Abstract
Biological function results from properly timed bio-molecular interactions that transduce external or internal signals, resulting in any number of cellular fates, including triggering of cell-state transitions (division, differentiation, transformation, apoptosis), metabolic homeostasis and adjustment to changing physical or nutritional environments, amongst many more. These bio-molecular interactions can be modulated by chemical modifications of proteins, nucleic acids, lipids and other small molecules. They can result in bio-molecular transport from one cellular compartment to the other and often trigger specific enzyme activities involved in bio-molecular synthesis, modification or degradation. Clearly, a mechanistic understanding of any given high level biological function requires a quantitative characterization of the principal bio-molecular interactions involved and how these may change dynamically. Such information can be obtained using fluctation analysis, in particular scanning number and brightness, and used to build and test mechanistic models of the functional network to define which characteristics are the most important for its regulation.
Keywords: Fluorescence microscopy, Fluctuation analysis, Number and birghtness, Bio-molecular interactions, Functional networks
Introduction
Biological function results from properly timed bio-molecular interactions that transduce external or internal signals, resulting in any number of cellular fates, including the triggering of cell-state transitions (division, differentiation, transformation, apoptosis), metabolic homeostasis and adjustment to changing physical or nutritional environments, amongst many more. These networks of bio-molecular interactions can be modulated by chemical modifications of the proteins, nucleic acids, lipids and other small molecules involved. They can result in bio-molecular transport from one cellular compartment to the other and often trigger specific enzyme activities involved in bio-molecular synthesis, modification or degradation. Clearly, a mechanistic understanding of any given high level biological function requires a quantitative characterization of the principal bio-molecular interactions involved and how these may change dynamically. Such information can be used to build and test mechanistic models of the functional network to define which characteristics are the most important for its regulation. As pointed out by Paul Nurse (Nurse and Hayles 2011),
“Biologists have a tendency to produce somewhat loosely formulated models summarized in the form of cartoons, and it is useful to subject these to the discipline of writing equations in the expectation that the thought imposed by equation writing will improve understanding of the model’s assumptions and dynamics. However, two major problems are often encountered when generating mathematical models for cell biology: the complexity of the pathways being modeled and the difficulty of estimating the appropriate values for rate constants and the concentration of components.”
In recent years, a host of significant achievements have been made in quantitative fluorescence microscopy, not only in terms of imaging modalities and analysis, but also in labeling strategies and fluorophore characteristics. These advances now allow for highly quantitative interrogation of the characteristics of biomolecules in their cellular context. This review will focus on how quantitative microscopy may be used to obtain one kind of the required information referred to by Paul Nurse, namely the concentrations of the components. We will discuss recent results from imaging methodologies based on fluctuations in fluorescence intensity. These approaches, essentially particle counting techniques, are rather numerous and include fluorescence correlation spectroscopy (FCS; Magde et al. 1974; Schwille et al. 1999), fluorescence cross-correlation spectroscopy (FCCS; Schwille and Rigler 1997), moment analysis (MA) (Qian and Elson 1990), fluorescence intensity distribution analysis (FIDA) (Kask et al. 1999), photon counting histogram analysis (PCH) (Chen et al. 1999), Q-Analysis (QA) (Sanchez-Andres et al. 2005), spatial intensity distribution analysis (spIDA) (Godin et al. 2011) and scanning number and brightness (sN&B) (Digman et al. 2008). Fundamentally, they all carry out the same analysis, which is to use the information inherent in the amplitude of fluorescence fluctuations to de-convolve fluorescence intensity into the number of observed fluorescent particles and their molecular brightness. These two parameters allow for calculation of the absolute concentration of fluorescently labeled bio-molecules. Knowledge of the absolute concentrations, as opposed to relative amounts, is paramount to developing testable quantitative models of biological function. Bio-molecular stoichiometry, another key ingredient for modeling biochemical networks in cells, is derived from the brightness values.
