Abstract
Chronic intractable neuropathic pain after central or peripheral nervous system injury remains refractory to therapeutic intervention. Using microarray and RT-qPCR methods, we found that Noggin mRNA is downregulated in the lumbar enlargement 2 weeks after chronic constriction injury (CCI) in rats.
Eight-week-old female Sprague Dawley rats were used for the CCI model. Two weeks after CCI, rats underwent a laminectomy at L5 under halothane anesthesia, and a silicone tube connected to an osmotic minipump was inserted intrathecally for 14 days. Rats were administered Noggin ranging from 10 ng/ml to 10 μg/ml. Phosphate buffered saline (PBS) was used as a control. The time course of mechanical allodynia was assessed for 5 weeks using von Frey filaments. An ANOVA showed that rats administered Noggin at 2 μg/ml had significantly less mechanical allodynia compared with controls.
We next compared the effect of intrathecal administration (14 days) of Noggin (2 μg/ml), bone morphogenetic protein 4 (BMP4; 2 μg/ml), or BMP4 (μg/ml) + Noggin (μg/ml) with controls. Only Noggin administration significantly reduced mechanical allodynia in the CCI model.
Fluorescence immunohistochemistry indicated that Noggin administration decreased astrocyte accumulation in the dorsal horn compared with PBS after administration for one week. BMP4-driven conversion of oligodendrocyte progenitor cells (OPCs) to type 2 astrocytes is inhibited by Noggin Hampton et al. (2007) . We speculated that Noggin administration inhibits the conversion of OPCs to astrocytes, and decreases glial fibrillar acidic protein expression. This histological condition could decrease neuropathic pain.
Keywords: Noggin, CCI, Microarray, RT-qPCR, Allodynia, GFAP
Highlights
-
•
Noggin mRNA is significantly down-regulated two weeks after CCI in rats.
-
•
The mechanical allodynia was decreased in Noggin administration at seven days.
-
•
Noggin administration influenced GFAP expression and reduced mechanical allodynia.
1. Introduction
Neuropathic pain is one of the most refractory sequela of neurological injury. The search for novel therapeutic agents including drugs for the treatment of neuropathic pain is an area of intense laboratory and clinical research [1]. Several models of peripheral nerve injury have been developed in rodents to help study the mechanisms responsible for neuropathic pain. Among these are the spared nerve injury (SNI) model, in which 2 of the 3 peripheral branches of the sciatic nerve are transected, producing a distal partial nerve lesion [2]; the chronic constriction injury (CCI) model, in which loose ligatures cause compression and inflammation of the sciatic nerve, injuring mainly myelinated axons [3]; and the spinal nerve ligation (SNL) model of proximal axonal injury [4]. All of these models generate prolonged peripheral hypersensitivity to noxious and innocuous mechanical and cold stimuli. A cellular mechanism of the neuropathic pain state in the CCI model was reported more than a decade ago [5]. On microarray analysis, CCI has the smallest number of uniquely regulated genes compared with SNL or SNI [6]. CCI was chosen over other nerve injury models as the most generalizable, and because the deviation of gene expression in the CCI model is less than in the other 2 models.
Although changes in gene expression associated with chronic pain have been studied by microarray profiling [7], [8], [9], [10], [21], studies of CCI are scant [6], [11]. In the present study, gene expression 14 days after CCI was compared with that in controls using an Agilent Rat Whole genome 4 × 44 K microarray, incorporated in the above study. On the microarray, 1136 genes were statistically significantly upregulated and 1709 genes were downregulated. After detailed consideration for gene ontology and gene function, we selected 6 upregulated genes and 2 downregulated genes for the following quantitative real-time RT-PCR experiments. IGF-1, tissue inhibitor of metalloproteinase-3 (TIMP-3), Pap, aquaporin-4 (Aqp4), CD38, and CD68 were significantly upregulated and Noggin and opioid receptor like-1 (Oprl1) were significantly downregulated. Generally, genes related with gliosis were upregulated, e.g., Timp-3 and Aqp4 are mainly upregulated on astrocytes [12], [13]. CD38 is a leukocyte related antigen and CD68 is well-known as an ED1 antigen on microglia.
