Abstract
Gene silencing via RNA interference (RNAi) is rapidly evolving as a personalized approach to cancer treatment. The effector molecules—small interfering RNAs (siRNAs) and microRNAs (miRNAs)—can be used to silence or “switch off” specific cancer genes. Currently, the main barrier to implementing siRNA- and miRNA-based therapies in clinical practice is the lack of an effective delivery system that can protect the RNA molecules from nuclease degradation, deliver to them to tumor tissue, and release them into the cytoplasm of the target cancer cells, all without inducing adverse effects. Here, we review the fundamentals of RNAi, cell membrane transport pathways, and factors that affect intracellular delivery. We discuss the advantages and disadvantages of the various types of nanoparticle delivery systems, with a focus on those that have been investigated in breast cancer in vivo.
Keywords: MicroRNA, siRNA, Cellular transport, Delivery, Nanoparticles, Breast cancer
Introduction
Breast cancer represents the leading cause of cancer-related deaths among women worldwide. Globally, breast cancer accounted for the highest number of new cancer cases in 2015 (Jemal et al. 2011; Global Burden of Disease Cancer Collaboration et al. 2017). It has been postulated that nearly 30% of newly diagnosed patients with early stage breast cancer will develop a distant metastasis despite receiving therapy (Redig and McAllister 2013; Morry et al. 2017). In routine clinical practice, breast cancer is traditionally classified as either non-invasive or invasive, and then according to stage and grade. The classification system is based on the histological features of breast tissue, location of abnormal tissues (e.g., milk ducts, lobules), and clinical symptoms presented by the patient (Goljan 2011; Eliyatkın et al. 2015). Non-invasive breast cancers remain localized and do not invade surrounding tissues. The two main types of non-invasive breast cancer are ductal carcinoma in situ and lobular carcinoma in situ. Most breast cancers are invasive and include infiltrating ductal carcinoma, Paget’s disease, medullary carcinoma, inflammatory carcinoma, invasive lobular carcinoma, tubular carcinoma, and colloid (mucinous) carcinoma (Goljan 2011). Breast cancers can become metastatic if the cancer cells spread to other parts of the body through the bloodstream and lymph nodes.
Breast cancers are further classified into four molecular subtypes (luminal A, luminal B, HER2-enriched, and basal-like) based on the level of expression of the estrogen receptor (ER), progesterone receptor (PR), and the human epidermal growth factor receptor 2 (HER2) (Mastoraki et al. 2014; Dai et al. 2016). Luminal A cancers overexpress the hormonal receptors only (ER+ and/or PR+ and HER2−), whereas the luminal B type overexpress all three receptors (ER+ and/or PR+ and HER2+). Luminal A and luminal B cancers are defined by the expression of genes in the luminal epithelial layer of the mammary gland. Approximately 70% of breast cancers are hormone receptor-positive and overexpress one or both ER and PR (Bae et al. 2015). Breast cancers that overexpress HER2 only (i.e., HER2+/ER−/PR−) are referred to as HER2-enriched or simply as HER2, and represent around 20% of all cases (Kittaneh et al. 2013). Basal-like breast cancers are based on a distinct gene signature in the basal cells that line the breast ducts. They are mostly triple-negative breast cancers (TNBCs), which do not express any of the receptors (i.e., ER−, PR−, and HER−). TNBCs account for about 10% of all breast cancers and are associated with high mortality (Badve et al. 2011; Lü et al. 2017; Peng et al. 2017). Approximately 50–80% of TNBCs are basal-like (Boyle 2012; Li et al. 2017). Further subclassification of breast cancer using more detailed molecular signatures has shown promise, but is yet to reach routine clinical practice (Eliyatkın et al. 2015).
Current therapy options for breast cancer include surgery, hormonal therapy, immunotherapy, chemotherapy, radiation therapy, or a combination of these (Parvani and Jackson 2017). Breast cancers that express any hormone receptors have targeted hormonal therapies available and have a more favorable prognosis than breast cancers that do not show receptor expression (Cho 2016). Hormonal therapies are often prescribed following surgery as an adjuvant treatment. Some block the interaction between receptors and hormones (such as tamoxifen), while others lower the level of hormones (such as aromatase inhibitors). Tamoxifen has been used for more than 30 years to treat hormone receptor-positive breast cancers (ER+ and/or PR+). It works by stopping cells from responding to estrogen (Chang 2012). Similarly, Herceptin (trastuzumab), a humanized anti-HER2 monoclonal antibody, works by binding to HER2 receptors on the surface of breast cancer cells and blocking growth factor signals (Maximiano et al. 2016). Tamoxifen and Herceptin can be administered alone or in conjunction with radiation therapy or chemotherapy drugs such as paclitaxel or doxorubicin (Ranftler and Strasser-Weippl 2017). There are currently no effective targeted treatments available for TNBC patients. The high genetic diversity and absence of ER, PR, and HER2 receptors makes it unresponsive to hormonal therapies. It is also frequently resistant to chemotherapy; the platinum drugs epirubicin and paclitaxel are currently prescribed for TNBC patients, but response rates are poor (Liu et al. 2014; Jing et al. 2016; Parvani and Jackson 2017; Peng et al. 2017).
The heterogeneity of breast cancer remains a key barrier to its accurate molecular classification and individualization of treatment (Eccles et al. 2013). Not all patients who overexpress the hormone receptors (ER+ and/or PR+) respond favorably to tamoxifen. Similarly, not all patients who overexpress HER2 respond to Herceptin alone (Liu et al. 2014; Jiang et al. 2015). The cut-offs to define luminal A and luminal B cancers are currently set in an arbitrary manner rather than from gene expression levels because of substantial ER+ tumor heterogeneity, which can add to the problems in selecting treatment (Eccles et al. 2013; Yersal and Barutca 2014). Several authors suggest that the Ki-67 index can help to differentiate luminal A from luminal B (Dowsett et al. 2011; Eroles et al. 2012; Guiu et al. 2012; Yersal and Barutca 2014; Cho 2016; Dai et al. 2016; Hennigs et al. 2016; Hon et al. 2016; Yip et al. 2016). Ki-67 is a protein that serves as a cellular marker for proliferation and has been used to suggest a fifth molecular subtype, “normal-like” breast cancer, which is similar to luminal A but with the additional characteristic of having low Ki-67, i.e., ER+/PR+/HER2−/Ki-67−. Luminal B can be further subdivided into two groups: ER+/PR+/HER2−/Ki-67+ and ER+/PR+/HER2+/Ki-67+ (Dai et al. 2016). Although the Ki-67 index has gained wide support in the literature as a prognostic and predictive biomarker, it is not yet established for use in the clinical management of breast cancer due to the lack of standardized procedures (Dowsett et al. 2011; Penault-Llorca and Radosevic-Robin 2017).
Several studies have attempted to further subclassify breast cancer based on gene mutations and/or gene expression signatures; however, the prognostic value is not yet clear. Known breast cancer susceptibility genes that make up 25–30% of the heritability include BRCA1, BRCA2, CHEK2, ATM, PALB2, BRIP1, TP53, PTEN, CDH1, and STK11 (Eccles et al. 2013). BRCA1 and BRCA2 are the most commonly mutated genes found in breast cancer, including in TNBC (Liang and Lam 2012; Gasparri et al. 2017). PLK1 expression has been reported as a potential genetic marker for TNBC (Maire et al. 2013; Morry et al. 2017), and BIRC5, MYBL2, IGFBP6, TP53, GAPDH, CCND1, HRAS, and PCNA were reported as the most significantly up- or downregulated differentially expressed genes in TNBC compared with normal tissue (Peng et al. 2017). Other studies have reported elevated expression of the AR and EGFR genes in TNBCs (Martin et al. 2012; Pietri et al. 2016).
A better understanding of the genetic mutations and gene expression patterns in breast cancer has the potential to help identify prognostic and predictive biomarkers. In addition, a deeper molecular understanding will also provide new therapeutic targets and candidates for gene silencing. In order to appreciate the potential therapeutic application of gene silencing, it is important to first understand the fundamentals of how this process regulates cell biology.
Gene silencing: history and discovery of microRNA and siRNA
Until recently, the role of non-coding RNAs in our DNA was unknown. The ability of RNA to inhibit gene expression was first observed in plants in the 1990s by a number of independent groups; however, the phenomenon was not fully understood and, so, it was not explained or reported. Then, in 1993, Ambros and coworkers discovered the first microRNA (miRNA) gene, lin-4, in the nematode worm Caenorhabditis elegans (Lee et al. 1993). They found that lin-4 physically binds to lin-14 RNA and prevents its translation. Their discovery was the first evidence that an miRNA can suppress protein production by inhibiting messenger RNAs (mRNAs). In normal cells, miRNAs play an important role in regulating functions such as proliferation and cell differentiation. In 2002, Calin et al. (2002) found that miRNAs are involved in cancer by showing that the miR-15a/16-1 locus is deleted or downregulated in 68% of chronic lymphocytic leukemias (Calin et al. 2002). Further research into miRNAs found that they comprise of a broad class of small RNA regulators with important functions in health and disease. Aberrant miRNA expression is frequently observed in various types of cancer, involving global upregulation and downregulation of miRNA expression (Sioud 2015; Mendelsohn et al. 2015).
In 1998, five years after the discovery of the first miRNA, Andrew Fire and Craig Mello discovered the process of RNA interference (RNAi) during their research on gene expressions in the nematode worm C. elegans. They found that, when worms were injected with double-stranded RNA coding for specific proteins, genes carrying the same sequence were switched off or silenced (Fire et al. 1998). Their findings resulted in the advent of RNAi as a central tool in modern molecular biology and were recognized by the Nobel Prize in 2006. Soon after the discovery of RNAi, David Baulcombe and Andrew Hamilton discovered naturally occurring small interfering RNAs (siRNAs) in plants (Hamilton and Baulcombe 1999), described as novel small antisense RNAs involved in gene silencing following transcription. Two years later, Thomas Tuschl and colleagues produced the first synthetic siRNAs able to silence genes in mammalian cells (Elbashir et al. 2001; Mattick 2001). This achievement heralded the widespread use of siRNAs to selectively knock down the activity of a specific gene. Currently, miRNAs and siRNAs are the two main classes of RNAs most widely employed for gene silencing. They have similar physicochemical properties but distinct functions and mechanisms of action, making their design requirements and therapeutic applications different.
Process of gene silencing by siRNAs
The first step to employing siRNAs in therapeutic applications is designing an siRNA sequence that is specific to the target mRNA, for which multiple algorithms are available. Once designed, siRNAs are produced by chemical synthesis or through gene expression. The former are completely synthetic RNAs that can be introduced into cells by various means, as will be discussed in the later parts of this review. The latter are transcribed inside the cell from expression constructs (such as plasmids and viral vectors) that express the short-hairpin RNA (shRNA) precursors of siRNAs (Siolas et al. 2005); these lie outside the scope of this review. Chemical synthesis of siRNA allows control over the amount and purity of siRNA and also enables chemical modifications to improve stability, an important aspect required for delivery. Chemically synthesized siRNAs can also be labeled for analyzing siRNA uptake or localization by fluorescence microscopy.
