Abstract
Cardiomyopathy, also known as heart muscle disease, is an unfavorable condition leading to alterations in myocardial contraction and/or impaired ability of ventricular filling. The onset and development of cardiomyopathy have not currently been well defined. Titin is a giant multifunctional sarcomeric filament protein that provides passive stiffness to cardiomyocytes and has been implicated to play an important role in the origin and development of cardiomyopathy and heart failure. Titin-based passive stiffness can be mainly adjusted by isoform switching and post-translational modifications in the spring regions. Recently, genetic mutations of TTN have been identified that can also contribute to variable passive stiffness, though the detailed mechanisms remain unclear. In this review, we will discuss titin isoform switching as it relates to alternative splicing during development stages and differences between species and muscle types. We provide an update on the regulatory mechanisms of TTN splicing controlled by RBM20 and cover the roles of TTN splicing in adjusting the diastolic stiffness and systolic compliance of the healthy and the failing heart. Finally, this review attempts to provide future directions for RBM20 as a potential target for pharmacological intervention in cardiomyopathy and heart failure.
Keywords: Titin isoform switching, Alternative splicing, Cardiomyopathy, RBM20
Introduction
Cardiomyopathy, or heart muscle disease, can result in either systolic dysfunction with decreased myocardial contraction or diastolic dysfunction with impaired ability of ventricular filling (Daughenbaugh 2007; Henry 2003; Peters 2016). A number of factors can contribute to the onset and development of cardiomyopathy, including abnormal energetic metabolism, intracellular calcium handling, and sarcomeric structure and protein changes (LeWinter 2005; Miyata et al. 2000). Among the sarcomeric proteins, titin protein is increasingly regarded as one of the molecular origins of cardiomyopathy and heart failure (Linke and Hamdani 2014; Yin et al. 2015). Titin protein is a giant multifunctional sarcomeric filament that plays a critical role in elastic recoil of the cardiac myocytes and contributes to diastolic function during the left ventricular filling phase (Granzier and Irving 1995; Granzier and Labeit 2004; Herman et al. 2012; LeWinter and Granzier 2010, 2014; Taylor et al. 2011). Titin can function as a “molecular blueprint”, “molecular spring”, and “molecular signaling mediator” (Granzier and Labeit 2004, 2005; Krüger and Linke 2011; Linke 2008). During the past decade, evidence has shown that titin elasticity is highly variable in the developing and the adult normal healthy heart, and that it can be pathologically modified in cardiomyopathy and heart failure. Pathological modifications can greatly compromise the extensibility and the diastolic passive stiffness of myocardium, and, presumably, the mechanosignaling transduction sensitivity (Makarenko et al. 2004; Opitz and Linke 2005). Variable titin elasticity is, in large part, attributed to titin isoform switching that results from alternative splicing (Granzier and Labeit 2004; Linke 2008). However, mechanistic study of titin isoform switching had never been addressed until the TTN splicing regulator, RNA binding motif 20 (RBM20), was cloned and identified recently in 2012 (Guo et al. 2012). This finding opened a new avenue for the identification of novel potential targets for therapeutic intervention for titin-based pathological alterations in cardiomyopathy and heart failure. In our review, we begin with an introduction to titin, including its structure, function, and arrangement in the sarcomere, and provide an update on known alternatively used exons and TTN splicing patterns. We then cover some details of TTN splicing mechanism(s) regulated by RBM20 and discuss the potential therapeutic strategies and future directions.
Titin structure, function, and arrangement in the sarcomere
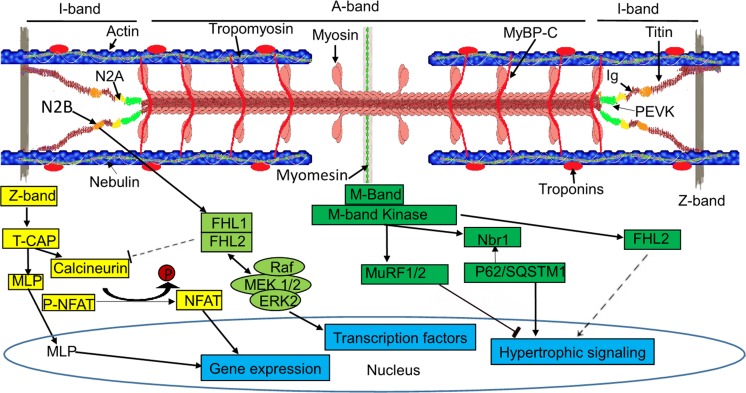
The sarcomere is the smallest contractile unit in myofibrils of striated muscle fibers and contains four major filament systems: actin-thin, myosin-thick, titin, and nebulin filaments (Trinick 1996; Tskhovrebova and Trinick 2003). Titin filament is the third one (after myosin and actin filaments), composed of a gigantic fibrous protein titin, which is the largest protein by far found in vertebrate animals (Gigli et al. 2016; Maruyama 1976; Wang et al. 1979; Yin et al. 2015). Titin spans half of a sarcomere and connects the Z-band to the M-band located in the center of the sarcomere. Its carboxy (C)-terminal cross-links with another titin’s C-terminal in the M-band, and its amino (N)-terminal attaches another titin’s N-terminal from adjacent sarcomere and anchors in the Z-band (Fürst et al. 1988; Labeit et al. 1990; Bang et al. 2001), which allows titin molecules to form a continuous system along the myofibril. Such structural arrangement of titin enables it to function as a molecular blueprint for the maintenance of sarcomere integrity and precise assembly of the regulatory, contractile, and structural proteins located in the sarcomere (Freiburg and Gautel 1996; Granzier and Labeit 2004; Labeit et al. 1992; Tskhovrebova and Trinick 2003) and transduce force generated during contraction at the Z-band (Gutierrez-Cruz et al. 2001; Labeit and Kolmerer 1995) (Fig. 1). Besides titin serving as a “molecular blueprint”, it also acts as a “molecular spring” and a “molecular signaling mediator” (Granzier and Labeit 2004, 2005, 2006; Kontrogianni-Konstantopoulos et al. 2009; Krüger and Linke 2011; Linke 2009). Titin contains both highly repetitive and non-repetitive domains. Approximately 90% of the titin sequence is repeating immunoglobulin (Ig) and fibronectin-3 (Fn3) domains, each around 100 residues. Each molecule contains between 240 and 300 repeats, depending on different isoforms. The remaining approximately 10% of non-repetitive sequence consists of unique sequences (e.g., N2B and N2A), 28–30 residue PEVK [proline (P), glutamate (E), valine (V), and lysine (K)] motifs, and a C-terminal Ser/Thr kinase domain interspersed between the Ig and Fn3 repeats (Guo et al. 2010; Kontrogianni-Konstantopoulos et al. 2009; Meyer and Wright 2013; Puchner et al. 2008; Tskhovrebova and Trinick 2004). The repeating Ig domains located in the I-band are not extensible, while the non-repetitive unique sequences N2B and the PEVK motifs are. These elastic segments in titin determine that titin is a spring-like protein and serves as a molecular spring (Granzier and Labeit 2002; Kellermayer et al. 1997; Linke et al. 1996, 1999; Trombitás et al. 1998; Watanabe et al. 2002) (Fig. 1). Titin’s modular structure with repetitive and non-repetitive domains provides diverse binding sites for a variety of proteins, which include myofibrillar proteins, membrane components, enzymes, and signaling molecules. Over 20 proteins have been found bound to titin, linking it to multiple potential mechanical signaling pathways (Krüger and Linke 2011; Linke 2008). Titin runs through distinct sarcomere regions: the Z-band, the I-band, and the A-band regions, each of which may transduce stress signals (Fig. 1). For example, at the Z-band, protein complexes with titin, telethonin/T-cap, MLP, and calsarcin-2 can activate the calcineurin/nuclear factor of activated T-cells (NFAT) signaling to induce hypotrophy, leading to heart failure (Fig. 1) (Hoshijima 2006; Lange et al. 2006; Linke 2008); at the I-band, the protein complexes of N2B, FHL1, FHL2, and MAPK may activate MAPK signaling cascade (raf/Mek1/2/Erk2) to induce hypertrophic signaling or repress calcineurin/NFAT signaling for pathological cardiac growth (Fig. 1) (Hojayev et al. 2012; Kötter et al. 2014; Lange et al. 2002; Scholl et al. 2000; Sheikh et al. 2008); at the M-band, titin can also mediate hypertrophic signaling through its kinase (TK) domain. Activated TK may interact with protein complexes of Nbr1, p62/SQSTM1, and MuRF1/2/3 to induce hypertrophic signaling (Fig. 1) (Kötter et al. 2014; Lange et al. 2005). Thus, this giant protein is believed to serve as a molecular signaling mediator, a central player for hypertrophic signaling.