Number and brightness analysis
While the dynamic properties of macromolecules in live cells are of great importance and interest, for the purposes of obtaining reliable values for the absolute concentrations and stoichiometry of bio-molecules in cells, the diffusion characteristics of the biomolecules are not directly relevant. Hence, in this review we focus on results of studies in live cells which have applied fluctuation analysis as a particle counting approach (MA, FIDA, spIDA, PCH, QA and sN&B), essentially focusing on the time-independent amplitude of the fluctuations. All of these approaches are based on the assumption that both the fluctuations in fluorescence and the detector shot noise follow Poisson statistics (rare events) (Qian and Elson 1990; Chen et al. 1999). Following this assumption, in the number and brightness (N&B) analysis, for example, the number of particles, N, within the observation volume [defined by the point spread function (PSF) of the microscope] can be calculated from the two moments of the distribution (which are nearly equal in a Poisson distribution), <F>, the average intensity, and σ2, the variance (Digman et al. 2008):
| 1 |
while the molecular brightness, B, is expressed as
| 2 |
One must also correct for detector shot noise. In this case the true number of particles, n, is:
| 3 |
while the true, shot noise-corrected molecular brightness (in counts per dwell time unit, τ, per molecule) is expressed as:
| 4 |
where
| 5 |
Once the value of n is known, dividing it by the value of the observation volume, Veff, and Avogadro’s number, NA, yields the average absolute concentration of molecules. Generally, in live cell studies, the fluorophores are fluorescent proteins fused genetically to the proteins of interest. In this case, one can measure with very high accuracy the molecular brightness of the free monomeric fluorescent protein, eF, expressed in the same cell line under the same imaging conditions as used for measurement of the labeled bio-molecule of interest, FP (fluorescent protein). Then, the absolute concentration of the bio-molecule fused to the fluorescent protein, (FP) (in units of monomeric fluorophore), can be calculated from its average intensity, <F > FP, using Eq. (6) (Fig. 1).
| 6 |
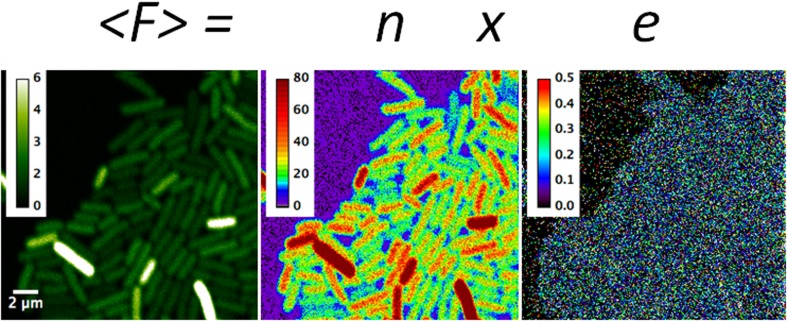
Fig. 1.
Example of scanning number and brightness (sN&B) analysis: expression of a highly fluorescent mutant of the green fluorescent protein (GFPmut2) from a promoter fusion in Bacillus subtilis. The average fluorescence from 50 rapid raster scans (<F>; in counts per dwell-time) at each pixel in the average image is equal to the number of molecules in the effective excitation volume at that pixel, n, times their molecular brightness, e [counts/(dwell-time × molecule)]. These values have been calculated using Eqs. (3) and (4) and are hence corrected for shot noise. Data were from Ferguson et al. (2011). The intensity scale on the left is 0–6 average photon counts per 40 μs dwell-time. The scale for n is 0–80 molecules present on average in the effective excitation volume. The scale in e is 0–0.5 photon counts per dwell-time per molecule. In this case the average for pixels inside bacterial cells (only the central 50% of pixels in each cell are used to avoid edge effects) was 0.05 counts per 40 μs dwell-time per molecule. Full x–y scale is 20 × 20 μm
Moreover, if the molecular brightness of the monomeric fluorescent protein, eF, is known, the stoichiometry of the bio-molecule of interest can be calculated as the ratio of its brightness, eFP, over that of monomer fluorescent proteins:
| 7 |
One of the first fluorescence fluctuation analyses carried out in live cells explored fluctuations in muscle contraction (Borejdo and Morales 1977). However, reports based on fluorescence fluctuations were rare until laser and detector technology caught up with the scientific ideas. Beginning in the late 1990s and into the new millennium, the number of such studies increased significantly, with many applications in live cells. The most numerous reports in the literature concern FCS, which involves measurement of fluorescence intensity fluctuations over a period of time, followed by calculation of the autocorrelation function of the intensity time traces. As noted above, the time-independent amplitude of the autocorrelation function in FCS is a measure of the amplitude of the fluctuations, which is inversely proportional to the number of molecules in the observation volume and directly proportional to their molecular brightness. Analysis of the time-dependence of the autocorrelation function yields the diffusion time and, if applicable, any fast conformational or photo-physical fluctuation rates. The theory and applications of FCS, particularly in live cells, have been reviewed extensively (see, for example, Schwille et al. 1999; Bacia and Schwille 2003; Haustein and Schwille 2007; and references therein).