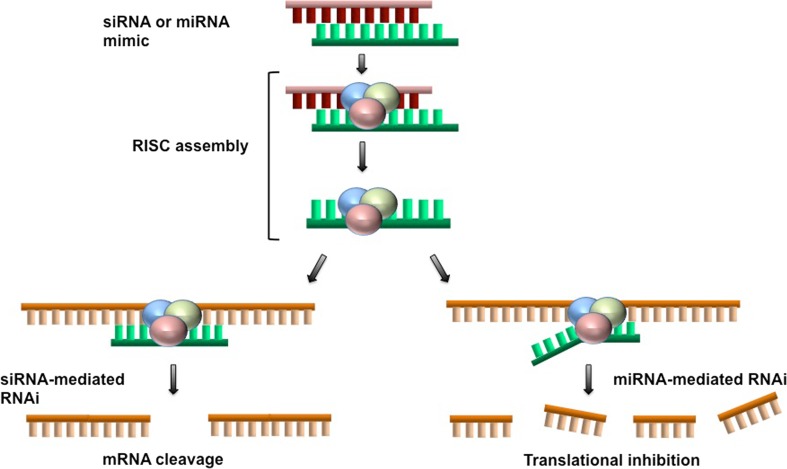
After the siRNA is introduced into the cell, the process of gene silencing is initiated and carried out by the endogenous RNAi machinery of the cell. Inside the cell, the duplex siRNA enters the RNAi pathway. The antisense strand is loaded into a protein complex called the RNA-induced silencing complex (RISC), and serves as the guide for the recognition of complementary mRNAs. After the target sequence is recognized, the mRNA is cleaved by Argonaute 2 of the RISC, resulting in reduced protein expression from the silenced gene (Finlay et al. 2015b; Kim et al. 2016; Parvani and Jackson 2017). Advantages of siRNAs over drug therapies include their high degree of specificity and low toxicity. Nevertheless, off-target effects do occur due to the miRNA-like activity of siRNAs acting through the seed-like sequence at the 5′ end and dsRNA-induced stimulation of the innate immune system, both reviewed recently (Suter et al. 2016; Meng and Lu 2017). A schematic of the siRNA gene silencing process is provided in Fig. 1.
Fig. 1.
Gene silencing using siRNA or miRNA mimics. Once the siRNA or miRNA mimic has been introduced into the cytoplasm of the cell, it is unwound and the active antisense strand (green) is incorporated into the RISC. This leads to gene silencing via two distinct mechanisms, depending on the extent of base pairing between the antisense strand and the target mRNA. With siRNA on the left, the complete homology between the antisense and target mRNA (yellow) leads to site-specific cleavage and degradation of the mRNA. In contrast, the partial sequence identity between the active miRNA strand and its mRNA target leads to inhibition of translation, decapping, and subsequent mRNA degradation
Process of gene silencing by miRNA
There are two ways of employing miRNAs for therapeutic applications: miRNA inhibition and miRNA replacement. miRNA inhibition is used when the target miRNA is overexpressed. It involves introducing synthetic single-stranded RNAs acting as miRNA antagonists that inhibit the action of the target miRNA. In this respect, miRNA inhibition is analogous to antisense inhibition with similar design and delivery considerations, and, as such, is outside the scope of this review. On the other hand, miRNA replacement is employed when the target miRNA is repressed or deactivated, and this tends to be more common in cancer. It involves introducing synthetic double-stranded miRNAs (called miRNA mimics) to mimic the function of the target miRNAs and bind to a target gene to initiate mRNA degradation and produce the gene silencing effect (Rothschild 2014; Wang et al. 2016a). The design of miRNA mimics is more straightforward than that of siRNA, as the sequence should be almost, if not entirely, identical to the endogenous miRNA. Synthetic miRNA mimics and siRNAs enter the same RNAi pathway and both are incorporated into RISC to create the final active complex.
Difference between siRNA and miRNA
While structurally and functionally similar, there are some important differences between siRNAs and miRNAs (Table 1). siRNAs are short double-stranded RNAs typically composed of 21–23 base pairs, often with two nucleotide overhangs at the 3′ ends, but multiple variations in length and overhangs are tolerated (Kim et al. 2016). They consist of an active (guide) strand and a complementary inactive (passenger) strand. siRNAs are regarded as exogenous RNAs that enter the endogenous RNAi pathway. miRNAs, on the other hand, are endogenous RNAs produced from within the cells, expressed as long primary miRNA transcripts (pri-miRs) from miRNA genes (Sioud 2015). After partial cleavage by the microprocessor complex in the nucleus, the stem-loop pre-miRNA is exported to the cytoplasm, where it is further processed by Dicer into a double-stranded RNA consisting of the active, or mature, strand and the inactive passenger strand. The mature miRNA is incorporated into the RISC to initiate gene silencing. In contrast to the perfect complementarity between an siRNA and its target mRNA, an miRNA has imperfect complementarity, mostly within the seed sequence at its 5′ end (Carthew and Sontheimer 2009).
Table 1.
Differences between siRNA and miRNA
| siRNA | miRNA | |
|---|---|---|
| Occurrence | Occurs naturally in plants and animals. It is currently unknown whether or not they occur naturally in mammals. | Occurs naturally in plants and animals. |
| Mean length | Approx. 21–22 nt | Approx. 19–25 nt |
| Complementarity to target mRNA | 100% perfect match; therefore, siRNAs knock down specific genes, with minor off-target exceptions. | Not exact; therefore, a single miRNA may target up to hundreds of mRNAs. |
| Biogenesis | Regulate the same genes that express them. | Expressed by genes whose purpose is to make miRNAs, but they regulate genes (mRNAs) other than the ones that expressed them. |
| Action | Cleave mRNA. | Inhibit or replace translation of mRNA. |
| Function | Act as gene silencing guardians in plants and animals that do not have antibody- or cell-mediated immunity. | Regulators (inhibitors) of genes (mRNAs) |
In terms of silencing and potential clinical use, a key difference between siRNAs and miRNAs is that an siRNA is specific for a single target site in a single mRNA, and, therefore, inhibits the expression of one target gene, whereas an miRNA can have multiple targets and can regulate multiple genes because mRNA recognition requires binding to the much shorter seed sequence rather than the entire 21-nucleotide sequence of an siRNA. To initiate RNAi, an siRNA must be fully complimentary to its target mRNA, whereas miRNA can be partially complimentary and bind to multiple mRNAs to inhibit their expression, and their mechanisms of action are different: siRNAs cleave mRNA, whereas miRNAs inhibit translation of mRNA (Carthew and Sontheimer 2009). To avoid repression of genes in normal cells, it is important to develop methods that will enable cell specificity, i.e., methods to deliver miRNA and siRNA to tumor cells at the target site (Lam et al. 2015; Takahashi et al. 2015).
In summary, in order to bring RNAi into clinical practice for cancer treatment, three key steps must be taken: (1) the target genes that are directly involved in cancer development must be identified; (2) an siRNA specific to the target gene must be designed and synthesized; (3) the synthetic siRNA must be delivered into the cytoplasm of the target cell. While methods for steps 1 and 2 are already effective, there are several challenges to the delivery of siRNA into the target cells. This last step is the major barrier to implementing siRNA therapy in clinical practice and is the subject of this review.
Current challenges of siRNA and microRNA therapies
The challenges associated with cellular delivery of both siRNA and miRNA are widely reported (Bouclier et al. 2008; Dahlman et al. 2014; Essex et al. 2015; Wang et al. 2016a; Arnold et al. 2017; Tatiparti et al. 2017; Parvani and Jackson 2017). Despite differences in their mechanism of action, siRNAs and miRNA mimics have similar chemical structure and face similar challenges in delivery to target cells. Therefore, the same delivery system can be employed to improve their cellular uptake (Lam et al. 2015). To date, most research on siRNAs and miRNAs as cancer therapies has focused on systemic delivery by intravenous injection. Other routes described include local delivery by intraocular and intratumoral injection, local delivery to the central nervous system (CNS), or intranasal delivery to the airway (Gao and Huang 2009).
When injected systemically, naked siRNAs and miRNAs can be easily degraded by serum nucleases, removed by cells of the immune system, and excreted through renal filtration. At the cell membrane level, naked siRNA and miRNA are negatively charged at normal pH, as is the cell membrane; therefore, they are repelled. They are also water-soluble, which makes it difficult for them to enter cells by passive diffusion (Essex et al. 2015). Other challenges include poor tissue penetration, instability, low efficiency of internalization, and non-specific immune stimulation (Tatiparti et al. 2017). The percentage actually taken up by cells is estimated to be 0.7% of the injected dose (Parvani and Jackson 2017). Naked siRNAs and miRNAs are, therefore, either degraded or entrapped before entering the target cell. There is a need for a delivery system that will not only protect the siRNA and miRNA molecules during systemic delivery, but also maximize delivery to the specific target tissues and promote entry into the target cell.
Delivery methods
Principles of transporting oligonucleotides into cells
There are two key barriers a delivery system must overcome to enable siRNA and miRNA molecules to enter the cell and target mRNA: (1) transport across the cell membrane and (2) escape from the endocytic pathway (Liang and Lam 2012). To determine the ideal characteristics of a delivery system, it is important to first understand the key processes taking place during cellular transport. The cell membrane is a selectively permeable phospholipid bilayer. Each phospholipid molecule consists of a hydrophilic (polar) phosphate head on the outside of the membrane and two hydrophobic (non-polar) fatty acid tails inside the membrane. Generally, the smaller and more hydrophobic a molecule is, the more rapidly it will diffuse across the lipid bilayer (Alberts et al. 2002; Prokop 2011).
Transport across the cell membrane primarily occurs through a process called endocytosis. There are two main categories of endocytosis: phagocytosis (cell eating) and pinocytosis (cell drinking), with particle size being one of the key factors that governs the mode of uptake (Prokop 2011). Phagocytosis involves the cell membrane extending outwards and engulfing the particle into a membrane-bound vesicle called a phagosome, which then fuses with lysosomes for degradation. Phagocytosis is performed by specialized cells such as neutrophils, macrophages, monocytes, and endothelial cells, and plays a role in the clearance of particles that have diameters generally greater than 0.5 μm, including larger particles of up to 10 μm (such as bacteria or cellular debris) (Hirota and Terada 2012). An in vitro study in phagocytic cells showed that macrophages can internalize IgG-coated polystyrene spherical particles that range from 200 nm to 2 μm (Koval et al. 1998). For this reason, delivery systems in this size range are considered undesirable for drug and RNA delivery (Hirota and Terada 2012; Bulbake et al. 2017; Tatiparti et al. 2017). On the other hand, pinocytosis occurs in virtually all cells, including cancer cells, and covers particle sizes ranging from 50 nm up to 5 μm. Particles to be imported by pinocytosis contact the exterior surface of the plasma membrane, triggering the membrane to fold inward, enveloping them. The enveloped membrane pinches off into a vesicle containing extracellular fluid and the imported particles. There are three main mechanisms of pinocytosis: receptor-mediated endocytosis, receptor-independent endocytosis, and macropinocytosis. Receptor-mediated endocytosis occurs when a particle interacts with a specific receptor and is selectively internalized. There are two main subtypes: clathrin-mediated endocytosis, which is initiated by the protein clathrin on the cell membrane and typically occurs for particles around 120 nm in size, and caveolin-mediated endocytosis, which is initiated by the cholesterol-binding protein caveolin and typically occurs for particles that are around 80 nm in size (Hirota and Terada 2012). Particles of 50 nm in size can be endocytosed without the involvement of clathrin or caveolin receptors (Hirota and Terada 2012). For particle sizes ranging from 100 nm to 5 μm, entry to the cell is via macropinocytosis, a process that involves non-selective uptake of particles by macropinosomes (Prokop 2011; Hirota and Terada 2012; Finlay et al. 2015b; Fan et al. 2016). In addition to controlling size, delivery systems can be designed to be active or passive. For active transport, the surface of the vehicle can be decorated with targeting moieties such as proteins, peptides, small molecules, or antibodies to trigger receptor-mediated endocytosis. Delivery systems for passive transport require the biophysical properties of the delivery vehicle to be manipulated, including its size, shape, and zeta potential (Dahlman et al. 2014).