Fig. 1.
The striated sarcomere structure and TTN-mediated signaling pathway. The upper schematic structure shows the layout of the sarcomere and the domains of TTN. The lower scheme shows the hypertrophic signaling pathways through TTN binding partners at the Z-band, I-band, and M-band. MLP, muscle LIM protein; NFAT, nuclear factor of activated T-cells; FHL1/2, four-and-a-half LIM domain protein 1/2; Raf, rapidly accelerated fibrosarcoma protein; MEK 1/2, mitogen-activated protein kinase kinase 1; ERK 1/2, extracellular signal-regulated kinase; Nbr1, neighbor-of-BRCA1-gene-1; p62, nucleoporin p62; QSTM1, sequestosome 1
TTN splicing
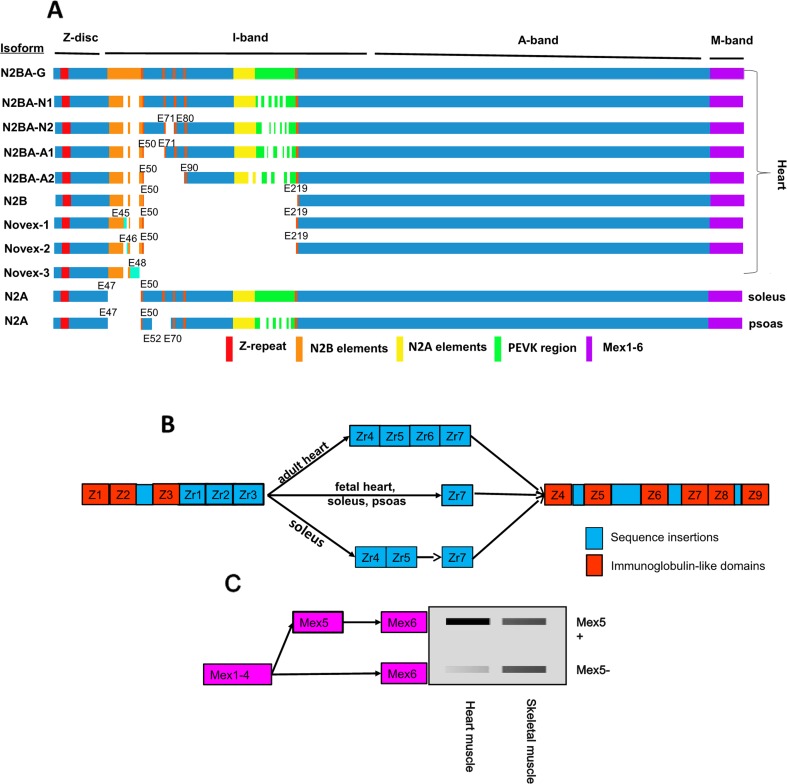
The human TTN gene is located on chromosome 2q31 and consists of 294-kilo base pairs (kb). This single mammalian gene contains 364 exons, with 363 coding ones (the first one is a non-coding exon) that can be transcribed to a more than 100-kb-long mRNA. Deduced from the mRNA sequence, a total of 38,138 amino acid residues with a molecular weight (MW) of 4.2 MDa could be produced (Bang et al. 2001). Interestingly, translation of full-length titin with 4.2 MDa has never been reported. Reported titin sizes range from approximately 3.0 to 3.9 MDa, resulting from alternatively used exons (Guo et al. 2010; Warren et al. 2004). Nearly all exons in the Z-band, A-band, and M-band regions are constitutively expressed in human striated muscle, while the I-band undergoes extensive alternative splicing with two splicing hot regions: the variable-length Ig region and the PEVK domain (Labeit and Kolmerer 1995; Linke and Kruger 2010). The splicing patterns of the PEVK domain are exceptionally complex. PCR results from a previous study indicated that at least 20 exons are absent in the PEVK region (Guo et al. 2010). If these 20 exons are independently spliced, the combinations of these splicing events could lead to millions of different splicing pathways or isoforms. However, using a well-recognized large protein electrophoresis gel detection system, vertical SDS-agarose gel electrophoresis, for titin isoform separation (Warren et al. 2003b), only six titin isoforms are detectable in rodent cardiac muscle, and one isoform is visible in most vertebrate skeletal muscles. The explanation for the few detectable isoforms could be that: (1) many isoforms have nearly equal sizes as the six isoforms and are overlapped at the same gel band as the six isoforms; (2) other potential isoforms are expressed in very low levels, which are undetectable; (3) both situations are combined. The six isoforms have been named as N2BA-N1 and -N2 (embryonic and neonatal forms) with apparent sizes of approximately 3.7 and 3.6 MDa, respectively; N2BA-A1 and -A2 (adult forms), with sizes of about 3.4 and 3.2 MDa, respectively; N2B (a cardiac-specific isoform), with a size of approximately 3.0 MDa (Warren et al. 2004); and N2BA-G (an unusual giant isoform), with a size of approximately 3.9 MDa, caused by a gene mutation (Greaser et al. 2005, 2008). In addition, one isoform in skeletal muscle is known as N2A, with variable sizes from approximately 3.2 to 3.7 MDa (Cazorla et al. 2000; Freiburg et al. 2000; Labeit and Kolmerer 1995) (Fig. 2a). These isoforms are called N2B, N2BA, and N2A because they contain either N2B or N2A or both unique domains. Finally, each isoform represents a class of titin isoforms resulting from the combinations of alternatively spliced exons, mainly in the I-band Ig domain and the PEVK region. The N2B unique domain is solely found in cardiac muscle and the N2A unique domain can be found in both skeletal and cardiac muscle. If the isoforms contain both N2B and N2A unique domains, they are called N2BA, which are also exclusively found in cardiac muscle (Guo et al. 2010; Labeit and Kolmerer 1995). The giant N2BA isoform (N2BA-G) is assumed to result from nearly all the alternatively used exons being spliced in. N2BA-N1 is expected to contain all the exons for expression of the middle Ig domain and N2BA-N2 is proposed to lack the sequence from exons 72 to 79. Likewise N2BA-A1 and N2BA-A2 are suggested to skip exons 51–70 and 51–89, respectively (Greaser et al. 2005; Guo et al. 2013). N2B skips exons 51–218 (Cazorla et al. 2000; Freiburg et al. 2000; Greaser et al. 2005; Guo et al. 2013). Human soleus N2A is proposed to skip exon 49 expressing for N2B unique sequence and rabbit psoas N2A is lacking exon 49 and exons 53–69 (Greaser et al. 2005; Guo et al. 2010) (Fig. 2a). Besides the N2B, N2BA, and N2A isoforms, three other isoforms have also been reported, which are Novex1, Novex2, and Novex3. The Novex1 and Novex2 isoforms result from alternative splicing and have similar size and splice patterns as the N2B isoform. Novex1 expresses exon 45, and Novex2 exon 46, while these two exons are not present in the major isoforms (N2B, N2BA, and N2A) (Bang et al. 2001; Linke 2008). Novex3 expresses an approximately 650 kDa protein and is produced from an alternative terminal coding exon 48 instead of alternative splicing, and the rest of the exons in Novex3 have the same splice pattern as the major isoforms (Bang et al. 2001; Chauveau et al. 2014; Linke 2008) (Fig. 2a).