Despite the importance of absolute bio-molecular concentrations for building testable models of functional bio-molecular networks, the vast majority of published studies have focused on bio-molecular stoichiometry. Membrane protein stoichiometry has proven to be of particular interest to researchers in the field. The oligomerization properties of several membrane receptors have been investigated using these fluctuation-based particle counting techniques (Patel et al. 2002; Hink et al. 2008; Nagy et al. 2010; Godin et al. 2011; Golebiewska et al. 2011; Hellriegel et al. 2011; Swift et al. 2011; Herrick-Davis et al. 2012; Ming et al. 2012; Sergeev et al. 2012a), as well as those of a number of other membrane or membrane associated proteins (Digman et al. 2009; Ross et al. 2011b; Vetri et al. 2011; Li et al. 2012; Sergeev et al. 2012b; James et al. 2014). Oligomerization of nuclear receptors (Chen and Müller 2007; Rosales et al. 2007; Savatier et al. 2010; Presman et al. 2012) and other nuclear proteins (Hinde et al. 2014, 2016; Hennen et al. 2017) have been reported as well. While the studies above were all carried out in mammalian cell lines, a few papers have been published which report protein stoichiometries of proteins in bacteria (Bourges et al. 2017) and yeast (Slaughter et al. 2007, 2008; García-Marcos et al. 2008). Only a very few of these studies report the concentration of the proteins of interest, in addition to their stoichiometry, and in a few cases oligomerization was shown to be concentration dependent (Ross et al. 2011a; James et al. 2012; Li et al. 2014). For more detailed information on the technical aspects of brightness analysis, the reader is referred to Digman et al. (2013) and MacDonald et al. (2013) and references therein.
In 2008, the Gratton group published a variation on the N&B analysis that we have found to be highly useful, called scanning number and brightness analysis (Digman et al. 2008). One important drawback to fluctuation analysis, as it had been applied to that point, arose from the photo-bleaching and the timescales inherent in the measurements. In most cases, the laser excitation was focused into the sample, for example a cell expressing a fluorescent protein fusion of a protein of interest, and the time traces for fluorescence intensity were acquired for a sufficient time to allow good signal to noise ratio in the FCS curves or in the photon counting statistics. If during the measurements, the proteins do not move in and out of the focal volume rapidly, they undergo photo-bleaching, either because they are confined in a small cellular compartment or exist in small cells, such as bacteria, or because their diffusion is slow, as in the case of membrane proteins. Moreover, no fluctuations other than shot noise are observed because the molecules are essentially immobile on the tme-scale of the measurements. Scanning FCS, in which the laser beam is scanned either back and forth or in a circular pattern, was implemented in order to mitigate these problems (Petersen 1986; Berland et al. 1995). However, the implementation proposed by Digman et al. (2008) goes even farther in resolving the photo-bleaching and diffusion time issues, while providing another essential piece of information, i.e. the image.
sN&B analysis
In sN&B analyses, advantage is taken of very rapid scanning mirrors in laser scanning confocal or two-photon microscopes. A field of view (FOV) is scanned multiple times, 50–100, providing 50–100 values for the fluorescence intensity at every pixel in the FOV. The key is to scan faster than diffusion, such that the molecules present in the excitation (two-photon excitation) or observation (confocal) volume at a given pixel do not average out during the acquisition. Thus, the pixel dwell-time is generally set at 40–50 μs, which is generally speaking much shorter than the diffusion time of proteins in live cells, even that of monomeric green fluorescent protein (GFP) (Medina and Schwille 2002). Then at each pixel, i, the average fluorescence intensity, <F > i, and the variance of the intensity distribution over the 50–100 values, σ2i, are calculated. Then using Eqs. (3) and (4), the shot noise-corrected number of molecules and molecular brightness at each pixel, ni and ei, respectively, are computed. A spatial map of the number of molecules and their molecular brightness are thus obtained. We note that because both the pixel dwell-time (40 μs) and the line time (~ 12 ms) are known, this hidden time information can be exploited to obtain the diffusion characteristics of the bio-molecules on these timescales (Digman et al. 2005, 2013), although the spatial information is lost. It has been shown recently that it is possible to filter the arrival time of photons from differently bright species between two locations (Hinde et al. 2016) and thus distinguish both the stoichiometry and the diffusion of different species in a cell.
Determination of absolute concentrations by sN&B
As noted above, most of the implementations of sN&B analyses, regardless of which type, have focused on the brightness aspect, i.e. determining bio-molecular complex stoichiometry. The vast majority of these studies have been carried out in mammalian cell lines using cells either stably or transiently transfected with the gene encoding the sequence for the fluorescent protein fusion of the protein of interest. In these instances, the absolute concentration of the fluorescent bio-molecule might be of interest to compare to endogenous levels (if known), but would not have any particular physiological significance. Hence, in many cases, the concentration was not reported in such studies.