Once a delivery vehicle passes the cell membrane via endocytosis, it is contained within an intracellular membrane-bound compartment called an endosome, where it enters the endocytic pathway and is recycled or degraded. The pH within the endosome plays an important role as the cell exposes the ingested substance to hydrolytic enzymes in a progressively acidic environment in order to facilitate degradation. The process occurs in stages as follows: (a) early endosomes are sorting organelles and the first compartments of the pathway located in the periphery of the cell, and receive most types of particles coming from the cell surface. They have a mildly acid pH from neutral to around pH 6. From here, many of the molecules are either recycled to the surface by exocytosis or sorted into late endosomes; (b) late endosomes receive the endocytosed materials from the early endosomes through transport carriers or by Rab conversion (Poteryaev et al. 2010). They are rapidly acidified to a pH of around 5.5 by the action of ATPase proton pumps which span the membrane; (c) lysosomes are the last compartment of the endocytic pathway, and have the primary function of breaking down cellular waste products, fats, carbohydrates, proteins, and other macromolecules into their respective subunits (Liang and Lam 2012; Arnold et al. 2017).
Lysosomes contain degradative hydrolytic enzymes and function in an acidic environment (further pH reduction to approximately 4.5). An inability of the delivery system to escape this compartment will lead to degradation of the particle and its cargo. Ideally, the delivery system should release its contents from the acidic compartment of the endosome and avoid exposure to the degrading enzymes contained within the lysosomes. Thus, escaping the late endosomes and being subjected to selective disassembly in cytoplasm are the key challenges to be addressed by successful siRNA and miRNA delivery systems within the cell.
Design considerations of delivery systems
As indicated above, size, shape, and surface charge are the key design considerations for nanoparticle delivery systems (Tatiparti et al. 2017). Delivery systems smaller than the renal filtration cut-off of 50 kDa (or 5–6 nm) are expected to be rapidly removed by renal filtration, whereas delivery systems larger than 100 nm in diameter can be trapped in the mononuclear phagocyte system (Wang et al. 2016a). According to Bedi et al. (2013), the ideal systemic delivery vehicle should be approximately 10–50 nm in size, whereas Wang et al. (2016a) suggest an optimal size range between 10 nm and 100 nm. Other studies suggest that the size should be within the range of 20–100 nm in diameter, with particles less than 100 nm showing higher tissue uptake and transfection efficiency (Young et al. 2016; Tatiparti et al. 2017). There is consensus among several authors that delivery systems should be less than 200 nm for efficient tissue and cellular uptake (Davis 1997; Govindarajan et al. 2012; Draz et al. 2014; Dong et al. 2015; Ngamcherdtrakul et al. 2015), although exceptions exist (Ahmad et al. 2016; Jing et al. 2016). Hirota and Terada (2012) suggested that the optimal size of particles for cellular uptake varies according to cell type, where, for cancer cells, a particle size of around 50 nm is favored (Hirota and Terada 2012).
Shape can also affect particle behavior and cellular uptake. There have been conflicting reports on the effect of particle shape on cellular uptake. Young et al. (2016) suggested that spherical structures have increased rates of endocytosis compared to rod-shaped structures because rods have a larger contact area that can block neighboring receptor sites on cells. Similarly, Li et al. (2015) suggested that spherical particles have the highest uptake and fastest internalization rate because they need to overcome a minimal membrane bending energy barrier compared with non-spherical systems. In contrast, He and Park (2016) showed that shapes with higher aspect ratio and sharper angular features have a higher chance of adhering to the cells and becoming internalized by cancer cells (He et al. 2016). To explain the variations in the cellular internalization of different shapes, Richards and Endres (2016) studied the effects of various shapes on receptor-mediated endocytosis and phagocytosis using 1-dimensional and 2-dimensional physical models. They showed that the orientation plays a critical role in the internalization rate, where non-spherical shapes can be internalized the fastest when the most highly curved tip is engulfed first (Richards and Endres 2016).
Since positively charged particles are taken up more readily than negatively charged particles, the surface charge is one of the most important properties of a delivery system that influences its uptake. The zeta potential has been used as an indicator of the surface charge (Honary and Zahir 2013). When a material is placed in a liquid, the zeta potential provides a measure of the magnitude of electrostatic repulsive interaction between particles, with a higher zeta potential indicating greater stability. Delivery systems with larger positive zeta potential bind strongly to the cell membrane and show higher cellular uptake, facilitated by the electrostatic interactions between the negatively charged cell membrane and positively charged particle surface (Honary and Zahir 2013; Dong et al. 2015).
Interestingly, negative zeta potentials have also been reported for some delivery systems. Although cell membranes should repel negatively charged particles, it has been reported that cells may adsorb negatively charged particles at the positively charged sites via electrostatic interaction, leading to localized neutralization and subsequent bending of the membrane to initiate endocytosis. Nevertheless, delivery vehicles with positive charge are preferentially taken up by tumors and have higher cellular uptake than negatively charged or neutral particles (Davis 1997; Honary and Zahir 2013; Young et al. 2016). It has been reported that cellular uptake is lowest for particles that have no surface charge as determined by the zeta potential (Hirota and Terada 2012). To improve specificity and cell uptake via receptor-mediated endocytosis, some investigators have suggested attaching cationic polymers such as poloxamers and polyethylene oxide (Draz et al. 2014; Dong et al. 2015; Arnold et al. 2017; Sun et al. 2017b).
Once a delivery system successfully passes the cell membrane and enters the cell, it must escape the endocytic pathway. Nanocarrier delivery systems can be designed to incorporate characteristics that facilitate endosomal–lysosomal escape. A commonly used approach is to make use of the proton sponge effect (Behr 1997), which is based on the hypothesis that pH buffering can facilitate escape from the endocytic pathway by preventing the acidification of the endosomes (Arnold et al. 2017; Dong et al. 2015; Gujrati et al. 2016; Li et al. 2016). When the endosomes struggle to become acidic, it is thought that the cell will continuously pump more protons into the endosomes in an attempt to lower the pH, followed by passive entry of chloride ions and, due to the increase in the ionic concentration, water influx. Eventually, this increase in osmotic pressure causes swelling and rupture of the endosome, releasing its contents into the cytosol (Liang and Lam 2012; Gujrati et al. 2016). The proton sponge effect has been observed with certain cationic polymers that have a high pH buffering capability over a wide pH range, such as polyamidoamine (PAMAM) dendrimers, lipopolyamines, and polyethylenimine (PEI). These polymers usually have secondary or tertiary amine groups with pKa close to the endosomal pH (Liang and Lam 2012). These polymers can be protonated to decrease the acidification of the endosomes. PEI is known for its extensive pH buffering capacity, ability to compact siRNAs into nano-scale complexes, and demonstrated ability to successfully transfect into a range of cells in vitro and in vivo (Dong et al. 2015; Essex et al. 2015). However, the clinical application of PEI has been limited due to reports of its toxicity, which is influenced by its molecular weight. The higher the molecular weight of PEI, the higher its transfection efficiency but also the higher its non-specific toxicity (Essex et al. 2015). Conversely, low molecular weight PEI displays low toxicity and low transfection efficiency (Essex et al. 2015; Li et al. 2016). Essex et al. (2015) found that chemical conjugation of low molecular weight PEI with dioleoylphosphatidylethanolamine (DOPE) improved the intracellular transfection efficiency of siRNA with low cytotoxicity levels.
Examples of polymers that do not have a pH buffering capability include polypeptides such as chitosan and polylysine. To improve the pH buffering capability of these polypeptides, molecules such as histidine can be added as a functional group or incorporated into the peptide sequence, as can pH-sensitive endosome-disruptive peptides such as GALA (Liang and Lam 2012). Bilayer disruption is another escape mechanism that involves destabilization of the cellular membrane via fusion of cationic polymers, peptides, or cationic lipids such as DOPE (Young et al. 2016).
In summary, multiple challenges must be overcome before siRNAs and miRNAs can be implemented as therapeutic agents (Table 2). An effective delivery system must be able to: (a) protect nucleic acid molecules from degradation during systemic delivery; (b) withstand prolonged circulation without being cleared by renal filtration; (c) accumulate into the target tissue from the circulation; (d) enter the target cells via a specific uptake mechanism; (e) initiate cellular uptake via endocytosis; (e) escape from the endosomal compartment to avoid being degraded in the lysosome; and (f) disassemble in the cytoplasm to release its siRNA or miRNA cargo (Prokop 2011; Bedi et al. 2013; Pittella and Kataoka 2013; Gujrati et al. 2016; Arnold et al. 2017). An ideal delivery system should also be biocompatible, biodegradable, non-toxic, and non-immunogenic (Young et al. 2016).
Table 2.
Barriers to the transport of siRNAs and miRNAs to target cells
| Level | Barriers |
|---|---|
| Circulation | Non-specific interactions with serum proteins, resulting in nanocarrier degradation, dissociation, or aggregation. Clearance by the renal system for particles less than 6–10 nm in size. Nanocarrier toxicity and immune response can be induced. |
| Tissue permeability | Endothelium penetration. Transport from the bloodstream to a desired tissue. |
| Extracellular | Extracellular stability and diffusion. Differences in the pH, enzymes, or ions of tissue microenvironment can damage the nanocarrier, causing dissociation before cellular entry. |
| Internalization | Cell specificity. Cellular uptake via endocytosis. |
| Intracellular | Escape from endosomal compartment. Dissociation from the nanocarrier in the cytoplasm to release siRNA or miRNA cargo. |
siRNA and miRNA delivery vehicles in breast cancer research
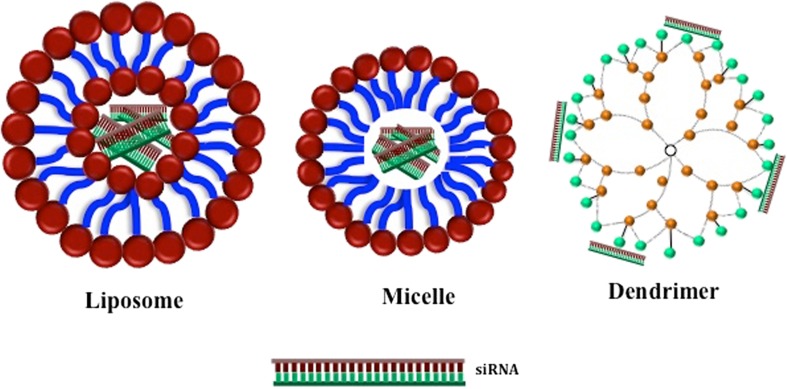
The most common chemical structures explored for gene silencing therapy are cell-penetrating peptides, liposomes, micelles, and polymeric nanoparticles, and all of them have been applied to experimental models of breast cancer (Fig. 2). To date, there are no reported clinical trials for breast cancer involving siRNA or miRNA therapies using non-viral chemical delivery systems. We have, therefore, focused on in vivo studies in rodent models.
Fig. 2.