Fig. 2.
Known TTN isoforms and TTN splicing patterns at the I-band, Z-band, and M-band. a Known TTN isoforms in striated muscles. b Nine Ig domains with insertions in the Z-band and overview of the Z-repeat isoforms expressed in different types of striated muscles. The repeats Zr1, 2, 3, and 7 express in all striated muscles, whereas Zr4, 5, and 6 are alternatively spliced, depending on the tissue type and the developmental stage. c Alternative splicing of the M-band. The right panel shows the approximate ratio of Mex5+:Mex5− in striated muscles. E, exon; Z, Ig domain repeats; Zr, Z-repeats between Z3 and Z4; Mex, M-band exon
In addition to the hot splicing region in the I-band, alternative splicing can also occur in the Z-band and M-band. The Z-band titin is encoded by the first 28 exons consisting of nine Ig domains (Z1–Z9) interspersed with large inter-domain insertions from Z2 to Z7 and between Z8 and Z9. The insertion between Z3 and Z4 contains alternatively spliced Z-repeats, termed Zr1 to Zr7. Alternative splicing leads to the differential expression of Z-repeats varying from 4 to 7. The repeats Zr1–3 and Zr7 are constitutively expressed in all striated muscles, while the repeats Zr4–6 are alternatively spliced depending on the developmental ages, the species, and the type of striated muscles. For example, in the adult heart, all seven repeats are included, whereas the fetal heart skips Zr4–6. In the slow muscle soleus, two isoforms could be identified, with one excluding Zr4–6 and the other skipping Zr6. However, in the fast twitch rabbit psoas muscle, only the isoform skipping Zr4–6 could be found (Fig. 2b) (Chauveau et al. 2014; Gautel et al. 1996; Guo et al. 2010; Linke 2008; Sorimachi et al. 1997). Kolmerer et al. (1996) searched for alternatively expressed exons in the M-band region of titin, and they found that there were six exons in the M-band, termed Mex1 to Mex6. Intriguingly, only Mex5 was found to undergo alternative splicing. Mex5 is mainly included in heart muscle with a higher ratio of Mex5+:Mex5−, but it seems that skeletal muscle contains equal or lower ratios of Mex5+:Mex5−. Alternative splicing of Mex5 was not observed during mouse development (Fig. 2c) (Granzier and Labeit 2002; Guo et al. 2010; Kolmerer et al. 1996).
TTN splicing patterns with development stages, species, and muscle types
The splicing patterns of individual alternative exons have not been well-characterized for different development stages, species, and muscle types (Lahmers et al. 2004; Ottenheijm et al. 2009; Opitz et al. 2004, Opitz and Linke 2005). In this section, we mainly focus on the discussion of the changes or switching of major classes of isoforms (N2B, N2BA, and N2A) resulting from alternative splicing. Patterns are primarily based on electrophoretic studies.
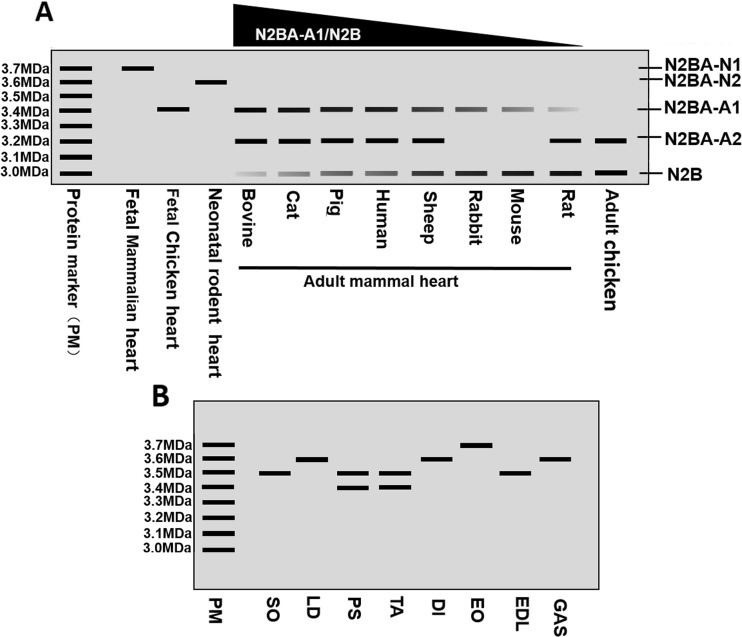
Titin plays a major role in determining passive tension in stretched muscle (Granzier and Labeit 2002; Linke and Fernandez 2002). Titin-based stiffness constitutes a major proportion of the total myocardial passive stiffness in the normal adult human heart and at approximately the time of birth. The heart modifies the mechanical properties of the titin spring to adjust to the global mechanical requirements (Opitz and Linke 2005). In cardiac muscle, N2B has a shorter spring-like domain than N2BA, and, thus, produces higher passive stiffness. Therefore, switching between these two isoforms provides variable titin-based passive stiffness in order to meet physiological requirements, depending on development stages and species. In the larger mammalian heart, including the cat, rabbit, pig, sheep, bovine, and human heart, all hearts express four major titin isoforms (N2BA-N1, N2BA-N2, N2BA-A1, and N2B). In the fetal stage, cardiac muscle expresses only the N2BA-N1 isoform (∼3.7 MDa). During development, the N2BA-N1 isoform is gradually replaced by one neonatal isoform (N2BA-N2, ∼3.6 MDa) and two or three adult isoforms, N2BA-A1/2 (∼3.2–3.4 MDa) and N2B (∼3.0 MDa). Nevertheless, the ratios of N2BA-A1 to N2B in the adult heart vary in distinct species. The estimated order is rabbit < sheep < human = pig < cat < bovine (Fig. 3a) (Freiburg et al. 2000; Fukuda et al. 2003; Neagoe et al. 2003; Opitz and Linke 2005; Opitz et al. 2004). By comparing to large animals, small mammals such as mouse and rat also express the N2BA-N1 isoform during prenatal development. The larger isoform is later replaced by the neonatal form N2BA-N2 (∼3.6 MDa) and adult forms N2BA-A1 (∼3.4 MDa), N2BA-A2 (∼3.2 MDa), and N2B (∼3.0 MDa) in rat heart, and N2BA-A1 and N2B in mouse heart (Fig. 3a) (Cazorla et al. 2000; Greaser et al. 2005). The rat heart contains a lower ratio of N2BA:N2B than that of mouse heart (rat < mouse), but small mammals have lower ratios than larger mammals (Cazorla et al. 2000; Neagoe et al. 2003). Moreover, in chicken heart, the largest embryonic titin isoform is about 3.4 MDa, similar to N2BA-A1 in large and small mammals, and the adult forms are approximately 3 MDa and 3.2 MDa, similar to the N2B and N2BA-A2 isoforms (Fig. 3a) (Opitz and Linke 2005).