However, from the perspective of attempting to characterize biological networks, and reveal their functional mechanisms, knowledge of the absolute concentration of the proteins in the network is essential. Biochemical equilibria are based on mass action. No in vitro binding study could be undertaken without knowledge of the concentrations of the components. Likewise, for a complete characterization of the biochemical network of interest in a live cell environment, the localization and local concentrations of the interacting molecules must be known. Moreover, for the values of intracellular concentrations of proteins of interest to be relevant to network function, the fluorescently labeled proteins in the sample must be expressed at endogenous levels, and no endogenous protein can be present, i.e., the fluorescent protein fusions must replace the endogenous unlabeled proteins. In addition, the fluorescent tag must not interfere with the protein’s function. All of these requirements place strong restrictions on which bio-molecular networks can be considered to be amenable to such studies. First and foremost, for endogenous level expression and replacement of any endogenous protein, the sequences for the fluorescent protein fusions replace the wild-type gene at the natural locus in the organism’s chromosome(s), limiting such studies at present, to genetically tractable organisms. While some higher eukaryotes, such as Drosophila melanogaster, Caenorhabditis elegans or the zebrafish Danio rerio, are genetically tractable in principle, the sheer number of control cell lines that must be constructed practically limits such studies to bacteria and yeast. Future progress in systematic gene editing approaches (Horvath and Barrangou 2010) will likely allow the extension of the sN&B approach for determining absolute concentrations to higher eukaryotes.
Stochastic gene expression—counting GFP expressed from promoter fusions
In bacteria, sN&B analysis has been used in the study of stochastic gene expression (Declerck and Royer 2013). In this application, the coding sequence for GFP is inserted downstream of a promoter of interest, and expression of free monomeric GFP from the promoter is monitored as a function of time or growth conditions by determining the absolute concentration of the free GFP expressed. Using an isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible promoter system in Bacillus subtilis, Ferguson and co-workers validated the sN&B approach for the study of stochastic gene expression and demonstrated that the level of biological noise in the repressed state, arising from stochastic dissociation of the LacI repressor from its operator sequence, was much larger than that for the induced promoter (Fig.1) (Ferguson et al. 2011). The absolute values obtained in sN&B analysis are highly useful for modeling gene expression networks because estimation of initial concentrations for ordinary differential equation (ODE) and partial differential equation (PDE) models is an ill-determined inverse problem.
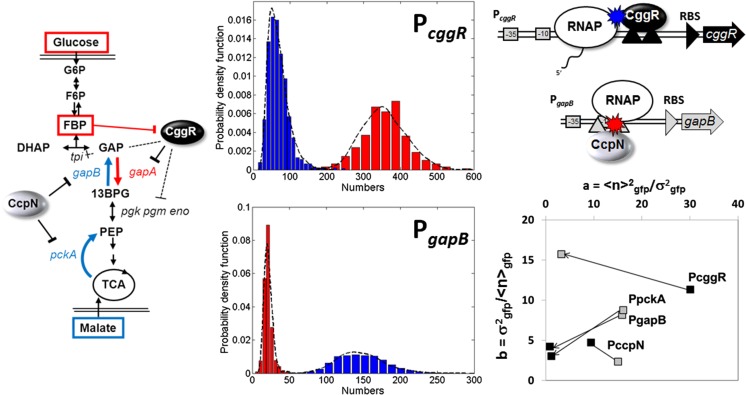
Further investigation of expression noise from two promoters implicated in the switch between glycolysis and gluconeogenesis in B. subtilis revealed how the known detailed biophysical mechanisms of repression by their specific repressor proteins at a given promoter impacted the expression variability (Fig. 2) (Ferguson et al. 2012). In this system, the CggR protein represses transcription from the gapA (also called cggR) promoter when the bacteria are grown on a non-glycolytic carbon source such as malate. This promoter controls expression of an entire operon, including other enzymes in the glycolytic pathway, as well as the repressor CggR itself, setting up a negative feedback loop. CggR is induced by the binding of fructose bis-phosphate, a degradation product of glucose, such that when glucose becomes available in the environment, the enzymes required to use it as an energy source are expressed. The target operator site for the CggR repressor is found downstream of the transcription start site, such that CggR works as a roadblock against elongating RNA polymerase molecules. In absence of glucose, very little CggR is expressed, such that it exists at extremely low concentrations. Thus, when CggR dissociates stochastically from its operator site, re-association is quite slow. This allows many RNA polymerase molecules to bind, initiate and elongate, prior to the rebinding of the repressor. Such a scenario results in a large size for the transcriptional bursts, with a low burst frequency.
Fig. 2.