Schematic representation of delivery vehicles. Liposomes and micelles encapsulate siRNA/miRNA within an aqueous compartment, whereas dendrimers condense RNA via cationic interactions with positive charges on the surface. For simplicity, the major differences between liposomes, micelles, and dendrimers are depicted; as discussed in the text, variations exist in the composition of all three, for example through PEGylation and addition of targeting moieties
Cell-penetrating peptides
Cell-penetrating peptides are known for their ability to facilitate cellular uptake of various “cargo” into cells, such as nano-size particles, chemical molecules, proteins, and large fragments of DNA (28). They are biologically or artificially manufactured short chains of amino acid monomers (up to 30 amino acids) that are linked by peptide bonds. They are usually positively charged and can bind to negatively charged siRNA to improve its stability in vivo (Jiang et al. 2015; Raucher and Ryu 2015; Jing et al. 2016). From a review of in vivo studies, we observed three design variations of delivery systems involving cell-penetrating peptides: cell-penetrating peptides modified with chitosan and then used to encapsulate the siRNA (Sun et al. 2017a); some studies entrapped or loaded a cell-penetrating peptide onto another primary delivery vehicle (such as liposomes or ultrasound-sensitive nanobubbles) (Jing et al. 2016; Xie et al. 2016); others conjugated a cell-penetrating peptide with other cationic polymers (such as PEG) or micelles to form a nanocomplex for siRNA encapsulation (Wang et al. 2015; Fang et al. 2016; Yang et al. 2016). All in vivo studies demonstrated positive outcomes by showing that tumor growth was inhibited by siRNA.
Cell-penetrating peptides have also demonstrated endosomal escape capabilities through the proton sponge effect (Sun et al. 2017a). The main challenge in the use of cell-penetrating peptides are the non-selective interactions of these peptides in vivo and their ability to translocate through any cell membrane, independent of receptors (Jing et al. 2016). Despite promising outcomes from in vivo studies, other challenges facing the use of cell-penetrating peptides are that their intracellular uptake and internalization pathways are not well defined (Raucher and Ryu 2015; Arnold et al. 2017) and they have short blood plasma half-life (Raucher and Ryu 2015). To our knowledge, there are currently no cell-penetrating peptide-based therapeutic delivery systems commercially available.
Liposomes
Liposomes are small artificial vesicles that are made of material similar to the cell membrane. They contain a lipid bilayer with an internal aqueous core and are characterized by their biocompatibility, biodegradability, low toxicity, and ability to trap both hydrophilic and lipophilic drugs. Liposomes fuse with other cell membranes so that their aqueous compartment becomes adjacent to the cytosol of the target cell to deliver the encapsulated material (Mikhaylova et al. 2009). They are commercially used as carriers for delivery of chemotherapy drugs such as doxorubicin and daunorubicin (Bulbake et al. 2017). It is noted that the majority of approved commercial liposome formulations for clinical use are less than 200 nm in size, with the exception of Myocet® (150–250 nm) and Visudyne® (150–300 nm) (Bulbake et al. 2017). Liposomes have been investigated as carriers of siRNAs primarily by encapsulation of siRNA either on its own or co-delivered with a drug. Cationic liposomes currently have the highest encapsulation efficiency (Young et al. 2016). On review of the in vivo studies in Table 3, it is noted that most of the studies used cationic lipid nanoparticles such as DOTAP (1,2-bis(oleoyloxy)-3-(trimethylammonio)propane), DOPE (dioleoylphosphatidylethanolamine), or DC-Chol (3β[N-(N′,N′-dimethylaminoethane)-carbamoyl]cholesterol). Cationic lipids mimic the chemical and physical attributes of biological lipids and are able to protect siRNAs from degradation; however. their application in gene delivery is limited by variations in their transfection efficiency and systemic toxicity when injected (Piao et al. 2013). Modification of the liposomal surface by coating with polyethylene glycol (PEG), through a process called PEGylation, has been shown to protect the positive charge, reduce immune response, and improve stability in vivo; however, it may compromise cellular uptake, endosomal escape, and resultant silencing efficacy (Ho et al. 2013; Ran et al. 2014; Essex et al. 2015; Gu et al. 2015; Li et al. 2016).
Table 3.
Nanoparticle delivery systems investigated in vivo in breast cancer cells
| Materials and design | Properties: size/zeta potential | Carrier/therapeutic agent | Cell line | Reported outcomes | References |
|---|---|---|---|---|---|
| Human serum albumin-coated lipid nanoparticles with cationic lipid (DDAB), hydrophilic surfactant, and cholesterol | 67.8 ± 3.5 nm/+22.1 mV | phrGFP siRNA | MCF7 | Tumor significantly decreased | Piao et al. (2013) |
| Dexamethasone (dex)-associated liposome | 272.1 ± 11.9 nm/23.6 ± 5.7 mV | Anti-cancer drug, ESC8, co-delivered with NRP-1 shRNA-encoded plasmid | ANV-1 | ~ 4-fold smaller tumor volume | Ahmad et al. (2016) |
| PEGylated lipid-based nanoparticles mPEG-DSPE/-DOTAP/DOPE |
80 to 90 nm/close to neutral surface charge | MDR1 | MCF7/ADR cells | Improved chemosensitivity to doxorubicin Downregulated P-glycoprotein Downregulated MDR1 expression by more than 80% |
Nourbakhsh et al. (2015 |
| PEGylated liposomal complex PEG covalently linked to hyaluronic acid and coated onto liposome complex (DOTAP and cholesterol) |
188.6 ± 10.8 nm and a dramatically declined zeta potential from + 34.9 ± 4.0 mV to − 18.2 ± 2.2 mV | Anti-P-glycoprotein siRNA | MCF-7/ADR cells | Downregulation of P-glycoprotein by 34% | Ran et al. (2014) |
| Cationic lipid liposomal nanoparticle formulation (DC-Chol, DSPC) containing two PEGylation steps: pre- and post-siRNA insertion | 111.3 ± 12.2 to 117.7 ± 10.3/average ~ 5 mV | Luciferase mRNA | MDA-MB-435/LCC6 W-GFP/luciferase cells | Downregulate luciferase mRNA expression by more than 50% | Ho et al. (2013) |
| Nanosized immunoliposome-based delivery complex (DOTAP:DOPE) | ~ 100 nm | Anti-HER-2 siRNA molecule | MDA-MB-435 | Silenced target gene Increased response to chemotherapy Tumor growth inhibition |
Pirollo et al. (2007) |
| Liposomal nanoplex (DOTAP:DOPE) | Not specified | Chemically modified short interfering hybrids (siHybrids) HER-2 expression |
MDA-MB-435 | Inhibited HER-2 expression | Hogrefe et al. (2006) |
| Cationic liposome, synthetic cationic cardiolipin analogue (CCLA) | 110–120 nm | siRNA | MDA-MB-231 cells | 73% of tumor growth suppression Transfection efficiency in mice was 7-fold higher than the commercially available DOTAP-based liposome |
Chien et al. (2005) |
| Polypeptide micelle complex, LA-CLss/siATG7 Lipoic acid (LA) conjugated with a cytosol localization and internalization peptide 6, cross-linked into cross-lined micelles (LA-CLss) |
< 200 nm/zeta potential ranging from 10 to 30 mV | ATG7 siRNA and docetaxel co-delivery | MCF-7 | At least a 1.84-fold greater tumor inhibition compared to that of DTX-loaded micelles in vivo | Gong et al. (2017) |
| Low-density lipoprotein-N-succinyl chitosan-cystamine-urocanic acid | Avg. 167.90 ± 1.46 nm/− 21.91 ± 1.25 mV | Co-delivery of siRNA and paclitaxel | MCF-7/taxol | Inhibited tumor growth | Zhu et al. (2017) |
| Pluronic P123-conjugated polypropylenimine (PPI) dendrimer (anti-CD44-P123-PPI) nanocarrier | Average diameter: 155.9 ± 8.9 nm and 182.3 ± 5.4 nm (with antibody conjugation) Zeta potential: 1.9 ± 0.8 to 28 ± 2.7 mV and 1.1 ± 0.5 to 25.8 ± 2.8 mV (with antibody) |
Anti-CD44-P123-PPI/pDNA-iMDR1-shRNA nanocomplexes combined with ADR to improve the effectiveness of chemotherapy | MCF-7/ADR | Tumor growth inhibition Knockdown of MDR1/P-gp |
Gu et al. (2015) |
| Modified poly(amidoamine) (PAMAM) dendrimer-siRNA complex | 100–200 nm | TWISTI gene | SUM1315 TNBC cells | High concentrating ability in xenograft model | Finlay et al. (2015b) |
| siRNA-loaded nanoparticle (mPEG-PLGA-PLL copolymer) with ultrasound-targeted microbubble destruction | 131.5 ± 6.5 nm | ABCG2-siRNA | MCF-7/S MCF-7/ADR |
Observed suppressed expression of the target gene in tumor xenografts ADR at 5 mg/kg was harmful to liver and kidney |
Bai et al. (2015) |
| Aqueous-core nanocapsules (NC) of PEG-co-poly(e-caprolactone-co-dodecyl-b-malate) (PEG-PCL/MA) | ~ 120 nm | siRNA-loaded nanoparticles targeting ER(alpha) | MCF-7 | Significant decrease in tumour growth and a decrease in ER(alpha) expression in tumour cell xenografts. | Bouclier et al. (2008) |
| Cholesterol-conjugated siRNA loaded onto reconstituted high-density lipoprotein (rHDL) | 100 nm Zeta potential ranging from − 16.85 mV to − 4.2 mV |
Cholesterol-conjugated siRNA targeting VEGF expression | MCF-7 tumor tissue | Tumor volume reduced by up to 54.5% | Ding et al. (2014) |
| PHD/PPF/siVEGF nanocomplex via a self-assembly process utilizing the pre-functionalized polymers: (1) a conjugate of polyethylenimine (PEI) with doxorubicin (DOX) via a pH-responsive hydrazone linkage (PEI–Hz–DOX, PHD) and (2) a tumor-targeting folate ligand conjugated to PEI using polyethylene glycol (PEG) as a linker (PEI–PEG–Folate, PPF) in tandem with VEGF siRNA | 120 nm/+ 3 mV | Co-delivery of siRNA and low-dose doxorubicin | MCF-7 cells | Up to 55% downregulation of VEGF mRNA expressed in vivo; 1.44-fold lower tumor volume | Dong et al. (2015) |
| Fusion protein of an anti-HER2 single-chain antibody fragment with a positively charged protamine, F5-P, complexed with siRNA | Not specified | siDNMT (siRNA targeting DNA methyltransferases 1 and/or 3b genes expressing HER2) | HER2-expressing BT474 breast cancer cells | Inhibited tumor growth and restored tumor suppressor gene expression | Dou et al. (2012) |
| Thioaptamer-conjugated CD44 Thioaptamer (TA)-modified nanoparticles consisting of ligand TA and dendritic polyamidoamine (PAMAM) |
~ 105-400 nm/~ − 5 mV to + 20 mV | TA-PEG-PAMAM/miRNA | CD44, a membrane protein that is overexpressed on the plasma membrane of metastatic breast cancer cells (MDA-MB-231 or LM2-4142) | Significant reduction in tumor weight | Fan et al. (2016) |
| Reduction-sensitive linear cationic click polymer/iMDR1–pDNA complex nanoparticles (RCPN) | 150 nm on average/0 mV to ~ 20 mV | To silence the expression of P-glycoprotein (P-gp) to reverse multidrug resistance | MCF-7/ADR cells | RCPNs inhibted expression of P-gp. RCPNs treatment did not show any inhibition effect on the growth of the tumor | Gao et al. (2011) |
