging-genomics reveals driving pathways of MRI derived volumetric tumor phenotype features in glioblastoma. BMC Cancer, 16:611; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo WT, Li H, Zhu YT, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging. 2015;2(4):041007. doi: 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutman DA, Cooper LAD, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology. 2013;267(2):560–569. doi: 10.1148/radiol.13120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybernetics. 1973;SMC-3(6):610–621. doi: 10.1109/TSMC.1973.4309314. [DOI] [Google Scholar]
- 46.Hawkins SH, Korecki JN, Balagurunathan Y, et al. Predicting outcomes of non-small cell lung cancer using CT image features. IEEE Access. 2014;2:1418–1426. doi: 10.1109/ACCESS.2014.2373335. [DOI] [Google Scholar]
- 47.He L, Huang YQ, Ma ZL, et al. Effects of contrast-enhancement, reconstruction slice thickness and convolution kernel on the diagnostic performance of radiomics signature in solitary pulmonary nodule. Sci Rep, 6:34921. 2016 doi: 10.1038/srep34921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horgan RP, Kenny LC. SAC review ‘Omic’ technologies: genomics, transcriptomics, proteomics and metabolomics. Obstet Gynaecol. 2011;13(3):189–195. [Google Scholar]
- 49.Huang YQ, Liang CH, He L, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 50.Hunter LA, Krafft S, Stingo F, et al. High-quality machine-robust image features: identification in non-small cell lung cancer computed tomography images. Med Phys. 2013;40(12):121916. doi: 10.1118/1.4829514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huynh E, Coroller TP, Narayan V, et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol. 2016;120(2):258–266. doi: 10.1016/j.radonc.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 52.Kass M, Witkin A, Terzopoulos D. Snakes: active contour models. Int J Comput Vision. 1988;1(4):321–331. doi: 10.1007/BF00133570. [DOI] [Google Scholar]
- 53.Kato H, Nakajima M. The efficacy of FDG-PET for the management of esophageal cancer: review article. Ann Thorac Cardiovasc Surg. 2012;18(5):412–419. doi: 10.5761/atcs.ra.12.01954. [DOI] [PubMed] [Google Scholar]
- 54.Khalvati F, Wong A, Haider MA. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging, 15:27. 2015 doi: 10.1186/s12880-015-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30(9):1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar YR, Muthukrishnan NM, Mahajan A, et al. Statistical parameter-based automatic liver tumor segmentation from abdominal CT scans: a potiential radiomic signature. Proced Comput Sci. 2016;93:446–452. doi: 10.1016/j.procs.2016.07.232. [DOI] [Google Scholar]
- 57.Kuo MD, Jamshidi N. Behind the numbers: decoding molecular phenotypes with radiogenomics–guiding principles and technical considerations. Radiology. 2014;270(2):320–325. doi: 10.1148/radiol.13132195. [DOI] [PubMed] [Google Scholar]
- 58.Lacroix M, Abi-Said D, Fourney DR, et al. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosur. 2001;95(2):190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 59.Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JS, Narang S, Martinez JJ, et al. Associating spatial diversity features of radiologically defined tumor habitats with epidermal growth factor receptor driver status and 12-month survival in glioblastoma: methods and preliminary investigation. J Med Imaging. 2015;2(4):041006. doi: 10.1117/1.JMI.2.4.041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leijenaar RTH, Carvalho S, Velazquez ER, et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52(7):1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leijenaar RTH, Nalbantov G, Carvalho S, et al. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep, 5:11075. 2015 doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lian CF, Ruan S, Denoeux T, et al. Selecting radiomic features from FDG-PET images for cancer treatment outcome prediction. Med Image Anal. 2016;32:257–268. doi: 10.1016/j.media.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 64.Liang CS, Huang YQ, He L, et al. The development and validation of a CT-based radiomics signature for the preoperative discrimination of stage I‒II and stage III‒IV colorectal cancer. Oncotarget. 2016;7(21):31401–31412. doi: 10.18632/oncotarget.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu W, Chen W. Positron emission tomography/computerized tomography for tumor response assessment–a review of clinical practices and radiomics studies. Transl Cancer Res. 2016;5(4):364–370. doi: 10.21037/tcr.2016.07.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu W, Wang J, Zhang HH. Computerized PET/CT image analysis in the evaluation of tumour response to therapy. Brit J Radiol. 2015;88(1048):20140625. doi: 10.1259/bjr.20140625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma J, Wu F, Jiang T, et al. Cascade convolutional neural networks for automatic detection of thyroid nodules in ultrasound images. Med Phys. 2017;44(5):1678–1691. doi: 10.1002/mp.12134. [DOI] [PubMed] [Google Scholar]
- 68.Ma J, Wu F, Zhu J, et al. A pre-trained convolutional neural network based method for thyroid nodule diagnosis. Ultrasonics. 2017;73:221–230. doi: 10.1016/j.ultras.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 69.Mackin D, Fave X, Zhang LF, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50(11):757–765. doi: 10.1097/RLI.0000000000000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malladi R, Sethian JA, Vemuri BC. Shape modeling with front propagation: a level set approach. IEEE Trans Pattern Anal Machine Intell. 1995;17(2):158–175. doi: 10.1109/34.368173. [DOI] [Google Scholar]