Noggin is a 26 kDa protein with a hydrophobic amino-terminal sequence and plays a role in normal dorsal development [14]. Mice lacking Noggin have defects in the projection of several groups of neurons, including initial ascending projections from the dorsal root ganglia (DRG) [15]. Overexpression of Noggin results in a significant increase in the number of neurons in the trigeminal and DRG [16]. Noggin plays a role in modulating sensory neuron number and axon guidance. These data suggest that Noggin administration modulates neuron number and axon outgrowth in the spinal cord, thereby reducing neuropathic pain. We also on focused Noggin antagonist, BMP4 from gliosis. BMP signaling suppresses oligodendrocyte development through a basic-helix–loop–helix transcription factor and promotes astrocytosis [20]. BMP promotes gliosis in demyelinating spinal cord lesions [25]. Inhibition of BMP4 by Noggin notably decreased the ratio of astrocytes to neuron numbers [24]. Therefore, we hypothesized that suppression of BMP signaling by Noggin suppresses astrocyte numbers.
In the present study, we focused our attention on Noggin, because Noggin is downregulated 14 days after CCI. Noggin is downregulated in the DRG after nerve injury [17]. These observations are consistent with the downregulation of Noggin after CCI. We hypothesized that administration of Noggin after CCI may ameliorate neuropathic pain. We administered Noggin intrathecally for 14 days after CCI using an osmotic minipump and evaluated mechanical allodynia using von Frey filaments weekly for 5 weeks. Our results showed that intrathecal administration of Noggin after CCI reduces mechanical allodynia, and thus may reduce neuropathic pain.
2. Materials and methods
2.1. Bennett chronic constriction injury (CCI) model
All rats were treated and cared for in accordance with the Chiba University School of Medicine guidelines pertaining to the treatment of experimental animals. The study was approved by the Animal Care and Use Committee of Chiba University Graduate School of Medicine (approval number 24–276). We used 18, 8-week-old adult female Sprague Dawley rats (Japan SLC, Hamamatsu, Japan), which were housed in individual cages and allowed food and water ad libitum. Rats were anesthetized with 1.5% of halothane in oxygen, delivered at 0.5 L/min. Sciatic nerve injury was induced using a previously reported CCI procedure with slight modifications. The right side biceps femoris and the gluteus muscles were divided to expose the sciatic nerve, around which 4 loose ligatures (6–0 nylon) were placed from a distal position on the femoral nerve to a proximal position. This model induces mechanical allodynia of the ipsilateral hind paw within the first week of the injury. Upon awakening, rats were housed in groups in our animal facility and maintained under conditions of constant temperature and humidity, and allowed food and water ad libitum.
2.2. Intrathecal Noggin injection
Fourteen days after CCI, rats were reanesthetized with 1.5% of halothane in oxygen, delivered at 0.5 L/min. After laminectomy at L5, a thin silicone tube was inserted into the subarachnoid space using a surgical microscope. The tube was connected to an osmotic minipump (model 2002; Alzet, Palo Alto, CA) containing 10 ng/mL, 2 μg/mL, or 10 μg/mL recombinant mouse Noggin (R & D Systems, Minneapolis, MN) based on previous reports [18] in phosphate-buffered saline (PBS), or PBS only. The infusion rate was 12 μL/day, resulting in delivery of 120 ng–120 μg Noggin/day. The infusion was continued for 14 days, and the total amount of Noggin administered was approximately 1.68 μg–1.68 mg/animal. Repeated-measures ANOVAs, and Bonferroni or Dunnett post hoc tests were applied to von Frey data and p < 0.05 was considered significant.