Mechanisms controlling biological noise in a gene expression network deduced from sN&B analysis. Left: Pathway for glycolysis and gluconeogenesis in B. subtilis. Expression of the gapA operon is repressed by CggR when cells are grown on malate, whereas expression of the pckA and gapB promoters is repressed by CcpN in cells grown in glucose medium. Middle: Histograms of the number of green fluorescent protein (GFP) molecules in the excitation volume expressed from the PgapB (bottom) and the PcggR(gapA) (top) promoters, as noted, in glucose (red) and in malate (blue). Note the difference in scales on the x-axes. Also, note that the histograms are not corrected for contributions from auto-fluorescence. Once this was done, the average number of molecules of GFP in the excitation volume expressed from PgapB was only 3 on average. Given the size of the volume (0.07 fL) this corresponds to a concentration of 20 nM. Top, right: The Road Block mechanism for Cggr repression of PcggR and the Hold Back mechanism for CcpN repression of PgapB . Schematics and data are from Ferguson et al. (2012). Bottom right: Noise characteristics for the three promoters, plus that of PccpN, which is known not to change between glucose and malate
The gapB promoter, on the other hand, is very strongly catabolite repressed, with almost no expression when B. subtilis is cultured in glucose medium. Deletion of the repressor CcpN leads to significant growth defects in glucose medium. The operator sequence for CcpN overlaps the promoter, and biochemical evidence suggests that CcpN interacts directly with the RNA polymerase. As a result, CcpN dissociates occasionally and stochastically from the DNA or the polymerase; however, its concentration is not particularly low, such that rebinding is rapid. Rather than a “road block” mechanism, CcpN represses transcription via a “hold back” mechanism in which CcpN interaction with the RNA polymerase prevents it from initiating transcription. This scenario leads not only to a low frequency of transcriptional bursts, but also to low burst size and to the overall extremely strong catabolite repression of this promoter. Such mechanistic insight into a gene expression network was possible in this study because absolute concentrations allowed determination of absolute biological noise levels. This information, combined with biophysical and biochemical information previously determined in vitro, yielded a mathematical model for the mechanisms of stochastic gene expression in the central carbon metabolism of this bacterium. Current work in this area involves determination of the concentrations, stoichiometries and heterologous interactions of the repressor molecules, themselves, as well as the time dependence of gene expression after nutrient switches (N. Declerck, personal communication). SN&B technology has been used in bacteria to determine the number of the sporulation-specific DNA translocase in clusters associated to the sporulation septum in B. subtilis (Fiche et al. 2013), for measuring the absolute concentration and stoichiometry of the type IV restriction endonuclease, Mrr, in Escherichia coli (Bourges et al. 2017) and to quantify expression from engineered gene circuits in E. coli (Guiziou et al. 2016).
Limitations of sN&B for measurement of absolute concentrations
As powerful and apparently straightforward as sN&B measurements may appear, there are a number of aspects that must be carefully controlled, if the values of the absolute concentrations are to be accurate and precise.
Dynamics of molecules and organelles
For fluctuations in fluorescence to be observed beyond the shot noise of the detector, the molecules of interest must move on the timescale of the sampling frequency. As noted above, in sN&B, this is quite slow, since each pixel is interrogated once every raster scan, which for a 40-μs pixel dwell-time and 256 × 256-pixel FOV is approximately every 3 s. Regions containing fluorescent molecules which are immobile on this timescale will yield a shot noise-corrected brightness value of zero (or an uncorrected brightness value of 1) (Digman et al. 2008). Likewise, if entire regions move, such as villi, membrane protrusions or entire nuclei, for example, then very large brightness values are measured around the edges of the moving organelle. Pixels or regions of interest displaying such behavior should be discarded, although the brightness can provide a gross estimate of how many molecules are in the organelle. Co-variance of fluorescence in two different channels for two different proteins (cross-brightness analysis) has been used to estimate the number of vinculin and paxillin molecules in focal adhesion complexes (Digman et al. 2009). Of course, the requirement for molecular motion on the seconds timescale also means that sN&B cannot be used to determine the stoichiometry or the concentration of immobile proteins, at least not directly from the fluctuations. Nonetheless, unlike point or even circular scanning FCS, since the timescale of molecular diffusion must be on the order of seconds or faster, even very slowly moving particles (such as membrane proteins, or proteins interacting with DNA and dissociating stochastically) can be measured by scanning N&B.