- 71.Mattonen SA, Tetar S, Palma DA, et al. Imaging texture analysis for automated prediction of lung cancer recurrence after stereotactic radiotherapy. J Med Imaging. 2015;2(4):041010. doi: 10.1117/1.JMI.2.4.041010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mattonen SA, Palma DA, Johnson C, et al. Detection of local cancer recurrence after stereotactic ablative radiation therapy for lung cancer: physician performance versus radiomic assessment. Int J Radiat Oncol Biol Phys. 2016;94(5):1121–1128. doi: 10.1016/j.ijrobp.2015.12.369. [DOI] [PubMed] [Google Scholar]
- 73.Mitra S, Shankar BU. Medical image analysis for cancer management in natural computing framework. Inform Sci. 2015;306:111–131. doi: 10.1016/j.ins.2015.02.015. [DOI] [Google Scholar]
- 74.Nair VS, Gevaert O, Davidzon G, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72(15):3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narang S, Lehrer M, Yang D, et al. Radiomics in glioblastoma: current status, challenges and potential opportunities. Transl Cancer Res. 2016;5(4):383–397. doi: 10.21037/tcr.2016.06.31. [DOI] [Google Scholar]
- 76.Nelson B. Data sharing: empty archives. Nature. 2009;461:160–163. doi: 10.1038/461160a. [DOI] [PubMed] [Google Scholar]
- 77.Nyflot MJ, Yang F, Byrd D, et al. Quantitative radiomics: impact of stochastic effects on textural feature analysis implies the need for standards. J Med Imaging. 2015;2(4):041002. doi: 10.1117/1.JMI.2.4.041002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oliver JA, Budzevich M, Zhang GG, et al. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol. 2015;8(6):524–534. doi: 10.1016/j.tranon.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1(2):207–226. doi: 10.1080/23808993.2016.1164013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Parmar C, Velazquez ER, Leijenaar R, et al. Robust radiomics feature quantification using semiautomatic volumetric segmentation. PLoS ONE. 2014;9(7):e102107. doi: 10.1371/journal.pone.0102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parmar C, Leijenaar RTH, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep, 5:11044. 2015 doi: 10.1038/srep11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Parmar C, Grossmann P, Rietveld D, et al. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front Oncol, 5:272. 2015 doi: 10.3389/fonc.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pizer SM, Amburn EP, Austin JD, et al. Adaptive histogram equalization and its variations. Comput Vision Graph Image Proc. 1987;39(3):355–368. doi: 10.1016/S0734-189X(87)80186-X. [DOI] [Google Scholar]
- 84.Raghavendra U, Acharya UR, Gudigar A, et al. Fusion of spatial gray level dependency and fractal texture features for the characterization of thyroid lesions. Ultrasonics. 2017;77:110–120. doi: 10.1016/j.ultras.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 85.Rahim MK, Kim SE, So H, et al. Recent trends in PET image interpretations using volumetric and texture-based quantification methods in nuclear oncology. Nucl Med Mol Imaging. 2014;48(1):1–15. doi: 10.1007/s13139-013-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sala E, Mema E, Himoto Y, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72(1):3–10. doi: 10.1016/j.crad.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scrivener M, de Jong EEC, van Timmeren JE, et al. Radiomics applied to lung cancer: a review. Transl Cancer Res. 2016;5(4):398–409. doi: 10.21037/tcr.2016.06.18. [DOI] [Google Scholar]
- 88.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25:675–680. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 89.Song JD, Dong D, Huang YQ, et al. Association between tumour heterogeneity and overall survival in patients with non-small cell lung cancer. 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). Prague, Czech Republic; 2016. pp. 1249–1252. [DOI] [Google Scholar]
- 90.Sonka M, Hlavac V, Boyle R, et al. Cengage Learning. 2007. Image processing, analysis, and machine vision. [Google Scholar]
- 91.Stoyanova R, Takhar M, Tschudi Y, et al. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res. 2016;5(4):432–447. doi: 10.21037/tcr.2016.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Szigeti K, Szabó T, Korom C, et al. Radiomics-based differentiation of lung disease models generated by polluted air based on X-ray computed tomography data. BMC Med Imaging, 16:14. 2016 doi: 10.1186/s12880-016-0118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Thie JA. Understanding the standardized uptake value, its methods, and implications for usage. J Nucl Med. 2004;45(9):1431–1434. [PubMed] [Google Scholar]
- 94.Tixier F, Hatt M, Cheze Le Rest C, et al. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012;53(5):693–700. doi: 10.2967/jnumed.111.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tixier F, Hatt M, Valla C, et al. Visual versus quantitative assessment of intratumor 18F-FDG PET uptake heterogeneity: prognostic value in non-small cell lung cancer. J Nucl Med. 2015;55(8):1235–1241. doi: 10.2967/jnumed.113.133389. [DOI] [PubMed] [Google Scholar]