Fig. 3.
Schematic diagram of TTN bands in the electrophoresis gel. a Cardiac TTN isoform migration and the ratio in the fetal and adult hearts in different species. b Skeletal muscle TTN migration in different skeletal muscle types. PM, protein marker; SO, soleus; LD, longissimus dorsi; PS, psoas; TA, tibialis anterior; DI, diaphragm; EO, extensor oblique; EDL, extensor digitorum longus; GAS, gastrocnemius
In addition to isoform switching in cardiac muscle, although skeletal muscles only express a single N2A titin isoform, the size of the N2A isoform vary from ∼3.3 MDa to ∼3.7 MDa (Granzier and Labeit 2002). Soleus (SO) and longissimus dorsi (LD) express a long single N2A isoform (∼3.7 MDa), while psoas (PS) and tibialis anterior (TA) express doublet bands, with ∼3.5 MDa and ∼3.4 MDa in psoas and ∼3.4 MDa and ∼3.3 MDa in TA (Granzier and Labeit 2002; Guo et al. 2012; Li et al. 2012; Neagoe et al. 2003; Wang et al. 1991). Li et al. (2012) have estimated the titin sizes in several different rat muscles, including diaphragm (DI), extensor oblique (EO), SO, extensor digitorum longus (EDL), PS, LD, TA, and gastrocnemius (GAS), and they confirmed previous evidence and found that all titin sizes are in the range of ∼3.3 MDa to ∼3.7 MDa (Fig. 3b).
TTN splicing mechanisms
TheTTN gene has been known to undergo alternative splicing since the original gene sequence report (Labeit and Kolmerer 1995). However, the mechanism of how TTN splicing is regulated remained completely unknown until RNA binding motif 20 (RBM20), a splicing factor, was cloned and identified (Guo et al. 2012). As discussed above, titin expresses various sizes as a result of alternative splicing varying with development, species, and different muscle types. In the absence of RBM20 in both rat and mouse models, titin expresses a consistently larger isoform (∼3.9 MDa) (Fig. 2a) under all conditions (developmental, species, and muscle types) (Greaser et al. 2008; Guo et al. 2012, 2013; Li et al. 2012; Methawasin et al. 2014). This suggested that RBM20 could function as a repressor of TTN splicing (Li et al. 2013). Our published data indicated that RBM20-mediated TTN splicing is dose-dependent. Cardiac muscle expresses mainly the smaller N2B isoform (N2BA:N2B = ∼20:80) in wild-type rats and high levels of medium-sized N2BA (N1 and N2) (N2BA:N2B = ∼60:40) in heterozygous RBM20 knockout (KO) rats, whereas only the large N2BA-G isoform is expressed in homozygous RBM20 KO rats (Guo et al. 2012). In wild-type rats or those with RBM20 expression, TTN exons undergo dynamic changes with development, with fewer exons skipped in the prenatal development stage and more exons spliced out after birth to adult. This implied that the RBM20 level should increase along with more exon skipping according to a repressing regulatory mechanism of RBM20 (Guo et al. 2012; Li et al. 2013). Unexpectedly, the RBM20 level not only increases with development, but also decreases. One explanation for the contradictory phenomena is that other splicing-associated factors may be involved in TTN splicing regulation cooperatively with RBM20 (Ito et al. 2016; Li et al. 2013; Yin et al. 2015; Zhu et al. 2016). More evidence is needed to support this hypothesis.
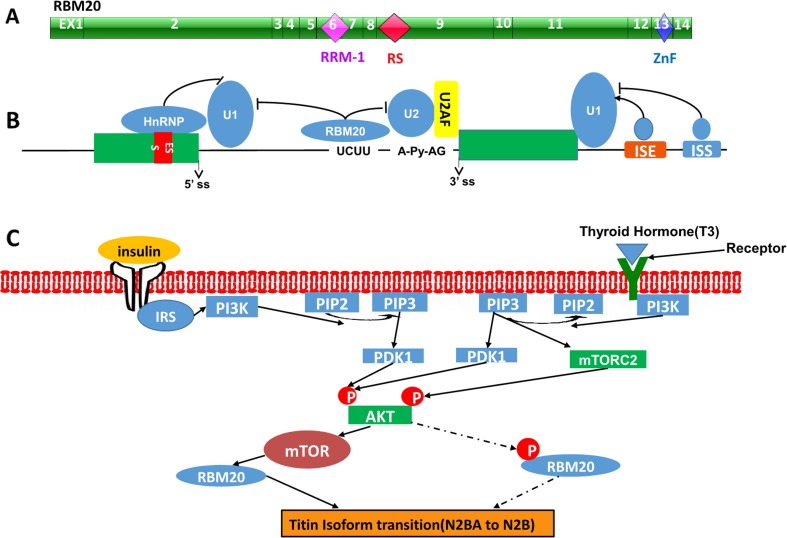
RBM20 consists of 14 exons and encodes an RNA-binding motif protein 20 with a prototypical RNA-recognition motif 1 (RRM-1), followed by an arginine/serine-rich (RS-rich) domain. These structural features are characteristic of a family of RNA-binding SR proteins that assemble in the spliceosome, a huge multiprotein complex that orchestrates constitutive and alternative splicing of pre-mRNA (Fig. 4a) (Brauch et al. 2009; Zhu et al. 2016). Usually, SR and SR-related proteins are frequently phosphorylated and function as splicing activators, recognizing exonic splicing enhancers (Lim et al. 2011; Manley and Tacke 1996). However, sporadic examples of SR proteins act as splicing repressors upon binding at intronic positions (Ibrahim et al. 2005; Kanopka et al. 1996; Shen and Mattox 2012; Wang et al. 2013). A recent study showed that RBM20 binds predominantly to introns near the 3′ and 5′ splice sites of repressed exons and in close proximity to the binding sites of U1 and U2 snRNPs. The binding sites of RBM20 to RNA may contain the UCUU core sequence (Figure 4b) (Maatz et al. 2014). In addition, RBM20 could regulate TTN splicing via cooperation with trans-acting factors and binding with cis-acting elements. Recent studies also indicated that TTN splicing can be regulated by external stimuli, such as hormones (thyroid hormone and insulin). With increased thyroid or insulin hormone, the ratio of N2BA to N2B is decreased (Krüger et al. 2008, 2010). However, the role RBM20 plays in hormone-modulated TTN splicing remains unclear. Currently, our studies demonstrated that RBM20 is the essential mediator linking hormone stimulation to TTN splicing via the PI3K/Akt/mTOR signaling pathway. Activated Akt increases the expression level of RBM20, and, thus, regulates titin isoform switching (Fig. 4c) (Zhu et al. 2015). Akt is one of four known kinase systems to regulate gene alternative splicing through the phosphorylation of SR proteins (Lynch 2007); therefore, whether phosphorylation of RBM20 plays a critical role in TTN splicing via the PI3K/Akt signaling pathway needs further study.
Fig. 4.