2.3. Intrathecal Noggin and bone morphogenetic protein 4 administration
Noggin is a potent inhibitor of bone morphogenetic protein (BMP) that exerts its function by binding to BMPs, preventing their interaction with their receptors. BMP4 production concomitantly decreases the BMP inhibitor Noggin, potentially resulting in a net increase in BMP signaling [19]. Conversely, recombinant human Noggin was used to suppress BMP action [20]. To block Noggin signaling in vivo, we selected BMP4 for the following experiments. Rats were divided into 3 groups, with intrathecal administration of either Noggin (2 μg/ml), BMP4 (2 μg/ml), or Noggin + BMP4 (2 μg/ml each) using an osmotic minipump for 14 days after CCI. Mechanical allodynia was measured for 42 days after implanting the minipump. The allodynic response was analyzed using repeated-measures ANOVAs with Bonferroni or Dunnett post hoc tests; p < 0.05 was considered significant.
2.4. Microarray experiments
Lacroix-Fralish et al. measured mechanical allodynia followed by analysis of global gene expression in the lumber spinal cord at two time points (7 days and 14 days) after surgery using the Affymetrix GeneChip. We decided to show microarray results at 14 days after surgery in the present study. The rats were deeply anesthetized with an intraperitoneal dose of pentobarbital (80 mg/kg; Abbott Laboratories, North Chicago, IL) and decapitated fourteen days after CCI. The lumbar enlargements of their spinal cords (1 cm) were rapidly excised and homogenized in TRizol (Invitrogen, Carlsbad, CA) to preserve RNA. The quality of the extracted RNA was checked using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, MA). The 260 nm/280 nm ratio was > 2.00 in all 6 samples. Samples were electrophoresed using a 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). An Agilent Low RNA Input Linear Amplification PLUS kit was used for RNA amplification and labeling, and an Agilent Gene Expression Hybridization Kit and Wash Buffer Kit was used for hybridization. An Agilent DNA microarray scanner was used to scan chips. Feature Extraction Software (Agilent Technology) was used for quantification. Data was imported into Agilent GeneSpring software.
Intensity dependent (LOWESS) normalization was applied in the present study. This option is recommended for use in most 2-color experiments. Intensity dependent normalization is used to eliminate dye-related artifacts in 2-color experiments that cause the cy5/cy3 ratio to be affected by the total intensity of the spot. This normalization process attempts to correct for artifacts caused by nonlinear rates of dye incorporation and inconsistencies in the relative fluorescence intensity between some red and green dyes. Reliable genes from 3 array experiments were selected based on the data quality flags in the original data files. To identify the significantly up- and downregulated genes, we used a t-test with differences from controls of p < 0.05 considered to be significant.
2.5. Scatter plot
A scatter plot view is useful for examining the levels of expression of genes in 2 distinct conditions, samples, or normalization schemes. For instance, a scatter plot can be used to identify genes that are differentially expressed in one sample versus another. The vertical position of each gene represents its expression level in the current conditions, and the horizontal position represents its control strength. Genes that fall above the diagonal are over expressed and genes that fall below the diagonal are under expressed compared with their median expression level over the course of the experiments.
2.6. Quantitative RT-PCR (RT-qPCR)
Qiagen (Venlo, The Netherlands) RNase-free DNase I was used for DNase-treatment. We subjected 1.0 μg RNA to reverse transcription using TaqMan reverse transcription reagent (ABI). DNA Engine PTC-200 (MJ Research: Bio-Rad, Hercules, CA) was used for reverse transcription; RT conditions were as follows: 25 °C for 10 min, 1 °C for 30 min, and 95 °C for 5 min. Rotor-Gene Q (Qiagen) and a Rotor-Gene SYBR Green RT-PCR kit (Qiagen) were used for qPCR. qPCR conditions were as follows: (95 °C for 10 min, R °C for 15 s, 72 °C for 20 s), 40 cycles, R: annealing temperature. The melting-point curve was checked for nonspecific amplification and primer–dimer complexes. A relative standard was incorporated and target quantity was determined from a standard curve and divided by the endogenous quantity control.
2.7. Mechanical allodynia
Mechanical allodynia was assessed using von Frey filaments according to a previously described protocol [22]. The von Frey filaments were applied to the central plantar surface of the ipsilateral hind paw in descending or ascending order of stiffness (0.7, 1.2, 1.5, 2.0, 3.6, 5.5, 8.5, 11.7, 15.1, and 29 g). Each filament was applied 3 times. When a rat showed a single withdrawal response to a given filament, the bending force for that filament was defined as the paw withdrawal threshold intensity. The median threshold intensity was calculated from the values following 1 descending and 2 ascending trials. The experimental conditions were identical for all groups of rats. This behavioral testing commenced one week after the surgery and continued for 6 consecutive weeks.