| Cell-penetrating peptide, TAT-MU (TM), and a targeting ligand, HER2 antibody mimetic-affibody (AF)-TMAF | ~ 185 nm | HER2 | MDA-MB-453 | Observed specific and significant reduction of tumor volume when injected intratumorally. | Govindarajan et al. (2012) |
| Chitosan hydrogel (Ch-hG) | Not specified | Alexa555 siRNa/Ch-hG into a375sM-bearing mice | MDa-MB231 | Higher localization into tumor cells 92% reduction in tumor growth |
Han et al. (2011) |
| HER2-scFv-arginine nonamer peptide fusion protein (e23sFv-9R) carrier | Not specified | CXCR4-siRNA for treatment of HER2+ breast cancers | MDA-MB-231 | Inhibited CXCR4 gene expression, suppressed tumor growth, reduced metastasis, and prolonged survival | Jiang et al. (2015) |
| Cell-penetrating peptide-loaded nanobubbles with ultrasound radiation | Average 582.80 ± 19.57 nm/+ 23.51 ± 0.32 mV | EGFR siRNA delivery (siEGFR) to TNBC cells | MDA-MB-231 | Downregulated expression of EGFR and inhibited growth of TNBC | Jing et al. (2016) |
| Magnetic nanoparticles (MN for magnetic resonance imaging), labeled with near-infrared dye Cy5.5 (for optical imaging) and conjugated to a peptide (EPPT) | Not specified | Tumor-specific antigen uMUC-1 and siRNA against gene birc5 (survivin) (siBIRC5) | BT-20 | Nearly 2-fold decrease in tumor growth rate | Kumar et al. (2010) |
| Cationic lipid-assisted PEG-PLA nanoparticles | Average diameter of 146.4 ± 5.9 nm | Cyclin-dependent kinase 1 (CDK1), siCDK1 | SUM149 and BT549 for c-Myc overexpressed TNBC | CDK1 expression inhibition | Liu et al. (2014) |
| PEG–PEI polyplexes | ~ 30–80 nm ~ − 10 mV to 15 mV |
STAT3 siRNA on breast cancer bone metastasis, targeting overexpressed E-selectin | MDA-MB-231 and MCF-7 | Knockdown of STAT3 expression in 48.7% of cancer cells inside the bone marrow Tumor growth inhibition |
Mai et al. (2014) |
| Micelleplex system based on an amphiphilic and cationic triblock copolymer, mPEG-b-PCL-b-PPEEA | 47.7 ± 0.4 nm/− 20.8 ± 2.8 mV | Targeting the acid ceramidase (AC) gene | BT474-luciferase cells | Significant gene knockdown Suppression of AC expression Inhibited tumor growth |
Mao et al. (2011) |
| Co-delivery of drug (doxorubicin) and siRNA in mesoporous silica nanoparticle functionalized by PEI–PEG copolymer | 50 nm | P-glycoprotein to overcome multidrug resistance | MCF-7/MDR | Significant Pgp knockdown Statistically lower Pgp expression (~ 50%) in the tumor from Pgp siRNA-Dox-MSNP-treated animals compared to saline controls Treatment with Pgp siRNA-Dox-MSNP led to ~ 70% reduction in mRNA expression compared to other treatment groups |
Meng et al. (2013) |
| Mesoporous silica nanoparticle core coated layer-by-layer with bioreducible cross-linked PEI and PEG polymers, conjugated with an antibody for selective uptake into cancer cells | 50 nm/8.10 − 0.25 mV | Polo-like kinase 1 siRNA (siPLK1) | TNBC cells BT549 and MDA-MB-231 |
Inhibited cancer migration and invasion in TNBC cells Knocked down about 80% of human PLK1 mRNA expression in metastatic breast cancer cells |
Morry et al. (2017) |
| Jet-PEI and ultrasound | Average diameter is 2.5 μm | ANT2 shRNA | MDA-MB-231 | Tumor regression and increased survival rate (80%), no significant toxicity | Park et al. (2015) |
| PEI substituted with linoleic acid and caprylic acid | Not specified | CDC20-1 DsiRNA | MDA-MB-435 MDA-MB-231 MCF7 |
Reduced tumor growth | Parmar et al. (2015) |
| Multilamellar gold niosomes: siRNA-Nio-Au-thymoquinone | 100–160 nm/1.14 mV | Akt-siRNA and thymoquinone MDM2 expression |
BALB/c (nu+/nu+) human breast cancer xenograft model | Decrease in tumor mass and volume | Rajput et al. (2015) |
| PEG-modified chitosan for siRNA encapsulation | 100 nm | Surviving | 4T1 | Significantly reduced growth of tumor in vivo | Sun et al. (2016) |
| Layer-by-layer films formed by alternately depositing siRNA and poly-L-arginine | 120 nm/− 55 mV | GFP-overexpressing cells in TNBC xenograft models | MDA-MB-468 | Reduced the target gene expression in the tumors by almost 80% | Deng et al. (2013) |
| Amphiphilic alkylated PEI/superparamagnetic iron oxide nanovehicles carrying siRNA on the outside through electrostatic interaction | ~ 88–130 nm/~ 37–44 mV | P-glycoprotein (P-gp) | MCF-7/ADR | Downregulated P-gp by 71.4% | Lin et al. (2014) |
All studies reported positive outcomes for their liposomal delivery system and observed downregulation of the target gene. Nourbakhsh et al. (2015) reported more than 80% downregulation of the drug resistance gene MDR1 in MCF-7/ADR breast cancer cells using a PEGylated DSPE/DOTAP/DOPE liposome delivery system. It is important to note that liposomes can present with high surface charge, which may induce toxic effects (Li et al. 2016). Other challenges reported for liposomes include their low solubility, high production costs, challenges with manufacturing scalability, short half-life (unless PEGylated), potential hypersensitivity reactions in vivo, and potential to form aggregates with negatively charged serum proteins that can accumulate in the lungs, liver, and spleen (Balazs and Godbey 2011; Bedi et al. 2013; Akbarzadeh et al. 2013; Sioud 2015). Whitehead et al. (2014) studied PEGylated lipid nanoparticles for siRNA delivery in mice. They established four design criteria to predict whether the delivery system will silence at least 95% of protein expression, which are that the scaffold has: (1) a COOC13H27 tail, (2) three or more tails, (3) is synthesized from an alkyl-amine precursor containing one or more tertiary amides, and (4) a surface pKa of 5.5 or higher (Whitehead et al. 2014).
Micelles
Micelles are closed spherical monolayers of phospholipids. They differ from liposomes in that they do not have an aqueous core. Polymeric micelles (having diameters ranging from 10 to 100 nm) have attracted attention for their versatile properties and ease of preparation (Yousefpour Marzbali and Yari Khosroushahi 2017). Reported benefits of micelles include simple preparation, low toxicity, long half-life, and good tissue penetration capabilities (Yousefpour Marzbali and Yari Khosroushahi 2017). However, similar to liposomes, they are subject to dilution following intravenous administration. Micelles are either made to encapsulate siRNA and drugs or chemically altered to form a nanocomplex (Falamarzian et al. 2012). They are typically modified at their core to improve encapsulation efficiency and modified at their shell to improve in vivo stability. For example, micelles have been coated with PEG by direct conjugation of PEG to siRNA and condensation of PEG-siRNA to a micelle structure (Wakaskar 2017).
Examples of micelles containing complexed siRNA include PEGPnBA–PDMAEMA (Draz et al. 2014) and PEG–PEI (Falamarzian et al. 2012). Yu et al. (2016) investigated a triple-layered pH-responsive micelleplex formed from PEG-b-PAGA-b-PDPA triblock copolymers. The micelleplex encapsulated a drug at its core and had siRNA loaded at the interlayer. The micelleplexes were delivered in vivo to 4T1 xenografts and were able to inhibit tumor growth. Polymeric micelles are advantageous in that their structure and the physiochemical and biological properties can be changed and multiple molecules can be integrated onto one micelle platform; however. the structure needs to be optimized to obtain a balance between the siRNA stability, cell uptake, and safety in vivo (Falamarzian et al. 2012).
Nanoparticles
Nanoparticles are generally 10–1000 nm in diameter (Wang et al. 2016a; Parvani and Jackson 2017). They possess several advantages to other systems, including small size, high surface area, stability in physiological media, and immunologically inert surfaces that gives them good in vivo retention (Bannunah et al. 2014; Draz et al. 2014; Young et al. 2016). The two main types of nanoparticles investigated for siRNA therapy in breast cancer are inorganic nanoparticles and polymeric nanoparticles (Young et al. 2016; Li et al. 2016; Wang et al. 2016b). Inorganic nanoparticles that have been used for delivery of siRNA include silica, calcium, gold, magnesium, strontium, metal oxides, and carbon nanotubes. Finlay et al. (2015a) delivered siRNA in PEI-coated mesoporous silica nanoparticles and observed knockdown of TWIST1 expression in vivo. Metal oxides such as iron oxide nanoparticles have magnetic resonance imaging properties, which enables them to offer combined imaging and delivery of siRNA to tumors (Kumar et al. 2010).
Cationic polymer-based nanoparticles are the dominant material used for delivery, owing to their strong electrostatic interactions with negatively charged siRNA and because they can be synthesized relatively easily compared to other types of nanoparticles (Sun et al. 2017b). The most common cationic polymer used for siRNA delivery systems is low molecular weight PEI due to its superior transfection efficiency (Arnold et al. 2017; Sun et al. 2017b). However, clinical application of unmodified PEI is hampered by its cytotoxic effects. Other common polymers include polylysine (PLL), poly(lactic-co-glycolic acid), and chitosan (Navarro et al. 2015).
Dendrimers are a relatively new class of cationic polymers. Dendrimers are hyper-branched globular structures composed of repeating units. They are typically uniform with a high degree of symmetry. Dendrimers have a large number of cavities as well as a large number of functional groups on the surface. The typical structure of a dendrimer consists of three parts: a central core, which defines the interior size and the quantity and direction of the branches; the repetitive branch units, which determine the molecular size and flexibility; and the terminal groups, which determine the chemical property and ability to interact (Fig. 2) (Wu et al. 2013). Dendrimers have positively charged amine functionalities at the surface that enable condensation of negatively charged siRNA molecules through electrostatic interactions. They also have tertiary amine groups in the interior that can be protonated in acidic endosomes to initiate the proton sponge effect (Liu et al. 2016). The most common dendrimer used for siRNA delivery systems is poly(amidoamine) (PAMAM) (Gu et al. 2015; Sun et al. 2017b), a cationic polymer that has good pH buffering capability. Limited in vivo studies were found for dendrimer-based delivery systems for breast cancer. Finlay et al. (2015b) investigated PAMAM-siRNA complexes in SUM1315 TNBC cells and found that the dendrimer–siRNA complexes were taken up by the cells and led to knockdown of TWIST1 expression in vivo. Dendrimers offer flexibility in being able to modify their structure and properties, as with liposomes and micelles. However, there are concerns with non-specific cytotoxicity, rapid clearance in vivo, and poor delivery efficiency (Biswas and Torchilin 2013; Wu et al. 2013; Finlay et al. 2015b).