RBM20 structure and potential TTN splicing mechanism(s) regulated by RBM20. a Schematic diagram of RBM20 structure. b RBM20 regulatory mechanism through binding to cis-regulatory elements in TTN. c TTN splicing mechanism via the PI3K/Akt/mTOR signaling pathway through regulation of RBM20. EX, exon; RRM, RNA recognition motif-1; RS, arginine/serine-rich domain; ZnF, zinc finger domain; ESS, exon splicing silencer; ISE, intron splicing enhancer; ISS, intron splicing silencer; ss, splice site; Py, polypyrimidine tract
TTN splicing and cardiomyopathies
Titin is emerging as a promising therapeutic molecular target by restoring compliance to the sarcomere and, thereby, improving diastolic function, or by increasing passive stiffness to the ventricular wall, and, thus, improving systolic function. Improving titin-based passive stiffness of the heart can take place in two ways. One is to switch the N2BA to N2B isoform ratio; the other can be tuned through post-translational modifications (PTMs) of these primary isoforms, particularly through the phosphorylation of titin’s spring domains. Switching titin isoform ratios could contribute to ventricular wall stiffness in large part (Guo et al. 2012; Hidalgo and Granzier 2013; Methawasin et al. 2014; Trombitás et al. 2000). Recently, variable titin isoform switching resulting from TTN splicing in animal models and human patients has lead to heart stiffness changes which have been associated with cardiomyopathies. In animal models, a dog tachycardia-induced model with dilated cardiomyopathy (DCM) showed an elevated ratio of N2BA:N2B under two-week mechanical challenge (Bell et al. 2000), while the ratio was decreased after four weeks of pacing (Wu et al. 2002). In addition, a hypertensive dog model with diastolic dysfunction demonstrated a reduced ratio of N2BA:N2B (Hamdani et al. 2013; Shapiro et al. 2007), and also a spontaneously hypertensive rat model (SHR) had slightly decreased ratio of N2BA:N2B in response to pressure overload (Warren et al. 2003a). In the mouse transverse aortic constriction (TAC) model, a commonly used experimental model for pressure overload-induced cardiac hypotrophy and heart failure, an elevated ratio of N2BA:N2B was observed (Hudson et al. 2011). In contrast, in a ligation of the left anterior descending coronary artery (LAD) rat model with ischemic cardiomyopathy, the rat heart expressed a reduced ratio of N2BA:N2B (Neagoe et al. 2002). However, in human patients, altered titin isoform ratios have also been detected. By comparing to nonfailing donor heart, the larger and compliant N2BA levels were increased in ischemic cardiomyopathy (Neagoe et al. 2002), non-ischemic DCM (Makarenko et al. 2004; Nagueh et al. 2004), and patients with heart failure with a reduced ejection fraction (HFrEF) (Borbély et al. 2009). Overall, increased compliant N2BA isoforms can be commonly found in eccentric remodeled hearts with systolic dysfunction, such as DCM, HFrEF, and chronic ischemic cardiomyopathy, while reduced N2BA isoforms can frequently be observed in concentric remodeled hearts (compensated hypertrophy) with diastolic dysfunction developed from hypertensive heart disease. Therefore, the manipulation of titin isoform ratios could be a potential therapeutic strategy for variable titin isoform ratios caused by cardiomyopathies and heart failure. A couple of very recent reports with mouse models of RBM20 manipulation showed the feasibility of such an approach. Reducing RBM20 levels in N2B knockout-induced diastolic dysfunction could improve diastolic stiffness (Hinze et al. 2016) and the inhibition of RBM20 in cardiac muscle can reduce ventricular wall stiffness induced in the transverse aortic constriction (TAC) mouse model, and, thus, improve diastolic function (Methawasin et al. 2016). Since RBM20 is a master regulator of titin isoform switching, it could be a promising therapeutic target for drug development to adjust the titin isoform ratio.
Future directions
Titin is increasingly recognized as a major human disease gene. Titin-associated heart disease is mainly based on the alteration of elastic force, which is highly variable between normal and diseased hearts. Titin-based passive stiffness in the heart can be affected by two main ways: titin isoform switching resulting from alternative splicing and post-translational modifications in TTN spring domains with particular phosphorylation. These two paths offer the therapeutic targets in titin to improve diastolic compliance or systolic stiffness and relieving symptoms. Titin isoform switching largely contributes to variable ventricular wall stiffness. As discussed earlier, such an approach could have the potential to adjust titin elasticity by the manipulation of TTN splicing under the control of the splicing factor RBM20. However, therapeutic modalities targeting RBM20 are currently largely theoretical due to the still less-defined mechanism(s) of how RBM20 regulates TTN splicing. Therefore, future work should aim to identify cooperative regulators, TTN pre-mRNA binding sites of RBM20, and the signaling pathways that control the expression and/or post-translational modifications of RBM20.
Acknowledgments
This work was supported by the National Institute of Health/National Institute of General Medical Sciences (NIGMSP20GM103432); the BGIA from the American Heart Association (16BGIA27790136 to WG); the USDA National Institute of Food and Agriculture (Hatch project 1009266 to WG). The authors would like to thank Dr. Marion Greaser and Dr. Rich McCormick for their helpful comments and proofreading of the manuscript.
Compliance with ethical standards
Conflict of interest
Wei Guo declares that he has no conflict of interest. Mingming Sun declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
This article is part of a Special Issue on “Titin and its Binding Proteins in Striated Muscles” edited by Amy Li and Cristobal G. dos Remedios.