Upper concentration limits
The amplitude of fluctuations in fluorescence intensity (relative to the mean fluorescence) are inversely proportional to the concentration of fluorescent molecules. This sets an upper limit to the concentrations which can be measured directly using Eqs. (1, 3). The larger the molecular brightness, the higher this limit will be. Generally, this limit is found to be in the range of 1–10 μM. However, high concentrations, even of immobile particles, can be determined (provided that photo-bleaching is largely avoided). This extension of the upper limits of sN&B is performed as per Eq. (6) as shown in Fig. 3. First, the molecular brightness of the fluorophore in question (GFP, for example, eF) is determined under ideal concentration conditions and in the same cell lines and imaging conditions as used for the fusion proteins of interest. This value can be determined from multiple FOV with millions of pixels and is thus extremely accurate and precise. The cells expressing the protein of interest are then imaged by sN&B, and the pixel-based values of the average fluorescence of the protein fusion, <F > FP, are determined. Generally, these pixel-based average intensity values are averaged over all pixels in a region of interest (ROI), which can be each cell or nucleus or even in all the cells of the FOV, providing the average intensity inside each ROI, <F > FP,ROI. Then, because the molecular brightness of a single GFP molecule is known, one can use Eq. (6), and the average intensity in the ROI, <F > FP,ROI, along with the value of the calibrated excitation volume, Veff, and Avogadro’s number, NA, to calculate the absolute concentration of molecules in the ROI (in GFP monomer units). This can be done for very high concentrations, to the extent that the detector response remains linear with the concentration (Ferguson et al. 2011). To extend this range, the excitation intensity can be decreased. Moreover, if photo-bleaching can be avoided, this approach can be used to determine the number of immobile particles as well (in monomeric units of the fluorophore).
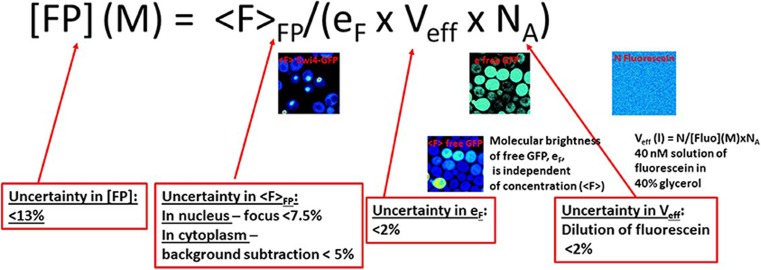
Fig. 3.
Schematics of the calibration method for measuring the absolute protein concentration. The absolute concentration of the fusion protein of interest ([FP]) is obtained from the average fluorescence intensity (in the appropriate region of interest) for the protein of interest (<F>FP) using the value of eF, the brightness of free monomeric GFP measured by sN&B analysis in the same background strain, on the same day, and under the same conditions, and the effective volume from which photons are collected, Veff, determined using a solution of fluorescein of known concentration in glycerol. These latter two parameters are highly precise and accurate. Thus, the error on the absolute value of the concentration of the protein of interest resides for > 98% in the ability to accurately and precisely measure the average intensity. NA Avogadro’s number
While Eq. (6) is valid at high concentrations, it is also valid across the entire detection range. Moreover, the uncertainty on the value of the average fluorescence in an ROI, <F > FP,ROI, is much lower than that of average number in an ROI, <n > ROI, calculated using Eq. (3), and averaged over all pixels in the ROI. This is especially true for low concentrations (low photon counts). Thus, the best approach to determining absolute concentrations in live cells using sN&B is to calibrate the system. First, one determines the molecular brightness of the monomeric form of the fluorophore, eF, in live cells under exactly the same imaging conditions as those used for the protein of interest. Then, in the absence of any specific geometric considerations (Macdonald et al 2010; Ferguson et al, 2011; Hur and Mueller 2015), the excitation volume is determined by sN&B (not FCS), using a solution of known concentration of fluorophore dissolved in at least 40% glycerol (to slow diffusion in to the sN&B timescale of >> 40 μs ) (Eq. 8, below).
| 8 |
Finally, eF, along with the value of the excitation volume, Veff, is used to calculate the absolute concentration of the protein of interest in units of moles per liter using Eq. 6. (Fig. 3). In bacteria, which are small relative to the effective volume, Brownian dynamics simulations were used to determine the value of Veff (Ferguson et al. 2011).
For example, if by sN&B, <F > F/eF = n = 12 molecules of a 40 nM solution of fluorescein in glycerol in the effective volume, Veff, then the effective volume is 0.5 fL. Figure 3 presents a schematic detailing the uncertainties in the determination of absolute protein concentrations in live cells by sN&B.
There are of course also lower concentration limits to sN&B quantification of proteins of interest. Theoretically, this limit should be a single molecule in a cell or ROI (not in the effective volume). We note that the concentration of one molecule in a volume of 1 fL (the volume of a bacterium) corresponds to 1.7 nM. Practically, however, the lower limit for reasonable quantification by sN&B is set by the level of cellular auto-fluorescence. We have found that using two-photon excitation at wavelengths between 0.95 and 1 μm, greatly reduces the contribution of auto-fluorescence. While this wavelength range is not optimal for GFP excitation, the ratio of GFP fluorescence to auto-fluorescence is optimized under these excitation conditions. Cellular auto-fluorescence originates primarily from enzyme co-factors NAD(P)H (in bound and free form, with different quantum yields) and FAD. The two-photon cross sections are significantly blue shifted with respect to double the one-photon excitation spectra, such that at 1 μm excitation, auto-fluorescence is minimized (Huang et al. 2002).