2.8. Tissue preparation
Tissue for histology was excised 6 weeks after surgery. Rats were anesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde in PBS. Tissue blocks of the spinal lumbar enlargement were excised, postfixed overnight in 4% paraformaldehyde, stored at 4 °C in 20% sucrose in PBS, and then cryoprotected by embedding in OCT compound (Sakura Finetechnical, Tokyo, Japan). The cryoprotected samples were frozen and stored at − 80 °C until use. The samples were cut into serial 20 °C for 15 μm transverse sections with a cryostat and mounted on amino-silane-coated slides (Matsunami, Tokyo, Japan).
2.9. Immunofluorescent labeling
For immunofluorescent labeling, sections were permeated with 0.3% Triton X-100 in PBS and treated for 1 h in blocking solution containing 1% bovine serum albumin and Block Ace (Dainippon Pharma, Japan). Sections were then incubated with the following primary antibodies: rabbit polyclonal anti-Iba-1 (1:400; Wako Pure Chemical Industries, Osaka, Japan) for microglia or mouse monoclonal anti-glial fibrillary acidic protein antibody (GFAP, 1:400; Sigma, St Louis, MO) for astrocytes. The sections were incubated with primary antibodies overnight at 4 °C, after which they were washed in PBS and then incubated for 30 min at room temperature with secondary antibodies, Alexa 594-labeled anti-rabbit IgG or Alexa 594-labeled anti-mouse IgG (both at 1:800 dilution; Invitrogen, Eugene, OR). Finally, the sections were washed twice in PBS and protected with coverslips. Positive labeling was observed using fluorescence microscopy. Negative controls were performed on control sections with the omission of primary or secondary antibodies. Positive immunofluorescent signals were counted for 3–5 randomly-selected transverse sections from the spinal lumbar enlargement using Scion Image computer analysis software (version beta 4.0.3; Scion Corporation, Frederick, MA). Averaged data from rats after Noggin administration and PBS administration were compared. A Student t-test was used to determine significant differences at a level of p < 0.05.
3. Results
3.1. Microarray
Microarrays (Agilent Rat Whole genome 4 × 44 K) contained 41,000 genes on each chip. After normalizing 41,000 genes, 14,775 genes were selected for statistical analysis. CCI/control ratio was analyzed for 3 arrays, and genes that were significantly up- or downregulated were selected. There were 1136 significantly upregulated genes and 1709 significantly downregulated genes (p < 0.05). After referring to the ontology for each gene, we selected IGF-1, Timp3, Pap, Aqp4, CD38, CD68 as upregulated genes, and Noggin and Oprl1 as downregulated genes to follow in a RT-qPCR study (Table 1).
Table 1.
Significantly up- or downregulated genes in microarray studies.

3.2. RT-qPCR
As described, we selected 6 upregulated genes and 2 downregulated genes for RT-qPCR. These 8 genes were also significantly up- or downregulated in RT-qPCR (Table 2). We hypothesized that administration of depleted gene products could reduce neuropathic pain.
Table 2.
Six significantly upregulated and 2 significantly downregulated genes detected in microarray studies were analyzed by RT-qPCR.

3.3. Pilot study for determining optimal Noggin concentration
We administered Noggin intrathecally using an osmotic minipump from 14 days after CCI. Noggin was administered at 10 ng/ml, 2 μg/ml, and 10 μg/ml and PBS was used as a control. One week after Noggin administration, mechanical allodynia decreased compared with PBS controls. Noggin at 2 μg/ml significantly reduced allodynia 7 days after administration. Thereafter, although rats that had been administered Noggin tended to have reduced allodynia compared with those administered PBS, there was no significant difference between the groups. We chose Noggin at 2 μg/ml for the following studies (Fig. 1).
Fig. 1.