References
- Bang ML, Centner T, Fornoff F, Geach AJ, Gotthardt M, McNabb M, Witt CC, Labeit D, Gregorio CC, Granzier H, Labeit S. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ Res. 2001;89:1065–1072. doi: 10.1161/hh2301.100981. [DOI] [PubMed] [Google Scholar]
- Bell SP, Nyland L, Tischler MD, McNabb M, Granzier H, LeWinter MM. Alterations in the determinants of diastolic suction during pacing tachycardia. Circ Res. 2000;87:235–240. doi: 10.1161/01.RES.87.3.235. [DOI] [PubMed] [Google Scholar]
- Borbély A, Falcao-Pires I, van Heerebeek L, Hamdani N, Edes I, Gavina C, Leite-Moreira AF, Bronzwaer JG, Papp Z, van der Velden J, Stienen GJ, Paulus WJ. Hypophosphorylation of the Stiff N2B titin isoform raises cardiomyocyte resting tension in failing human myocardium. Circ Res. 2009;104:780–786. doi: 10.1161/CIRCRESAHA.108.193326. [DOI] [PubMed] [Google Scholar]
- Brauch KM, Karst ML, Herron KJ, de Andrade M, Pellikka PA, Rodeheffer RJ, Michels VV, Olson TM. Mutations in ribonucleic acid binding protein gene cause familial dilated cardiomyopathy. J Am Coll Cardiol. 2009;54:930–941. doi: 10.1016/j.jacc.2009.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazorla O, Freiburg A, Helmes M, Centner T, McNabb M, Wu Y, Trombitás K, Labeit S, Granzier H. Differential expression of cardiac titin isoforms and modulation of cellular stiffness. Circ Res. 2000;86:59–67. doi: 10.1161/01.RES.86.1.59. [DOI] [PubMed] [Google Scholar]
- Chauveau C, Rowell J, Ferreiro A. A rising titan: TTN review and mutation update. Hum Mutat. 2014;35:1046–1059. doi: 10.1002/humu.22611. [DOI] [PubMed] [Google Scholar]
- Daughenbaugh LA. Cardiomyopathy: an overview. J Nurse Pract. 2007;3:248–258. doi: 10.1016/j.nurpra.2007.01.015. [DOI] [Google Scholar]
- Freiburg A, Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur J Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- Freiburg A, Trombitas K, Hell W, Cazorla O, Fougerousse F, Centner T, Kolmerer B, Witt C, Beckmann JS, Gregorio CC, Granzier H, Labeit S. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ Res. 2000;86:1114–1121. doi: 10.1161/01.RES.86.11.1114. [DOI] [PubMed] [Google Scholar]
- Fukuda N, Wu Y, Farman G, Irving TC, Granzier H. Titin isoform variance and length dependence of activation in skinned bovine cardiac muscle. J Physiol. 2003;553:147–154. doi: 10.1113/jphysiol.2003.049759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M, Goulding D, Bullard B, Weber K, Fürst DO. The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J Cell Sci. 1996;109(Pt 11):2747–2754. doi: 10.1242/jcs.109.11.2747. [DOI] [PubMed] [Google Scholar]
- Gigli M, Begay RL, Morea G, Graw SL, Sinagra G, Taylor MR, Granzier H, Mestroni L. A review of the giant protein titin in clinical molecular diagnostics of cardiomyopathies. Front Cardiovasc Med. 2016;3:21. doi: 10.3389/fcvm.2016.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier HL, Irving TC. Passive tension in cardiac muscle: contribution of collagen, titin, microtubules, and intermediate filaments. Biophys J. 1995;68:1027–1044. doi: 10.1016/S0006-3495(95)80278-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier H, Labeit S. Cardiac titin: an adjustable multi-functional spring. J Physiol. 2002;541:335–342. doi: 10.1113/jphysiol.2001.014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granzier HL, Labeit S. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ Res. 2004;94:284–295. doi: 10.1161/01.RES.0000117769.88862.F8. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Labeit S. Titin and its associated proteins: the third myofilament system of the sarcomere. Adv Protein Chem. 2005;71:89–119. doi: 10.1016/S0065-3233(04)71003-7. [DOI] [PubMed] [Google Scholar]
- Granzier HL, Labeit S. The giant muscle protein titin is an adjustable molecular spring. Exerc Sport Sci Rev. 2006;34:50–53. doi: 10.1249/00003677-200604000-00002. [DOI] [PubMed] [Google Scholar]
- Greaser ML, Krzesinski PR, Warren CM, Kirkpatrick B, Campbell KS, Moss RL. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J Muscle Res Cell Motil. 2005;26:325–332. doi: 10.1007/s10974-005-9039-0. [DOI] [PubMed] [Google Scholar]
- Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, Krzesinski PR, Norman HS, Dunning S, Fitzsimons DP, Moss RL. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. J Mol Cell Cardiol. 2008;44:983–991. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Bharmal SJ, Esbona K, Greaser ML. Titin diversity—alternative splicing gone wild. J Biomed Biotechnol. 2010;2010:753675. doi: 10.1155/2010/753675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, Dauksaite V, Vakeel P, Klaassen S, Gerull B, Thierfelder L, Regitz-Zagrosek V, Hacker TA, Saupe KW, Dec GW, Ellinor PT, MacRae CA, Spallek B, Fischer R, Perrot A, Özcelik C, Saar K, Hubner N, Gotthardt M. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18:766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Pleitner JM, Saupe KW, Greaser ML. Pathophysiological defects and transcriptional profiling in the RBM20−/− rat model. PLoS One. 2013;8:e84281. doi: 10.1371/journal.pone.0084281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Cruz G, Van Heerden AH, Wang K. Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J Biol Chem. 2001;276:7442–7449. doi: 10.1074/jbc.M008851200. [DOI] [PubMed] [Google Scholar]
- Hamdani N, Bishu KG, von Frieling-Salewsky M, Redfield MM, Linke WA. Deranged myofilament phosphorylation and function in experimental heart failure with preserved ejection fraction. Cardiovasc Res. 2013;97:464–471. doi: 10.1093/cvr/cvs353. [DOI] [PubMed] [Google Scholar]
- Henry LB. Left ventricular systolic dysfunction and ischemic cardiomyopathy. Crit Care Nurs Q. 2003;26:16–21. doi: 10.1097/00002727-200301000-00003. [DOI] [PubMed] [Google Scholar]
- Herman DS, Lam L, Taylor MR, Wang L, Teekakirikul P, Christodoulou D, Conner L, DePalma SR, McDonough B, Sparks E, Teodorescu DL, Cirino AL, Banner NR, Pennell DJ, Graw S, Merlo M, Di Lenarda A, Sinagra G, Bos JM, Ackerman MJ, Mitchell RN, Murry CE, Lakdawala NK, Ho CY, Barton PJ, Cook SA, Mestroni L, Seidman JG, Seidman CE. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo C, Granzier H. Tuning the molecular giant titin through phosphorylation: role in health and disease. Trends Cardiovasc Med. 2013;23:165–171. doi: 10.1016/j.tcm.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinze F, Dieterich C, Radke MH, Granzier H, Gotthardt M. Reducing RBM20 activity improves diastolic dysfunction and cardiac atrophy. J Mol Med (Berl) 2016;94:1349–1358. doi: 10.1007/s00109-016-1483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojayev B, Rothermel BA, Gillette TG, Hill JA. FHL2 binds calcineurin and represses pathological cardiac growth. Mol Cell Biol. 2012;32:4025–4034. doi: 10.1128/MCB.05948-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima M. Mechanical stress–strain sensors embedded in cardiac cytoskeleton: Z disk, titin, and associated structures. Am J Physiol Heart Circ Physiol. 2006;290:H1313–H1325. doi: 10.1152/ajpheart.00816.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson B, Hidalgo C, Saripalli C, Granzier H. Hyperphosphorylation of mouse cardiac titin contributes to transverse aortic constriction-induced diastolic dysfunction. Circ Res. 2011;109(8):858–866. doi: 10.1161/CIRCRESAHA.111.246819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim EC, Schaal TD, Hertel KJ, Reed R, Maniatis T. Serine/arginine-rich protein-dependent suppression of exon skipping by exonic splicing enhancers. Proc Natl Acad Sci U S A. 2005;102:5002–5007. doi: 10.1073/pnas.0500543102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Iijima M, Yoshimoto N, Niimi T, Kuroda S, Maturana AD. RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Lett. 2016;590:2262–2274. doi: 10.1002/1873-3468.12251. [DOI] [PubMed] [Google Scholar]