The following types of designs were observed in nanoparticle delivery systems for breast cancer in vivo: the majority of delivery systems were composed of multiple polymers mixed or conjugated to form a nanocomplex; siRNAs loaded onto the surface of a nanocomplex (Lin et al. 2014); and layer-by-layer nanoparticles (Deng et al. 2013). It is noted that PEG and PEI are incorporated in the majority of the siRNA nanocomplexes, providing strong support for cationic polymeric nanoparticles as part of a delivery system. Furthermore, almost all delivery systems were less than 200 nm. Although to our knowledge none have progressed to clinical trials for breast cancer, some such as the commercially available jet PEI have been trialed for other indications (Walther et al. 2008).
A critical consideration in the design of delivery vehicles for breast cancer is a specific targeting capability. In vivo studies have shown that specific gene silencing of breast tumors can be achieved by combining siRNAs with cell type-specific ligands such as antibody fragments or chemokine receptors, which are known to affect protein kinase pathways that control key signaling steps involved in the invasion and growth of the breast tumor cells. These ligands help the delivery vehicles enter target breast cells through receptor-mediated endocytosis. Examples of ligands that have been shown to facilitate specific targeting of breast tumors in in vivo xenograft models include fusion protein containing single-chain antibody fragment (scFv) (Jiang et al. 2015), stromal cell-derived factor (SDF-1α), CXC chemokine receptor 4 (CXCR4) (Jiang et al. 2015), neuropilin-1 (NRP-1), heat shock protein 90 (Hsp90) (Ahmad et al. 2016), c-raf (Chien et al. 2005), and tissue transglutaminase (TG2) (Han et al. 2011). In general, antibody fragments with well-defined cell-type specificity and favorable pharmacokinetics and biodistributions are desired.
Conclusion
RNA interference (RNAi) using small interfering RNA (siRNA) and microRNA (miRNA) is an evolving field for cancer treatment, particularly for cancers that display highly heterogeneous characteristics such as breast cancer. A better understanding of the genetic basis of the molecular subtypes of breast cancer is required to identify the target genes for silencing. There has been considerable progress in employing siRNA in breast cancer through various delivery systems. On review of delivery systems using studies in vivo, we noted that there has been considerable effort directed towards optimizing the design by modifying the core and surface of the delivery vehicles to address the various challenges encountered in circulation and at the tissue and cellular levels. We observed that the surface of the delivery vehicle is primarily where ligands have been attached for cell-specific recognition in breast tumors. The majority of studies employed PEGylation of the surface to protect the delivery system from nuclease degradation. The core of the delivery system has been mainly modified to enhance its ability to escape the endosome, normally through incorporating polymers with proton-sponge effect capabilities such as polyethylenimine (PEI). We also observed that the size of almost all nanoparticle delivery vehicles was less than 200 nm. Concerning translation to the clinic, large-scale production of nanoparticles can be labor-intensive and, therefore, costly. The delivery system must consider careful selection of materials, solvents, and procedures for nanoparticle development, batch-to-batch consistency, reproducibility, and acceptability of the final product during scale-up. Despite these hurdles, gene silencing through RNAi has significant clinical implications for cancer treatment and continued research into the design of an effective delivery vehicle for siRNAs and miRNAs will accelerate the trend towards precision medicine.
Compliance with ethical standards
Conflict of interest
Tamkin Ahmadzada declares that she has no conflict of interest. Glen Reid declares that he has no conflict of interest. David McKenzie declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Ahmad A, Mondal SK, Mukhopadhyay D, Banerjee R, Alkharfy KM. Development of liposomal formulation for delivering anticancer drug to breast cancer stem-cell-like cells and its pharmacokinetics in an animal model. Mol Pharm. 2016;13(3):1081–1088. doi: 10.1021/acs.molpharmaceut.5b00900. [DOI] [PubMed] [Google Scholar]
- Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, et al. Liposome: classification, preparation, and applications. Nanoscale Res Lett. 2013;8(1):102. doi: 10.1186/1556-276X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Principles of membrane transport. In: Molecular biology of the cell, 4th edn. Garland Science, New York. https://www.ncbi.nlm.nih.gov/books/NBK26815. Accessed 29 Jul 2017
- Arnold AE, Czupiel P, Shoichet M. Engineered polymeric nanoparticles to guide the cellular internalization and trafficking of small interfering ribonucleic acids. J Control Release. 2017;259:3–15. doi: 10.1016/j.jconrel.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Badve S, Dabbs DJ, Schnitt SJ, Baehner FL, Decker T, Eusebi V, et al. Basal-like and triple-negative breast cancers: a critical review with an emphasis on the implications for pathologists and oncologists. Mod Pathol. 2011;24(2):157–167. doi: 10.1038/modpathol.2010.200. [DOI] [PubMed] [Google Scholar]
- Bae SY, Kim S, Lee JH, Lee HC, Lee SK, Kil WH, et al. Poor prognosis of single hormone receptor-positive breast cancer: similar outcome as triple-negative breast cancer. BMC Cancer. 2015;15:138. doi: 10.1186/s12885-015-1121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M, Shen M, Teng Y, Sun Y, Li F, Zhang X, et al. Enhanced therapeutic effect of Adriamycin on multidrug resistant breast cancer by the ABCG2-siRNA loaded polymeric nanoparticles assisted with ultrasound. Oncotarget. 2015;6(41):43779–43790. doi: 10.18632/oncotarget.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs DA, Godbey WT. Liposomes for use in gene delivery. J Drug Deliv. 2011;2011:326497. doi: 10.1155/2011/326497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannunah AM, Vllasaliu D, Lord J, Stolnik S. Mechanisms of nanoparticle internalization and transport across an intestinal epithelial cell model: effect of size and surface charge. Mol Pharm. 2014;11(12):4363–4373. doi: 10.1021/mp500439c. [DOI] [PubMed] [Google Scholar]
- Bedi D, Gillespie JW, Petrenko VA, Jr, Ebner A, Leitner M, Hinterdorfer P, et al. Targeted delivery of siRNA into breast cancer cells via phage fusion proteins. Mol Pharm. 2013;10(2):551–559. doi: 10.1021/mp3006006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behr JP. The proton sponge: a trick to enter cells the viruses did not exploit. Chimia. 1997;51:34–36. [Google Scholar]
- Biswas S, Torchilin VP. Dendrimers for siRNA delivery. Pharmaceuticals (Basel) 2013;6(2):161–183. doi: 10.3390/ph6020161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouclier C, Moine L, Hillaireau H, Marsaud V, Connault E, Opolon P, et al. Physicochemical characteristics and preliminary in vivo biological evaluation of nanocapsules loaded with siRNA targeting estrogen receptor alpha. Biomacromolecules. 2008;9(10):2881–2890. doi: 10.1021/bm800664c. [DOI] [PubMed] [Google Scholar]
- Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–vi12. doi: 10.1093/annonc/mds187. [DOI] [PubMed] [Google Scholar]
- Bulbake U, Doppalapudi S, Kommineni N, Khan W. Liposomal formulations in clinical use: an updated review. Pharmaceutics. 2017;9(2):12. doi: 10.3390/pharmaceutics9020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136(4):642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. Tamoxifen resistance in breast cancer. Biomol Ther (Seoul) 2012;20(3):256–267. doi: 10.4062/biomolther.2012.20.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien PY, Wang J, Carbonaro D, Lei S, Miller B, Sheikh S, et al. Novel cationic cardiolipin analogue-based liposome for efficient DNA and small interfering RNA delivery in vitro and in vivo. Cancer Gene Ther. 2005;12(3):321–328. doi: 10.1038/sj.cgt.7700793. [DOI] [PubMed] [Google Scholar]
- Cho N. Molecular subtypes and imaging phenotypes of breast cancer. Ultrasonography. 2016;35(4):281–288. doi: 10.14366/usg.16030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlman JE, Kauffman KJ, Langer R, Anderson DG. Nanotechnology for in vivo targeted siRNA delivery. Adv Genet. 2014;88:37–69. doi: 10.1016/B978-0-12-800148-6.00003-1. [DOI] [PubMed] [Google Scholar]
- Dai X, Xiang L, Li T, Bai Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer. 2016;7(10):1281–1294. doi: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis SS. Biomedical applications of nanotechnology—implications for drug targeting and gene therapy. Trends Biotechnol. 1997;15(6):217–224. doi: 10.1016/S0167-7799(97)01036-6. [DOI] [PubMed] [Google Scholar]
- Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano. 2013;7(11):9571–9584. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Wang Y, Zhou J, Gu X, Wang W, Liu C, et al. Direct cytosolic siRNA delivery by reconstituted high density lipoprotein for target-specific therapy of tumor angiogenesis. Biomaterials. 2014;35(25):7214–7227. doi: 10.1016/j.biomaterials.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Dong D, Gao W, Liu Y, Qi XR. Therapeutic potential of targeted multifunctional nanocomplex co-delivery of siRNA and low-dose doxorubicin in breast cancer. Cancer Lett. 2015;359(2):178–186. doi: 10.1016/j.canlet.2015.01.011. [DOI] [PubMed] [Google Scholar]
- Dou S, Yao YD, Yang XZ, Sun TM, Mao CQ, Song EW, et al. Anti-Her2 single-chain antibody mediated DNMTs-siRNA delivery for targeted breast cancer therapy. J Control Release. 2012;161(3):875–883. doi: 10.1016/j.jconrel.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Dowsett M, Nielsen TO, A’Hern R, Bartlett J, Coombes RC, Cuzick J, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. doi: 10.1093/jnci/djr393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draz MS, Fang BA, Zhang P, Hu Z, Gu S, Weng KC, et al. Nanoparticle-mediated systemic delivery of siRNA for treatment of cancers and viral infections. Theranostics. 2014;4(9):872–892. doi: 10.7150/thno.9404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15(5):R92. doi: 10.1186/bcr3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411(6836):494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Eliyatkın N, Yalçın E, Zengel B, Aktaş S, Vardar E. Molecular classification of breast carcinoma: from traditional, old-fashioned way to a new age, and a new way. J Breast Health. 2015;11(2):59–66. doi: 10.5152/tjbh.2015.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eroles P, Bosch A, Pérez-Fidalgo JA, Lluch A. Molecular biology in breast cancer: intrinsic subtypes and signaling pathways. Cancer Treat Rev. 2012;38(6):698–707. doi: 10.1016/j.ctrv.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Essex S, Navarro G, Sabhachandani P, Chordia A, Trivedi M, Movassaghian S, et al. Phospholipid-modified PEI-based nanocarriers for in vivo siRNA therapeutics against multidrug-resistant tumors. Gene Ther. 2015;22(3):257–266. doi: 10.1038/gt.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falamarzian A, Xiong X-B, Uludag H, Lavasanifar A. Polymeric micelles for siRNA delivery. J Drug Deliv Sci Technol. 2012;22:43–54. doi: 10.1016/S1773-2247(12)50004-3. [DOI] [Google Scholar]