Pilot study for determining optimal Noggin concentration Noggin was administered at 10 ng/ml, 2 mg/ml, and 10 mg/ml and PBS was used as a control. One week after Noggin administration, mechanical allodynia decreased compared with PBS controls. Noggin at 2 mg/ml significantly reduced allodynia 7 days after administration.
3.4. Noggin and BMP4 administration for CCI rats
We determined the optimal concentration of Noggin to reduce neuropathic pain using the intrathecal osmotic minipump administration of 2 μg/ml. Rats were divided into 3 groups administered either Noggin (2 μg/ml), BMP4 (2 μg/ml), or Noggin + BMP4 (2 μg/ml each). Each rat had the agent(s) or controls administered intrathecally for 14 days using an osmotic minipump from 14 days after CCI. Mechanical allodynia significantly decreased one week (3 weeks after CCI) after Noggin administration compared with BMP4 or Noggin + BMP4 administration (n = 4 each, p < 0.05). Thereafter, there was no significant difference between the 3 groups (Fig. 2).
Fig. 2.
Intrathecal administration of Noggin only alleviates mechanical allodynia. Mechanical allodynia was significantly decreased one week (3 weeks after CCI) after Noggin administration compared with BMP4 or Noggin + BMP4 injection (n = 4 each, p < 0.05).
3.5. Immunohistochemistry
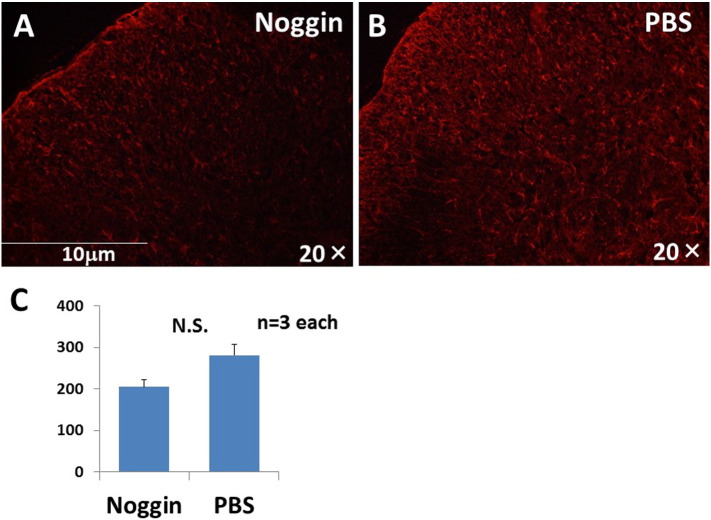
Because the decrease in mechanical allodynia was no longer significant more than one week after Noggin administration (3 weeks after CCI), we decided to time the immunohistochemistry study for one week after Noggin administration. We counted GFAP-immunoreactive astrocytes and Iba-1-immunoreactive microglia one week after implanting the minipump. There were 205.5 ± 16.7 GFAP-immunoreactive astrocytes after Noggin administration and 281.3 ± 25.5 after PBS administration (n = 3 each). The number of astrocytes with GFAP-like immunoreactivity tended to be decreased after Noggin administration, but the decrease was not significant compared with PBS-treated controls (p = 0.068) (Fig. 3). The number of microglia with Iba-1-like immunoreactivity also tended to be decreased after Noggin administration (1157 ± 273) compared with PBS administration (2479 ± 983), but the difference between groups was not significant (p = 0.265).
Fig. 3.
The number of astrocytes with GFAP-like immunoreactivity were decreased after Noggin administration. We counted the number of astrocytes with GFAP-like immunoreactivity one week after implanting the minipump. The number of astrocytes with GFAP-like immunoreactivity tended to be decreased after Noggin administration (A) compared with PBS administration (B), but the difference was not significant (p = 0.068).