- Kanopka A, Mühlemann O, Akusjärvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- Kellermayer MS, Smith SB, Granzier HL, Bustamante C. Folding–unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- Kolmerer B, Olivieri N, Witt CC, Herrmann BG, Labeit S. Genomic organization of M line titin and its tissue-specific expression in two distinct isoforms. J Mol Biol. 1996;256:556–563. doi: 10.1006/jmbi.1996.0108. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Ackermann MA, Bowman AL, Yap SV, Bloch RJ. Muscle giants: molecular scaffolds in sarcomerogenesis. Physiol Rev. 2009;89:1217–1267. doi: 10.1152/physrev.00017.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötter S, Andresen C, Krüger M. Titin: central player of hypertrophic signaling and sarcomeric protein quality control. Biol Chem. 2014;395:1341–1352. doi: 10.1515/hsz-2014-0178. [DOI] [PubMed] [Google Scholar]
- Krüger M, Linke WA. The giant protein titin: a regulatory node that integrates myocyte signaling pathways. J Biol Chem. 2011;286:9905–9912. doi: 10.1074/jbc.R110.173260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger M, Sachse C, Zimmermann WH, Eschenhagen T, Klede S, Linke WA. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/AKT pathway. Circ Res. 2008;102:439–447. doi: 10.1161/CIRCRESAHA.107.162719. [DOI] [PubMed] [Google Scholar]
- Krüger M, Babicz K, von Frieling-Salewsky M, Linke WA. Insulin signaling regulates cardiac titin properties in heart development and diabetic cardiomyopathy. J Mol Cell Cardiol. 2010;48:910–916. doi: 10.1016/j.yjmcc.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 1995;270:293–296. doi: 10.1126/science.270.5234.293. [DOI] [PubMed] [Google Scholar]
- Labeit S, Barlow DP, Gautel M, Gibson T, Holt J, Hsieh CL, Francke U, Leonard K, Wardale J, Whiting A, Trinick J. A regular pattern of two types of 100-residue motif in the sequence of titin. Nature. 1990;345:273–276. doi: 10.1038/345273a0. [DOI] [PubMed] [Google Scholar]
- Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11:1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circ Res. 2004;94(4):505–513. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- Lange S, Auerbach D, McLoughlin P, Perriard E, Schäfer BW, Perriard JC, Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- Lange S, Xiang F, Yakovenko A, Vihola A, Hackman P, Rostkova E, Kristensen J, Brandmeier B, Franzen G, Hedberg B, Gunnarsson LG, Hughes SM, Marchand S, Sejersen T, Richard I, Edström L, Ehler E, Udd B, Gautel M. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308:1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- Lange S, Ehler E, Gautel M. From A to Z and back? Multicompartment proteins in the sarcomere. Trends Cell Biol. 2006;16:11–18. doi: 10.1016/j.tcb.2005.11.007. [DOI] [PubMed] [Google Scholar]
- LeWinter MM. Functional consequences of sarcomeric protein abnormalities in failing myocardium. Heart Fail Rev. 2005;10:249–257. doi: 10.1007/s10741-005-5254-4. [DOI] [PubMed] [Google Scholar]
- LeWinter MM, Granzier H. Cardiac titin: a multifunctional giant. Circulation. 2010;121:2137–2145. doi: 10.1161/CIRCULATIONAHA.109.860171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeWinter MM, Granzier HL. Cardiac titin and heart disease. J Cardiovasc Pharmacol. 2014;63:207–212. doi: 10.1097/FJC.0000000000000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Guo W, Schmitt BM, Greaser ML. Comprehensive analysis of titin protein isoform and alternative splicing in normal and mutant rats. J Cell Biochem. 2012;113:1265–1273. doi: 10.1002/jcb.23459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Guo W, Dewey CN, Greaser ML. Rbm20 regulates titin alternative splicing as a splicing repressor. Nucleic Acids Res. 2013;41:2659–2672. doi: 10.1093/nar/gks1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A. 2011;108:11093–11098. doi: 10.1073/pnas.1101135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke WA. Sense and stretchability: the role of titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. Cardiovasc Res. 2008;77:637–648. doi: 10.1016/j.cardiores.2007.03.029. [DOI] [PubMed] [Google Scholar]
- Linke WA. Titin and titin-associated proteins in myocardial stress-sensing and mechanical dysfunction. In: Kamkin A, Kiseleva I, editors. Mechanosensitivity of the heart. Dordrecht: Springer; 2009. pp. 3–34. [Google Scholar]
- Linke WA, Fernandez JM. Cardiac titin: molecular basis of elasticity and cellular contribution to elastic and viscous stiffness components in myocardium. J Muscle Res Cell Motil. 2002;23:483–497. doi: 10.1023/A:1023462507254. [DOI] [PubMed] [Google Scholar]
- Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circ Res. 2014;114:1052–1068. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- Linke WA, Krüger M. The giant protein titin as an integrator of myocyte signaling pathways. Physiology (Bethesda) 2010;25:186–198. doi: 10.1152/physiol.00005.2010. [DOI] [PubMed] [Google Scholar]
- Linke WA, Ivemeyer M, Olivieri N, Kolmerer B, Rüegg JC, Labeit S. Towards a molecular understanding of the elasticity of titin. J Mol Biol. 1996;261:62–71. doi: 10.1006/jmbi.1996.0441. [DOI] [PubMed] [Google Scholar]
- Linke WA, Rudy DE, Centner T, Gautel M, Witt C, Labeit S, Gregorio CC. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J Cell Biol. 1999;146:631–644. doi: 10.1083/jcb.146.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KW. Regulation of alternative splicing by signal transduction pathways. Adv Exp Med Biol. 2007;623:161–174. doi: 10.1007/978-0-387-77374-2_10. [DOI] [PubMed] [Google Scholar]
- Maatz H, Jens M, Liss M, Schafer S, Heinig M, Kirchner M, Adami E, Rintisch C, Dauksaite V, Radke MH, Selbach M, Barton PJ, Cook SA, Rajewsky N, Gotthardt M, Landthaler M, Hubner N. RNA-binding protein RBM20 represses splicing to orchestrate cardiac pre-mRNA processing. J Clin Invest. 2014;124:3419–3430. doi: 10.1172/JCI74523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenko I, Opitz CA, Leake MC, Neagoe C, Kulke M, Gwathmey JK, del Monte F, Hajjar RJ, Linke WA. Passive stiffness changes caused by upregulation of compliant titin isoforms in human dilated cardiomyopathy hearts. Circ Res. 2004;95:708–716. doi: 10.1161/01.RES.0000143901.37063.2f. [DOI] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Maruyama K. Connectin, an elastic protein from myofibrils. J Biochem. 1976;80:405–407. doi: 10.1093/oxfordjournals.jbchem.a131291. [DOI] [PubMed] [Google Scholar]
- Methawasin M, Hutchinson KR, Lee EJ, Smith JE, 3rd, Saripalli C, Hidalgo CG, Ottenheijm CA, Granzier H. Experimentally increasing titin compliance in a novel mouse model attenuates the Frank–Starling mechanism but has a beneficial effect on diastole. Circulation. 2014;129(19):1924–1936. doi: 10.1161/CIRCULATIONAHA.113.005610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methawasin M, Strom JG, Slater RE, Fernandez V, Saripalli C, Granzier H. Experimentally increasing the compliance of titin through RNA binding motif-20 (RBM20) inhibition improves diastolic function in a mouse model of heart failure with preserved ejection fraction. Circulation. 2016;134:1085–1099. doi: 10.1161/CIRCULATIONAHA.116.023003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer LC, Wright NT. Structure of giant muscle proteins. Front Physiol. 2013;4:368. doi: 10.3389/fphys.2013.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata S, Minobe W, Bristow MR, Leinwand LA. Myosin heavy chain isoform expression in the failing and nonfailing human heart. Circ Res. 2000;86:386–390. doi: 10.1161/01.RES.86.4.386. [DOI] [PubMed] [Google Scholar]