The lower detection limit in sN&B—endogenous levels of a scarce protein in bacteria
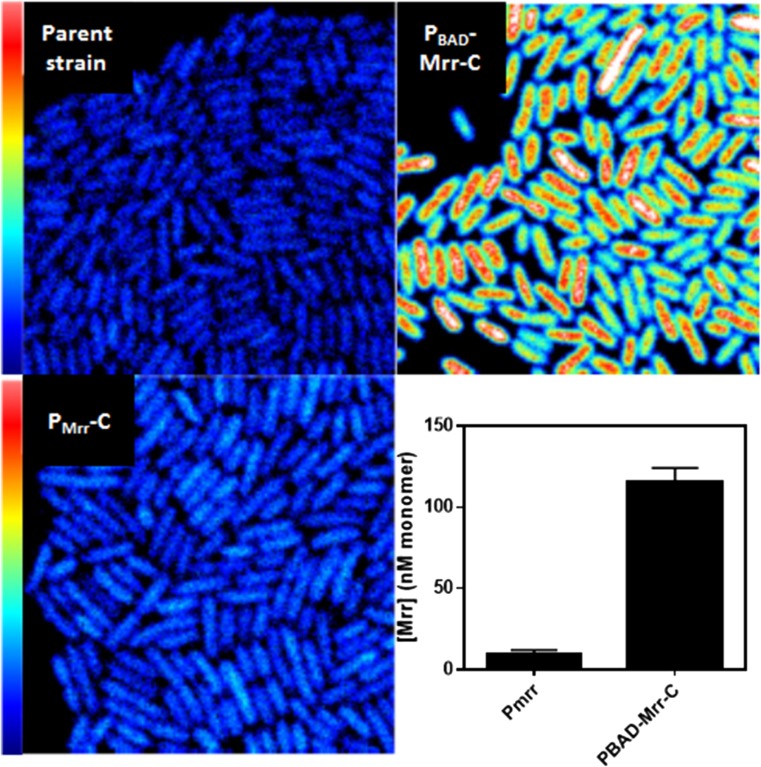
In a recent study aimed at determining the molecular mechanisms underlying the effect of pressure on the activity of a type IV restriction endonuclease, Mrr, in certain strains of E. coli, we compared the concentrations of Mrr–GFPmut2 (highly fluorescent GFP mutant protein) fusions expressed from the natural promoter, Pmrr, in the E. coli chromosome, and from the replacement of the natural promoter with an arabinose-inducible (PBAD) promoter. From Fig. 4 it is clear that Mrr–GFP could be detected above the auto-fluorescence of the parent strain, although the levels obtained when the PBAD promoter was induced with 0.002% arabinose were clearly much higher. To obtain average concentrations, the average intensity from all pixels in all the cells from eight FOV of the background strain were subtracted from the average intensity of all pixels in all cells from eight FOV of the two strains expressing Mrr–GFP to yield the background corrected average intensity of Mrr, <F > Mrr_cor. Then the corrected molecular brightness, <e > GFP, sample, and the corrected number of GFP molecules, <n > GFP, sample, were calculated as follows:
| 9 |
| 10 |
where Fsample and esample are the uncorrected average fluorescence and brightness from all cells and all pixels from the strains of interest, and ebg and fbg are the brightness and the average fluorescence. respectively, from the parent strain expressing no GFP. The detection limit in this case is 6 nM, although this value represents the average concentration over all cells. Individual cell information is lost. This method of background auto-fluorescence subtraction is based on the assumption that the auto-fluorescence and that of the GFP are both independent Poisson distributions. Current work aimed at developing methodology for subtraction of auto-fluorescence at the single cell level will be published in the near future (Dorses et al., submitted). The sN&B studies on Mrr–GFP in E. coli MG1655 strains before and after pressure treatment (Bourges et al. 2017) demonstrated that its endonuclease function is activated via pressure-induced tetramer to dimer dissociation, followed by subsequent interaction with and cleavage of the E. coli chromosome, leading to the well-documented pressure-induced SOS response (Aertsen and Michiels 2005).
Fig. 4.