4. Discussion
In the present study, the RNA quality was reliable because lumbar enlargement of the CCI model is constantly stable and nonvariant. Three rats were assigned to the CCI group and another 3 rats were assigned to the control group. Using a NanoDrop spectrophotometer the absorbance ratio of 260 nm/280 nm was > 2.00 in all 6 samples. CCI and control rats were comparatively hybridized into 3 microarrays. The microarrays (Agilent Rat Whole genome 4 × 44 K) contain 44,000 genes on each chip. After normalizing 41,000 genes, 14,775 genes were selected for analysis. There were 1136 significantly upregulated genes and 1709 significantly downregulated genes. The number of downregulated genes was larger than that of upregulated genes. We previously reported the gene expression profiling of mice spinal cord injury [23]. We pooled spinal cord samples for RNA extraction. NIA 15 K cDNA microarray was used and within about 15,000 genes, only 864 genes were considered suitable for analysis. There were 84 upregulated genes and no downregulated genes in that study. The RNA quality determined by the absorbance ratio of 260 nm/280 nm was 1.6 to 1.7. The quality of RNA is important for reliable data.
The RNA experiments of the present study consistently showed that Noggin is downregulated 14 days after CCI. Intrathecal administration of Noggin for 14 days one week after CCI reduced mechanical allodynia one week after beginning administration. Although the number of astrocytes with GFAP-like immunoreactivity tended to be reduced one week after Noggin administration, the difference from controls was not significant (p = 0.068). Intrathecal administration of Noggin in the injured spinal cord failed to attenuate GFAP expression even though it effectively reduced pSmad expression. Noggin treatment did not block phosphorylation of Stat3 and the induction of GFAP in the injured spinal cord, suggesting that in addition to the BMP/Smad pathway, the JAK/STAT pathway may also be involved in the regulation of GFAP expression after spinal cord injury [24]. BMP4 or BMP4 + Noggin administration did not reduce mechanical allodynia. Cultured mature astrocytes respond directly to BMPs with Smad1 translocation to the nucleus, increased phosphorylated Smad1/5/8, and increases in GFAP and CSPG expression [25]. We speculated that Noggin administration partially influenced BMP-induced GFAP expression (p = 0.068) and reduced mechanical allodynia in the present study, suggesting that it may reduce neuropathic pain.
Why is Noggin downregulated in the rat CCI model? Is the Noggin downregulated because of relatively upregulated BMP reduced Noggin expression? We could not find any change in BMP-related gene expression in the present microarray study. BMP4 and BMP7 increase rapidly at the site of demyelination in CCI and astrocytes surrounding the lesion increase expression of phosphorylated Smad1/5/8 [25]. Sciatic nerve sections showed a marked degeneration of myelinated fibers in rats with CCI [26]. We speculate that Noggin downregulation is relative to overall BMP upregulation in degenerated myelinated fibers in rats with CCI. There are many subtypes of BMPs. In the present study, none of the changes in genes for BMPs were significant and this may be related to subtle changes in the regulation of their genes.
Noggin administration consistently attenuated mechanical allodynia in the present study. In a deafferented dorsal horn following rhizotomy, Noggin-FbAb treatment induced significant increases in the density of substance P, calcitonin gene-related peptide (CGRP)- and 5-hydroxytryptamine-immunoreactive axons [27]. That is to say, Noggin depletion may increase the expression of pain-related peptides. We speculated that Noggin depletion in the present study increased hypersensitivity as seen previously. By contrast, increased CGRP-like immunoreactivity was detected in epidermal keratinocytes of transgenic mice with keratin-14 promoter driven overexpression of Noggin, which blocks BMP4 signaling [28]. In the present study, reduction in mechanical allodynia was only achieved for one week after Noggin administration. Continuous administration of Noggin (14 days, 336 μg in total) may upregulate CGRP expression, returning the rat to a hypersensitive state. Immunohistochemistry to detect CGRP- and GAP43-like immunoreactivity one week after Noggin injection did not show any significant changes in the present study (data not shown). Neuropathic pain is mediated not only by the amount of pain-related neuropeptides, but may also be mediated by as yet uncharacterized neuronal plasticity in the spinal cord or degenerated sciatic nerves in CCI models.