- Fan W, Wang X, Ding B, Cai H, Wang X, Fan Y, et al. Thioaptamer-conjugated CD44-targeted delivery system for the treatment of breast cancer in vitro and in vivo. J Drug Target. 2016;24(4):359–371. doi: 10.3109/1061186X.2015.1077850. [DOI] [PubMed] [Google Scholar]
- Fang WB, Yao M, Brummer G, Acevedo D, Alhakamy N, Berkland C, et al. Targeted gene silencing of CCL2 inhibits triple negative breast cancer progression by blocking cancer stem cell renewal and M2 macrophage recruitment. Oncotarget. 2016;7(31):49349–49367. doi: 10.18632/oncotarget.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay J, Roberts CM, Dong J, Zink JI, Tamanoi F, Glackin CA. Mesoporous silica nanoparticle delivery of chemically modified siRNA against TWIST1 leads to reduced tumor burden. Nanomedicine. 2015;11(7):1657–1666. doi: 10.1016/j.nano.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay J, Roberts CM, Lowe G, Loeza J, Rossi JJ, Glackin CA. RNA-based TWIST1 inhibition via dendrimer complex to reduce breast cancer cell metastasis. Biomed Res Int. 2015;2015:382745. doi: 10.1155/2015/382745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gao K, Huang L. Nonviral methods for siRNA delivery. Mol Pharm. 2009;6(3):651–658. doi: 10.1021/mp800134q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Chen L, Zhang Z, Chen Y, Li Y. Reversal of multidrug resistance by reduction-sensitive linear cationic click polymer/iMDR1-pDNA complex nanoparticles. Biomaterials. 2011;32(6):1738–1747. doi: 10.1016/j.biomaterials.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Gasparri ML, Casorelli A, Bardhi E, Besharat AR, Savone D, Ruscito I, et al. Beyond circulating microRNA biomarkers: urinary microRNAs in ovarian and breast cancer. Tumor Biol. 2017;39(5):1010428317695525. doi: 10.1177/1010428317695525. [DOI] [PubMed] [Google Scholar]
- Global Burden of Disease Cancer Collaboration. Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3(4):524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goljan EF. Rapid review pathology. 3. Philadelphia: Mosby/Elsevier; 2011. [Google Scholar]
- Gong C, Hu C, Gu F, Xia Q, Yao C, Zhang L, et al. Co-delivery of autophagy inhibitor ATG7 siRNA and docetaxel for breast cancer treatment. J Control Release. 2017;266:272–286. doi: 10.1016/j.jconrel.2017.09.042. [DOI] [PubMed] [Google Scholar]
- Govindarajan S, Sivakumar J, Garimidi P, Rangaraj N, Kumar JM, Rao NM, et al. Targeting human epidermal growth factor receptor 2 by a cell-penetrating peptide–affibody bioconjugate. Biomaterials. 2012;33(8):2570–2582. doi: 10.1016/j.biomaterials.2011.12.003. [DOI] [PubMed] [Google Scholar]
- Gu J, Fang X, Hao J, Sha X. Reversal of P-glycoprotein-mediated multidrug resistance by CD44 antibody-targeted nanocomplexes for short hairpin RNA-encoding plasmid DNA delivery. Biomaterials. 2015;45:99–114. doi: 10.1016/j.biomaterials.2014.12.030. [DOI] [PubMed] [Google Scholar]
- Guiu S, Michiels S, Andre F, Cortes J, Denkert C, Di Leo A, et al. Molecular subclasses of breast cancer: how do we define them? The IMPAKT 2012 Working Group Statement. Ann Oncol. 2012;23(12):2997–3006. doi: 10.1093/annonc/mds586. [DOI] [PubMed] [Google Scholar]
- Gujrati M, Vaidya A, Lu ZR. Multifunctional pH-sensitive amino lipids for siRNA delivery. Bioconjug Chem. 2016;27(1):19–35. doi: 10.1021/acs.bioconjchem.5b00538. [DOI] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286(5441):950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Han HD, Mora EM, Roh JW, Nishimura M, Lee SJ, Stone RL, et al. Chitosan hydrogel for localized gene silencing. Cancer Biol Ther. 2011;11(9):839–845. doi: 10.4161/cbt.11.9.15185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Park K (2016) Effects of the Microparticle Shape on Cellular Uptake. Mol Pharm 13(7):2164. 10.1021/acs.molpharmaceut.5b00992. [DOI] [PMC free article] [PubMed]
- Hennigs A, Riedel F, Gondos A, Sinn P, Schirmacher P, Marmé F, et al. Prognosis of breast cancer molecular subtypes in routine clinical care: a large prospective cohort study. BMC Cancer. 2016;16(1):734. doi: 10.1186/s12885-016-2766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Terada H (2012) Endocytosis of particle formulations by macrophages and its application to clinical treatment. In: Ceresa B (ed) Molecular regulation of endocytosis. InTech. 10.5772/45820. Available online at: https://www.intechopen.com/books/molecular-regulation-of-endocytosis/endocytosis-of-particle-formulations-by-macrophages-and-its-application-to-clinical-treatment
- Ho EA, Osooly M, Strutt D, Masin D, Yang Y, Yan H et al (2013) Characterization of long-circulating cationic nanoparticle formulations consisting of a two-stage PEGylation step for the delivery of siRNA in a breast cancer tumor model. J Pharm Sci 102(1):227–236. 10.1002/jps.23351 [DOI] [PubMed]
- Hogrefe RI, Lebedev AV, Zon G, Pirollo KF, Rait A, Zhou Q, et al. Chemically modified short interfering hybrids (siHYBRIDS): nanoimmunoliposome delivery in vitro and in vivo for RNAi of HER-2. Nucleosides Nucleotides Nucleic Acids. 2006;25(8):889–907. doi: 10.1080/15257770600793885. [DOI] [PubMed] [Google Scholar]
- Hon JD, Singh B, Sahin A, Du G, Wang J, Wang VY, et al. Breast cancer molecular subtypes: from TNBC to QNBC. Am J Cancer Res. 2016;6(9):1864–1872. [PMC free article] [PubMed] [Google Scholar]
- Honary S, Zahir F (2013) Effect of zeta potential on the properties of Nano-drug delivery systems—a review (part 2). Trop J Pharm Res 12(2):265–273. 10.4314/tjpr.v12i2.20
- Jemal A, Bray F, Center MM. Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Jiang K, Li J, Yin J, Ma Q, Yan B, Zhang X, et al. Targeted delivery of CXCR4-siRNA by scFv for HER2(+) breast cancer therapy. Biomaterials. 2015;59:77–87. doi: 10.1016/j.biomaterials.2015.04.030. [DOI] [PubMed] [Google Scholar]
- Jing H, Cheng W, Li S, Wu B, Leng X, Xu S, et al. Novel cell-penetrating peptide-loaded nanobubbles synergized with ultrasound irradiation enhance EGFR siRNA delivery for triple negative breast cancer therapy. Colloids Surf B Biointerfaces. 2016;146:387–395. doi: 10.1016/j.colsurfb.2016.06.037. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim A, Miyata K, Kataoka K. Recent progress in development of siRNA delivery vehicles for cancer therapy. Adv Drug Deliv Rev. 2016;104:61–77. doi: 10.1016/j.addr.2016.06.011. [DOI] [PubMed] [Google Scholar]
- Kittaneh M, Montero AJ, Glück S. Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer. 2013;5:61–70. doi: 10.4137/BIC.S9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koval M, Preiter K, Adles C, Stahl PD, Steinberg TH. Size of IgG-opsonized particles determines macrophage response during internalization. Exp Cell Res. 1998;242(1):265–273. doi: 10.1006/excr.1998.4110. [DOI] [PubMed] [Google Scholar]
- Kumar M, Yigit M, Dai G, Moore A, Medarova Z. Image-guided breast tumor therapy using a small interfering RNA nanodrug. Cancer Res. 2010;70(19):7553–7561. doi: 10.1158/0008-5472.CAN-10-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- Li Y, Kröger M, Liu WK. (2015) Shape effect in cellular uptake of PEGylated nanoparticles: comparison between sphere, rod, cube and disk. Nanoscale 7(40):16631-46. 10.1039/c5nr02970h. [DOI] [PubMed]
- Li J, Xue S, Mao Z-W. Nanoparticle delivery systems for siRNA-based therapeutics. J Mater Chem B. 2016;4(41):6620–6639. doi: 10.1039/C6TB01462C. [DOI] [PubMed] [Google Scholar]
- Li X, Oprea-Ilies GM, Krishnamurti U. New developments in breast cancer and their impact on daily practice in pathology. Arch Pathol Lab Med. 2017;141(4):490–498. doi: 10.5858/arpa.2016-0288-SA. [DOI] [PubMed] [Google Scholar]
- Liang W, Lam JKW (2012) Endosomal escape pathways for non-viral nucleic acid delivery systems. In: Ceresa B (ed) Molecular regulation of endocytosis. InTech. 10.5772/46006. Available online at: https://www.intechopen.com/books/molecular-regulation-of-endocytosis/endosomal-escape-pathways-for-non-viral-nucleic-acid-delivery-systems
- Lin G, Zhu W, Yang L, Wu J, Lin B, Xu Y, et al. Delivery of siRNA by MRI-visible nanovehicles to overcome drug resistance in MCF-7/ADR human breast cancer cells. Biomaterials. 2014;35(35):9495–9507. doi: 10.1016/j.biomaterials.2014.07.049. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhu YH, Mao CQ, Dou S, Shen S, Tan ZB, et al. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. J Control Release. 2014;192:114–121. doi: 10.1016/j.jconrel.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Liu H, Chang H, Lv J, Jiang C, Li Z, Wang F, et al. Screening of efficient siRNA carriers in a library of surface-engineered dendrimers. Sci Rep. 2016;6:25069. doi: 10.1038/srep25069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü L, Mao X, Shi P, He B, Xu K, Zhang S, et al. MicroRNAs in the prognosis of triple-negative breast cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2017;96(22):e7085. doi: 10.1097/MD.0000000000007085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai J, Huang Y, Mu C, Zhang G, Xu R, Guo X, et al. Bone marrow endothelium-targeted therapeutics for metastatic breast cancer. J Control Release. 2014;187:22–29. doi: 10.1016/j.jconrel.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maire V, Némati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, et al. Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res. 2013;73(2):813–823. doi: 10.1158/0008-5472.CAN-12-2633. [DOI] [PubMed] [Google Scholar]
- Mao CQ, Du JZ, Sun TM, Yao YD, Zhang PZ, Song EW, et al. A biodegradable amphiphilic and cationic triblock copolymer for the delivery of siRNA targeting the acid ceramidase gene for cancer therapy. Biomaterials. 2011;32(11):3124–3133. doi: 10.1016/j.biomaterials.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L et al (2012). Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol 27(6):785. 10.14670/HH-27.785. [DOI] [PubMed]
- Mastoraki A, Kazani A, Mastoraki S, Konstantiadou I, Kokoropoulos P, Smyrniotis V, et al. Breast-cancer subtyping in clinical practice: Clinicopathologic features and outcomes. J Gynecol Surg. 2014;30(5):260–264. doi: 10.1089/gyn.2014.0022. [DOI] [Google Scholar]
- Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2(11):986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maximiano S, Magalhães P, Guerreiro MP, Morgado M. Trastuzumab in the treatment of breast cancer. BioDrugs. 2016;30(2):75–86. doi: 10.1007/s40259-016-0162-9. [DOI] [PubMed] [Google Scholar]
- Mendelsohn J, Howley PM, Israel MA, Gray JW, Thompson CB. The molecular basis of cancer. 4. Philadelphia, PA: Saunders/Elsevier; 2015. [Google Scholar]