- Nagueh SF, Shah G, Wu Y, Torre-Amione G, King NM, Lahmers S, Witt CC, Becker K, Labeit S, Granzier HL. Altered titin expression, myocardial stiffness, and left ventricular function in patients with dilated cardiomyopathy. Circulation. 2004;110:155–162. doi: 10.1161/01.CIR.0000135591.37759.AF. [DOI] [PubMed] [Google Scholar]
- Neagoe C, Kulke M, del Monte F, Gwathmey JK, de Tombe PP, Hajjar RJ, Linke WA. Titin isoform switch in ischemic human heart disease. Circulation. 2002;106:1333–1341. doi: 10.1161/01.CIR.0000029803.93022.93. [DOI] [PubMed] [Google Scholar]
- Neagoe C, Opitz CA, Makarenko I, Linke WA. Gigantic variety: expression patterns of titin isoforms in striated muscles and consequences for myofibrillar passive stiffness. J Muscle Res Cell Motil. 2003;24:175–189. doi: 10.1023/A:1026053530766. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Linke WA. Plasticity of cardiac titin/connectin in heart development. J Muscle Res Cell Motil. 2005;26:333–342. doi: 10.1007/s10974-005-9040-7. [DOI] [PubMed] [Google Scholar]
- Opitz CA, Leake MC, Makarenko I, Benes V, Linke WA. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circ Res. 2004;94:967–975. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- Ottenheijm CA, Knottnerus AM, Buck D, Luo X, Greer K, Hoying A, Labeit S, Granzier H. Tuning passive mechanics through differential splicing of titin during skeletal muscle development. Biophys J. 2009;97:2277–2286. doi: 10.1016/j.bpj.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. Ion channel diseases as a part in the definition and classification of cardiomyopathies recently confirmed in Brugada syndrome. Int J Cardiol. 2016;207:103. doi: 10.1016/j.ijcard.2016.01.136. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Alexandrovich A, Kho AL, Hensen U, Schäfer LV, Brandmeier B, Gräter F, Grubmüller H, Gaub HE, Gautel M. Mechanoenzymatics of titin kinase. Proc Natl Acad Sci U S A. 2008;105:13385–13390. doi: 10.1073/pnas.0805034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl FA, McLoughlin P, Ehler E, de Giovanni C, Schäfer BW. DRAL is a p53-responsive gene whose four and a half LIM domain protein product induces apoptosis. J Cell Biol. 2000;151:495–506. doi: 10.1083/jcb.151.3.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro BP, Lam CS, Patel JB, Mohammed SF, Kruger M, Meyer DM, Linke WA, Redfield MM. Acute and chronic ventricular-arterial coupling in systole and diastole: insights from an elderly hypertensive model. Hypertension. 2007;50:503–511. doi: 10.1161/HYPERTENSIONAHA.107.090092. [DOI] [PubMed] [Google Scholar]
- Sheikh F, Raskin A, Chu PH, Lange S, Domenighetti AA, Zheng M, Liang X, Zhang T, Yajima T, Gu Y, Dalton ND, Mahata SK, Dorn GW, 2nd, Brown JH, Peterson KL, Omens JH, McCulloch AD, Chen J. An FHL1-containing complex within the cardiomyocyte sarcomere mediates hypertrophic biomechanical stress responses in mice. J Clin Invest. 2008;118:3870–3880. doi: 10.1172/JCI34472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Mattox W. Activation and repression functions of an SR splicing regulator depend on exonic versus intronic-binding position. Nucleic Acids Res. 2012;40(1):428–437. doi: 10.1093/nar/gkr713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio CC, Labeit D, Linke WA, Suzuki K, Labeit S. Tissue-specific expression and alpha-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J Mol Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Taylor M, Graw S, Sinagra G, Barnes C, Slavov D, Brun F, Pinamonti B, Salcedo EE, Sauer W, Pyxaras S, Anderson B, Simon B, Bogomolovas J, Labeit S, Granzier H, Mestroni L. Genetic variation in titin in arrhythmogenic right ventricular cardiomyopathy—overlap syndromes. Circulation. 2011;124:876–885. doi: 10.1161/CIRCULATIONAHA.110.005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick J. Titin as a scaffold and spring. Cytoskeleton. Curr Biol. 1996;6:258–260. doi: 10.1016/S0960-9822(02)00472-4. [DOI] [PubMed] [Google Scholar]
- Trombitás K, Greaser M, Labeit S, Jin JP, Kellermayer M, Helmes M, Granzier H. Titin extensibility in situ: entropic elasticity of permanently folded and permanently unfolded molecular segments. J Cell Biol. 1998;140:853–859. doi: 10.1083/jcb.140.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombitás K, Redkar A, Centner T, Wu Y, Labeit S, Granzier H. Extensibility of isoforms of cardiac titin: variation in contour length of molecular subsegments provides a basis for cellular passive stiffness diversity. Biophys J. 2000;79:3226–3234. doi: 10.1016/S0006-3495(00)76555-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat Rev Mol Cell Biol. 2003;4:679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L, Trinick J. Properties of titin immunoglobulin and fibronectin-3 domains. J Biol Chem. 2004;279:46351–46354. doi: 10.1074/jbc.R400023200. [DOI] [PubMed] [Google Scholar]
- Wang K, McClure J, Tu A. Titin: major myofibrillar components of striated muscle. Proc Natl Acad Sci U S A. 1979;76:3698–3702. doi: 10.1073/pnas.76.8.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, McCarter R, Wright J, Beverly J, Ramirez-Mitchell R. Regulation of skeletal muscle stiffness and elasticity by titin isoforms: a test of the segmental extension model of resting tension. Proc Natl Acad Sci U S A. 1991;88:7101–7105. doi: 10.1073/pnas.88.16.7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao X, Zhang J, Choudhury R, Robertson A, Li K, Ma M, Burge CB, Wang Z. A complex network of factors with overlapping affinities represses splicing through intronic elements. Nat Struct Mol Biol. 2013;20:36–45. doi: 10.1038/nsmb.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CM, Jordan MC, Roos KP, Krzesinski PR, Greaser ML. Titin isoform expression in normal and hypertensive myocardium. Cardiovasc Res. 2003;59:86–94. doi: 10.1016/S0008-6363(03)00328-6. [DOI] [PubMed] [Google Scholar]
- Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003;24:1695–1702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- Warren CM, Krzesinski PR, Campbell KS, Moss RL, Greaser ML. Titin isoform changes in rat myocardium during development. Mech Dev. 2004;121:1301–1312. doi: 10.1016/j.mod.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Nair P, Labeit D, Kellermayer MS, Greaser M, Labeit S, Granzier H. Molecular mechanics of cardiac titin’s PEVK and N2B spring elements. J Biol Chem. 2002;277:11549–11558. doi: 10.1074/jbc.M200356200. [DOI] [PubMed] [Google Scholar]
- Wu Y, Bell SP, Trombitas K, Witt CC, Labeit S, LeWinter MM, Granzier H. Changes in titin isoform expression in pacing-induced cardiac failure give rise to increased passive muscle stiffness. Circulation. 2002;106:1384–1389. doi: 10.1161/01.CIR.0000029804.61510.02. [DOI] [PubMed] [Google Scholar]
- Yin Z, Ren J, Guo W. Sarcomeric protein isoform transitions in cardiac muscle: a journey to heart failure. Biochim Biophys Acta. 2015;1852:47–52. doi: 10.1016/j.bbadis.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Yin Z, Ren J, McCormick RJ, Ford SP, Guo W. RBM20 is an essential factor for thyroid hormone-regulated titin isoform transition. J Mol Cell Biol. 2015;7:88–90. doi: 10.1093/jmcb/mjv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Chen Z, Guo W (2016) Pre-mRNA mis-splicing of sarcomeric genes in heart failure. Biochim Biophys Acta (in press). pii: S0925-4439(16)30290-3. doi:10.1016/j.bbadis.2016.11.008 [DOI] [PMC free article] [PubMed]