5. Conclusions
In the microarray and RT-qPCR study, Noggin RNA is significantly down-regulated compared with sham surgery two weeks after CCI in the rat lumber enlargements. We administered Noggin intrathecally two weeks after CCI and continued for 14 days. The mechanical allodynia using von Frey filament was decreased significantly in Noggin administration compared with PBS administration at seven days after Noggin administration. From fluorescent immunohistochemistry, we speculated that Noggin administration partially influenced BMP-induced GFAP expression and reduced mechanical allodynia.
Conflict of interest
This research was supported by grant-in-aid for Japanese Scientific Research grant 24592186.
References
- 1.Backonja M.M. Neuropathic pain therapy: from bench to bedside. Semin Neurol. 2012;32:264–268. doi: 10.1055/s-0032-1329204. [DOI] [PubMed] [Google Scholar]
- 2.Decosterd I., Woolf C.J. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87(2):149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 3.Bennett G.J., Xie Y.K. A peripheral mononeuropathy in rat that produces disorders of pain sensation like those seen in man. Pain. 1988;33(1):87–107. doi: 10.1016/0304-3959(88)90209-6. [DOI] [PubMed] [Google Scholar]
- 4.Kim S.H., Chung J.M. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. 1992;50(3):355–363. doi: 10.1016/0304-3959(92)90041-9. [DOI] [PubMed] [Google Scholar]
- 5.Mayer D.J., Mao J., Holt J. Cellular mechanisms of neuropathic pain, morphine tolerance, and their interactions. Proc Natl Acad Sci U S A. 1999;96(14):7731–7736. doi: 10.1073/pnas.96.14.7731. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin R.S., Costigan M., Brenner G.J. Complement induction in spinal cord microglia results in anaphylatoxin C5a-mediated pain hypersensitivity. J Neurosci. 2007;27(32):8699–8708. doi: 10.1523/JNEUROSCI.2018-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai S., Ikegami D., Yamashita A. Epigenetic transcriptional activation of monocyte chemotactic protein 3 contributes to long-lasting neuropathic pain. Brain. 2013;136(Pt 3):828–843. doi: 10.1093/brain/aws330. [DOI] [PubMed] [Google Scholar]
- 8.Levin M.E., Jin J.G., Ji R.R. Complement activation in the peripheral nervous system following the spinal nerve ligation model of neuropathic pain. Pain. 2008;137(1):182–201. doi: 10.1016/j.pain.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Sun H., Xu J., Della Penna K.B. Dorsal horn-enriched genes identified by DNA microarray, in situ hybridization and immunohistochemistry. BMC Neurosci. 2002;3:11. doi: 10.1186/1471-2202-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nesic O., Lee J., Johnson K.M. Transcriptional profiling of spinal cord injury-induced central neuropathic pain. J Neurochem. 2005;95(4):998–1014. doi: 10.1111/j.1471-4159.2005.03462.x. (Epub 2005 Oct 10) [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez Parkitna J., Korostynski M., Kaminska-Chowaniec D. Comparison of gene expression profiles in neuropathic and inflammatory pain. J Physiol Pharmacol. 2006;57:401–414. [PubMed] [Google Scholar]
- 12.Liu W., Furuichi T., Miyake M. Differential expression of tissue inhibitor of metalloproteinases-3 in cultured astrocytes and neurons regulates the activation of matrix metalloproteinase-2. J Neurosci Res. 2007;85(4):829–836. doi: 10.1002/jnr.21179. [DOI] [PubMed] [Google Scholar]
- 13.Satoh J., Tabunoki H., Yamamura T. Human astrocytes express aquaporin-1 and aquaporin-4 in vitro and in vivo. Neuropathology. 2007;27(3):245–256. doi: 10.1111/j.1440-1789.2007.00774.x. [DOI] [PubMed] [Google Scholar]
- 14.Smith W.C., Harland R.M. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell. 1992;70(5):829–840. doi: 10.1016/0092-8674(92)90316-5. [DOI] [PubMed] [Google Scholar]