- Meng Z, Lu M. RNA interference-induced innate immunity, off-target effect, or immune adjuvant? Front Immunol. 2017;8:331. doi: 10.3389/fimmu.2017.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Mai WX, Zhang H, Xue M, Xia T, Lin S, et al. Codelivery of an optimal drug/siRNA combination using mesoporous silica nanoparticles to overcome drug resistance in breast cancer in vitro and in vivo. ACS Nano. 2013;7(2):994–1005. doi: 10.1021/nn3044066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova M, Stasinopoulos I, Kato Y, Artemov D, Bhujwalla ZM. Imaging of cationic multifunctional liposome-mediated delivery of COX-2 siRNA. Cancer Gene Ther. 2009;16(3):217–226. doi: 10.1038/cgt.2008.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morry J, Ngamcherdtrakul W, Gu S, Reda M, Castro DJ, Sangvanich T, et al. Targeted treatment of metastatic breast cancer by PLK1 siRNA delivered by an antioxidant nanoparticle platform. Mol Cancer Ther. 2017;16(4):763–772. doi: 10.1158/1535-7163.MCT-16-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G, Pan J, Torchilin VP. Micelle-like nanoparticles as carriers for DNA and siRNA. Mol Pharm. 2015;12(2):301–313. doi: 10.1021/mp5007213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngamcherdtrakul W, Morry J, Gu S, Castro DJ, Goodyear SM, Sangvanich T, et al. Cationic polymer modified mesoporous silica nanoparticles for targeted SiRNA delivery to HER2+ breast cancer. Adv Funct Mater. 2015;25(18):2646–2659. doi: 10.1002/adfm.201404629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourbakhsh M, Jaafari MR, Lage H, Abnous K, Mosaffa F, Badiee A, Behravan J. Nanolipoparticles-mediated MDR1 siRNA delivery reduces doxorubicin resistance in breast cancer cells and silences MDR1 expression in xenograft model of human breast cancer. Iranian J Basic Med Sci. 2015;18(4):385–392. [PMC free article] [PubMed] [Google Scholar]
- Park DH, Jung BK, Lee YS, Jang JY, Kim MK, Lee JK, et al. Evaluation of in vivo antitumor effects of ANT2 shRNA delivered using PEI and ultrasound with microbubbles. Gene Ther. 2015;22(4):325–332. doi: 10.1038/gt.2014.120. [DOI] [PubMed] [Google Scholar]
- Parmar MB, Aliabadi HM, Mahdipoor P, Kucharski C, Maranchuk R, Hugh JC, Uludağ H. Targeting cell cycle proteins in breast cancer cells with siRNA by using lipid-substituted polyethylenimines. Front Bioeng Biotechnol. 2015;3:14. doi: 10.3389/fbioe.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvani JG, Jackson MW. Silencing the roadblocks to effective triple-negative breast cancer treatments by siRNA nanoparticles. Endocr Relat Cancer. 2017;24(4):R81–R97. doi: 10.1530/ERC-16-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, Radosevic-Robin N. Ki67 assessment in breast cancer: an update. Pathology. 2017;49(2):166–171. doi: 10.1016/j.pathol.2016.11.006. [DOI] [PubMed] [Google Scholar]
- Peng C, Ma W, Xia W, Zheng W. Integrated analysis of differentially expressed genes and pathways in triple-negative breast cancer. Mol Med Rep. 2017;15(3):1087–1094. doi: 10.3892/mmr.2017.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao L, Li H, Teng L, Yung BC, Sugimoto Y, Brueggemeier RW, et al. Human serum albumin-coated lipid nanoparticles for delivery of siRNA to breast cancer. Nanomedicine. 2013;9(1):122–129. doi: 10.1016/j.nano.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri E, Conteduca V, Andreis D, Massa I, Melegari E, Sarti S et al (2016) Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr Relat Cancer 23(10):R485-98. 10.1530/ERC-16-0190. [DOI] [PubMed]
- Pirollo KF, Rait A, Zhou Q, Hwang SH, Dagata JA, Zon G, et al. Materializing the potential of small interfering RNA via a tumor-targeting nanodelivery system. Cancer Res. 2007;67(7):2938–2943. doi: 10.1158/0008-5472.CAN-06-4535. [DOI] [PubMed] [Google Scholar]
- Pittella F, Kataoka K (2013) Polymeric micelles for siRNA delivery. In: RNA interference from biology to therapeutics, pp 161–184. 10.1007/978-1-4614-4744-3_8
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141(3):497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Prokop A. Intracellular delivery: fundamentals and applications. Dordrecht: Springer; 2011. [Google Scholar]
- Rajput S, Puvvada N, Kumar BP, Sarkar S, Konar S, Bharti R, et al. Overcoming Akt induced therapeutic resistance in breast cancer through siRNA and thymoquinone encapsulated multilamellar gold niosomes. Mol Pharm. 2015;12(12):4214–4225. doi: 10.1021/acs.molpharmaceut.5b00692. [DOI] [PubMed] [Google Scholar]
- Ran R, Liu Y, Gao H, Kuang Q, Zhang Q, Tang J, et al. Enhanced gene delivery efficiency of cationic liposomes coated with PEGylated hyaluronic acid for anti P-glycoprotein siRNA: a potential candidate for overcoming multi-drug resistance. Int J Pharm. 2014;477(1–2):590–600. doi: 10.1016/j.ijpharm.2014.11.012. [DOI] [PubMed] [Google Scholar]
- Ranftler M, Strasser-Weippl K. New chemotherapies in breast cancer. Memo. 2017;10(3):127–131. doi: 10.1007/s12254-017-0348-y. [DOI] [Google Scholar]
- Raucher D, Ryu JS. Cell-penetrating peptides: strategies for anticancer treatment. Trends Mol Med. 2015;21(9):560–570. doi: 10.1016/j.molmed.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274(2):113–126. doi: 10.1111/joim.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards DM, Endres RG. Target shape dependence in a simple model of receptor-mediated endocytosis and phagocytosis. Proc Natl Acad Sci U S A. 2016;113(22):6113–6118. doi: 10.1073/pnas.1521974113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothschild SI. microRNA therapies in cancer. Mol Cell Ther. 2014;2:7. doi: 10.1186/2052-8426-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, et al. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23(2):227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sioud M. RNA interference: mechanisms, technical challenges, and therapeutic opportunities. Methods Mol Biol. 2015;1218:1–15. doi: 10.1007/978-1-4939-1538-5_1. [DOI] [PubMed] [Google Scholar]
- Sun P, Huang W, Jin M, Wang Q, Fan B, Kang L, et al. Chitosan-based nanoparticles for survivin targeted siRNA delivery in breast tumor therapy and preventing its metastasis. Int J Nanomedicine. 2016;11:4931–4945. doi: 10.2147/IJN.S105427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P, Huang W, Kang L, Jin M, Fan B, Jin H, et al. siRNA-loaded poly(histidine-arginine)6-modified chitosan nanoparticle with enhanced cell-penetrating and endosomal escape capacities for suppressing breast tumor metastasis. Int J Nanomedicine. 2017;12:3221–3234. doi: 10.2147/IJN.S129436. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sun H, Yarovoy I, Capeling M, Cheng C. Polymers in the co-delivery of siRNA and anticancer drugs for the treatment of drug-resistant cancers. Top Curr Chem. 2017;375(2):24. doi: 10.1007/s41061-017-0113-z. [DOI] [PubMed] [Google Scholar]
- Suter SR, Sheu-Gruttadauria J, Schirle NT, Valenzuela R, Ball-Jones AA, Onizuka K, et al. Structure-guided control of siRNA off-target effects. J Am Chem Soc. 2016;138(28):8667–8669. doi: 10.1021/jacs.6b06137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi R, Miyazaki H, Ochiya T. The roles of microRNAs in breast cancer. Cancers. 2015;7(2):598–616. doi: 10.3390/cancers7020598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatiparti K, Sau S, Kashaw SK, Iyer AK. siRNA delivery strategies: a comprehensive review of recent developments. Nanomaterials. 2017;7(4):77. doi: 10.3390/nano7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakaskar RR. Polymeric micelles for drug delivery. Int J Drug Dev Res. 2017;9:01–02. [Google Scholar]
- Walther W, Siegel R, Kobelt D, Knösel T, Dietel M, Bembenek A, et al. Novel jet-injection technology for nonviral intratumoral gene transfer in patients with melanoma and breast cancer. Clin Cancer Res. 2008;14(22):7545–7553. doi: 10.1158/1078-0432.CCR-08-0412. [DOI] [PubMed] [Google Scholar]
- Wang S, Cao M, Deng X, Xiao X, Yin Z, Hu Q, et al. Degradable hyaluronic acid/protamine sulfate interpolyelectrolyte complexes as miRNA-delivery nanocapsules for triple-negative breast cancer therapy. Adv Healthcare Mater. 2015;4(2):281–290. doi: 10.1002/adhm.201400222. [DOI] [PubMed] [Google Scholar]
- Wang K, Kievit FM, Zhang M. Nanoparticles for cancer gene therapy: recent advances, challenges, and strategies. Pharmacol Res. 2016;114:56–66. doi: 10.1016/j.phrs.2016.10.016. [DOI] [PubMed] [Google Scholar]
- Wang F, Li C, Cheng J, Yuan Z. Recent advances on inorganic nanoparticle-based cancer therapeutic agents. Int J Environ Res Public Health. 2016;13(12):1182. doi: 10.3390/ijerph13121182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KA, Dorkin JR, Vegas AJ, Chang PH, Veiseh O, Matthews J, et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang W, He Z. Dendrimers as carriers for siRNA delivery and gene silencing: a review. Sci World J. 2013;2013:630654. doi: 10.1155/2013/630654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Lin W, Li M, Yang Y, Deng J, Liu H, et al. Efficient siRNA delivery using novel cell-penetrating peptide-siRNA conjugate-loaded nanobubbles and ultrasound. Ultrasound Med Biol. 2016;42(6):1362–1374. doi: 10.1016/j.ultrasmedbio.2016.01.017. [DOI] [PubMed] [Google Scholar]
- Yang Y, Xia X, Dong W, Wang H, Li L, Ma P, et al. Acid sensitive polymeric micelles combining folate and bioreducible conjugate for specific intracellular siRNA delivery. Macromol Biosci. 2016;16(5):759–773. doi: 10.1002/mabi.201500389. [DOI] [PubMed] [Google Scholar]
- Yersal O, Barutca S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J Clin Oncol. 2014;5(3):412–424. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip CH, Bhoo-Pathy N, Daniel JM, Foo YC, Mohamed AK, Abdullah MM, et al. Roles of Ki67 in breast cancer—important for management? Asian Pac J Cancer Prev. 2016;17(3):1077–1082. doi: 10.7314/APJCP.2016.17.3.1077. [DOI] [PubMed] [Google Scholar]
- Young SWS, Stenzel M, Yang J. Nanoparticle-siRNA: a potential cancer therapy? Crit Rev Oncol Hematol. 2016;98:159–169. doi: 10.1016/j.critrevonc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Yousefpour Marzbali M, Yari Khosroushahi A. Polymeric micelles as mighty nanocarriers for cancer gene therapy: a review. Cancer Chemother Pharmacol. 2017;79(4):637–649. doi: 10.1007/s00280-017-3273-1. [DOI] [PubMed] [Google Scholar]
- Yu MZ, Pang WH, Yang T, Wang JC, Wei L, Qiu C, et al. Systemic delivery of siRNA by T7 peptide modified core-shell nanoparticles for targeted therapy of breast cancer. Eur J Pharm Sci. 2016;92:39–48. doi: 10.1016/j.ejps.2016.06.020. [DOI] [PubMed] [Google Scholar]
- Zhu WJ, Yang SD, Qu CX, Zhu QL, Chen WL, Li F, et al. Low-density lipoprotein-coupled micelles with reduction and pH dual sensitivity for intelligent co-delivery of paclitaxel and siRNA to breast tumor. Int J Nanomedicine. 2017;12:3375–3393. doi: 10.2147/IJN.S126310. [DOI] [PMC free article] [PubMed] [Google Scholar]