- 96.Upadhaya T, Morvan Y, Stindel E, et al. A framework for multimodal imaging-based prognostic model building: preliminary study on multimodal MRI in glioblastoma multiforme. IRBM. 2015;36(6):345–350. doi: 10.1016/j.irbm.2015.08.001. [DOI] [Google Scholar]
- 97.Upadhaya T, Morvan Y, Stindel E, et al. Prognostic value of multimodal MRI tumor features in glioblastoma multiforme using textural features analysis. 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), New York, NY, USA. IEEE; 2015. [DOI] [Google Scholar]
- 98.Vallières X, Freeman CR, Skamene SR, et al. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60:5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 99.van den Burg EL, van Hoof M, Postma AA, et al. An exploratory study to detect Ménière’s disease in conventional MRI scans using radiomics. Front Neurol, 7:190. 2016 doi: 10.3389/fneur.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van Rossum PSN, Xu C, Fried DV, et al. The emerging field of radiomics in esophageal cancer: current evidence and future potential. Transl Cancer Res. 2016;5(4):410–423. doi: 10.21037/tcr.2016.06.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Velden FHP, Kramer GM, Frings V, et al. Repeatability of radiomic features in non-small-cell lung cancer [18F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18(5):788–795. doi: 10.1007/s11307-016-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Velazquez ER, Parmar C, Jermoumi M, et al. Volumetric CT-based segmentation of NSCLC using 3D-Slicer. Sci Rep, 3:3529. 2013 doi: 10.1038/srep03529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Velazquez ER, Meier R, Dunn Jr WD, et al. Fully automatic GBM segmentation in the TCGA-GBM dataset: prognosis and correlation with VASARI features. Sci Rep, 5:16822. 2015 doi: 10.1038/srep16822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang H, Schabath MB, Liu Y, et al. Semiquantitative computed tomography characteristics for lung adenocarcinoma and their association with lung cancer survival. Clin Lung Cancer. 2015;16(6):e141–e163. doi: 10.1016/j.cllc.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang H, Xu ZS, Fujita H, et al. Towards felicitous decision making: an overview on challenges and trends of Big Data. Inform Sci. 2016;367-368:747–765. doi: 10.1016/j.ins.2016.07.007. [DOI] [Google Scholar]
- 106.Wang X, Wong BS, Guan TC. Image enhancement for radiography inspection. Proceedings Volume 5852, Third International Conference on Experimental Mechanics and Third Conference of the Asian Committee on Experimental Mechanics, Singapore; 2005. [DOI] [Google Scholar]
- 107.Wanichthanarak K, Fahrmann JF, Grapov D. Genomic, proteomic, and metabolomic data integration strategies. Biomark Insights. 2015;10(Suppl 4):1–6. doi: 10.4137/BMI.S29511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Diagnostic imaging. WHO (World Health Organization); https://www.who.int/diagnostic_imaging/en [accessed on May 13, 2017].2017. [Google Scholar]
- 109.Wong AJ, Kanwar A, Mohamed AS, et al. Radiomics in head and neck cancer: from exploration to application. Transl Cancer Res. 2016;5(4):371–382. doi: 10.21037/tcr.2016.07.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu WM, Parmar C, Grossmann P, et al. Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol, 6:71. 2016 doi: 10.3389/fonc.2016.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamamoto S, Korn RL, Oklu R, et al. ALK molecular phenotype in non-small cell lung cancer: CT radiogenomic characterization. Radiology. 2014;272(2):568–576. doi: 10.1148/radiol.14140789. [DOI] [PubMed] [Google Scholar]
- 112.Yamamoto S, Han W, Kim Y, et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275(2):384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 113.Yan J, Chu-Shern JL, Loi HY, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015;56(11):1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 114.Yang JZ, Zhang LF, Fave XJ, et al. Uncertainty analysis of quantitative imaging features extracted from contrast-enhanced CT in lung tumors. Comput Med Imaging Graph. 2016;48:1–8. doi: 10.1016/j.compmedimag.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yip SSF, Aerts HJWL. Applications and limitations of radiomics. Phys Med Biol. 2016;61(13):R150–R166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoon HJ, Sohn I, Cho JH, et al. Decoding tumor phenotypes for ALK, ROS1, and RET fusions in lung adenocarcinoma using a radiomics approach. Medicine. 2015;94(41):e1753. doi: 10.1097/MD.0000000000001753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ypsilantis PP, Siddique M, Sohn H, et al. Predicting response to neoadjuvant chemotherapy with PET imaging using convolutional neural networks. PLoS ONE. 2015;10(9):e0137036. doi: 10.1371/journal.pone.0137036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao BS, Tan YQ, Tsai WY, et al. Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep, 6:23428. 2016 doi: 10.1038/srep23428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou M, Hall LO, Goldgof DB. Exploring brain tumor heterogeneity for survival time prediction. 2014 22nd International Conference on Pattern Recognition (ICPR), Stockholm, Sweden. IEEE; 2014. pp. 580–585. [DOI] [Google Scholar]
- 120.Zinn PO, Majadan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS ONE. 2011;6(10):e25451. doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]