- 15.Dionne M.S., Brunet L.J., Eimon P.M. Noggin is required for correct guidance of dorsal root ganglion axons. Dev Biol. 2002;251(2):283–293. doi: 10.1006/dbio.2002.0829. [DOI] [PubMed] [Google Scholar]
- 16.Guha U., Gomes W.A., Samanta J. Target-derived BMP signaling limits sensory neuron number and the extent of peripheral innervation in vivo. Development. 2004;131(5):1175–1186. doi: 10.1242/dev.01013. [DOI] [PubMed] [Google Scholar]
- 17.Ma C.H., Brenner G.J., Omura T. The BMP coreceptor RGMb promotes while the endogenous BMP antagonist noggin reduces neurite outgrowth and peripheral nerve regeneration by modulating BMP signaling. J Neurosci. 2011;31(50):18391–18400. doi: 10.1523/JNEUROSCI.4550-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Rosa I., Camargo L., Pereira M.M. Effects of bone morphogenic protein 4 (BMP4) and its inhibitor, Noggin, on in vitro maturation and culture of bovine preimplantation embryos. Reprod Biol Endocrinol. 2011;9:18. doi: 10.1186/1477-7827-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lassová L., Niu Z., Golden E.B. Thyroid hormone treatment of cultured chondrocytes mimics in vivo stimulation of collagen X mRNA by increasing BMP 4 expression. J Cell Physiol. 2009;219(3):595–605. doi: 10.1002/jcp.21704. [DOI] [PubMed] [Google Scholar]
- 20.Dummula K., Vinukonda G., Chu P. Bone morphogenetic protein inhibition promotes neurological recovery after intraventricular hemorrhage. J Neurosci. 2011;31(34):12068–12082. doi: 10.1523/JNEUROSCI.0013-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacroix-Fralish M.L., Tawfik V.L., Tanga F.Y. Differential spinal cord gene expression in rodent models of radicular and neuropathic pain. Anesthesiology. 2006;104(6):1283–1292. doi: 10.1097/00000542-200606000-00025. [DOI] [PubMed] [Google Scholar]
- 22.Koda M., Furuya T., Kato K. Delayed granulocyte colony-stimulating factor treatment in rats attenuates mechanical allodynia induced by chronic constriction injury of the sciatic nerve. Spine (Phila Pa 1976) 2014;39(3):192–197. doi: 10.1097/BRS.0000000000000108. [DOI] [PubMed] [Google Scholar]
- 23.Hashimoto M., Koda M., Ino H. Gene expression profiling of cathepsin D, metallothioneins-1 and -2, osteopontin, and tenascin-C in a mouse spinal cord injury model by cDNA microarray analysis. Acta Neuropathol. 2005;109(2):165–180. doi: 10.1007/s00401-004-0926-z. [DOI] [PubMed] [Google Scholar]
- 24.Xiao Q., Du Y., Wu W. Bone morphogenetic proteins mediate cellular response and, together with Noggin, regulate astrocyte differentiation after spinal cord injury. Exp Neurol. 2010;221(2):353–366. doi: 10.1016/j.expneurol.2009.12.003. Epub 2009 Dec 11. [DOI] [PubMed] [Google Scholar]
- 25.Fuller M.L., DeChant A.K., Rothstein B. Bone morphogenetic proteins promote gliosis in demyelinating spinal cord lesions. Ann Neurol. 2007;62(3):288–300. doi: 10.1002/ana.21179. [DOI] [PubMed] [Google Scholar]
- 26.Costa B., Trovato A.E., Colleoni M. Effect of the cannabinoid CB1 receptor antagonist, SR141716, on nociceptive response and nerve demyelination in rodents with chronic constriction injury of the sciatic nerve. Pain. 2005;116(1–2):52–61. doi: 10.1016/j.pain.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 27.Hampton D.W., Steeves J.D., Fawcett J.W. Spinally upregulated noggin suppresses axonal and dendritic plasticity following dorsal rhizotomy. Exp Neurol. 2007;204(1):366–379. doi: 10.1016/j.expneurol.2006.11.017. [DOI] [PubMed] [Google Scholar]
- 28.Hou Q., Barr T., Gee L. Keratinocyte expression of calcitonin gene-related peptide β: implications for neuropathic and inflammatory pain mechanisms. Pain. 2011;152(9):2036–2051. doi: 10.1016/j.pain.2011.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]