Detection limits for GFPmut2 in 2-photon sN&B. Top left: Average auto-fluorescence image from 50 sN&B scans of the parent MG1655 strain of Escherichia coli. Bottom left: Average fluorescence image from 50 sN&B scans of the MG1655 strain of E. coli expressing a GFPmut2 fusion of the Mrr (type IV restriction endonuclease) protein from its natural promoter at its natural locus in the E. coli chromosome. The endogenous gene has been replaced at that locus by the sequence encoding the FP-fusion of Mrr. Top right: Average fluorescence image from 50 sN&B scans of the MG1655 strain of E. coli expressing a GFPmut2 fusion of the Mrr protein from an arabinose-inducible promoter replacing the natural promoter at its natural locus in the E. coli chromosome. Arabinose concentration was 0.002%. Bottom right: Bar graphs of the absolute concentration of Mrr in the GFPmut2 monomer units expressed from its natural promoter, PMrr (left) and the PBAD promoter (right). The full scale in all three fluorescence images is 1.5 photon counts. Full x–y scale is 20 × 20 μm. Data are taken from Bourges et al. (2017)
Conclusions
With proper controls and careful measurements, sN&B constitutes a powerful methodology for obtaining some of the most important and elusive quantitative information concerning bio-molecules in live cells, namely their concentrations. Using cross-sN&B (Digman et al. 2009) in two-channel detection mode, eluded to here briefly, can yield the concentration of heterologous bio-molecular complexes. The fact that sN&B returns an image, rather than numerical values at a few individual points within cells, opens the possibilities of measuring cell-to-cell variations in protein levels and stochastic gene expression, and provides an overview of the localization of proteins of interest and how this changes with conditions. Making such measurements as a function of growth conditions or internal or external signals provides quantitative input to mathematical models of bio-molecular circuits, for which only qualitative parts-level epistasis descriptions are currently available. While such studies are currently limited to simple, genetically tractable organisms such as bacteria and yeast, much insight will be gained by complete quantitative characterization of the mechanisms underlying any number of functional networks in these model organisms, particularly in light of the existence of homologous systems in higher organisms. Nonetheless, we predict that as gene editing methodology improves, sN&B will provide information on concentration and stoichiometry of the key proteins involved in important cell state transitions, such as differentiation and transformation, in higher eukaryotic cell lines. Such insight will be invaluable for developing new strategies to combat a large number of human diseases.
Compliance with ethical standards
Conflict of interest
Christina Papini declares that she has no conflict of interest. Catherine A. Royer declares that she has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- Aertsen A, Michiels CW. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol Microbiol. 2005;58:1381–1391. doi: 10.1111/j.1365-2958.2005.04903.x. [DOI] [PubMed] [Google Scholar]
- Bacia K, Schwille P. A dynamic view of cellular processes by in vivo fluorescence auto- and cross-correlation spectroscopy. Methods. 2003;29:74–85. doi: 10.1016/S1046-2023(02)00291-8. [DOI] [PubMed] [Google Scholar]
- Berland KM, So PT, Gratton E. Two-photon fluorescence correlation spectroscopy: method and application to the intracellular environment. Biophys J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borejdo J, Morales MF. Fluctuations in tension during contraction of single muscle fibers. Biophys J. 1977;20:315–334. doi: 10.1016/S0006-3495(77)85552-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourges AC, Torres Montaguth OE, Ghosh A, Tadesse WM, Declerck N, Aertsen A, Royer CA (2017) High pressure activation of the Mrr restriction endonuclease in Escherichia coli involves tetramer dissociation. Nucleic Acids Res 45(9):5323–5332 [DOI] [PMC free article] [PubMed]
- Chen Y, Müller JD. Determining the stoichiometry of protein heterocomplexes in living cells with fluorescence fluctuation spectroscopy. Proc Natl Acad Sci USA. 2007;104:3147–3152. doi: 10.1073/pnas.0606557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Müller JD, So PT, Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Declerck N, Royer C (2013) Interactions in gene expression networks studied by two-photon fluorescence fluctuation spectroscopy. Methods Enzymol 519:203–30 [DOI] [PubMed]
- Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E. Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J. 2005;89:1317–1327. doi: 10.1529/biophysj.105.062836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Dalal R, Horwitz AF, Gratton E. Mapping the number of molecules and brightness in the laser scanning microscope. Biophys J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Wiseman PW, Choi C, Horwitz AR, Gratton E. Stoichiometry of molecular complexes at adhesions in living cells. Proc Natl Acad Sci USA. 2009;106:2170–2175. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Stakic M, Gratton E. Raster image correlation spectroscopy and number and brightness analysis. Methods Enzymol. 2013;518:121–144. doi: 10.1016/B978-0-12-388422-0.00006-6. [DOI] [PubMed] [Google Scholar]
- Ferguson ML, Le Coq D, Jules M, Aymerich S, Declerck N, Royer CA. Absolute quantification of gene expression in individual bacterial cells using two-photon fluctuation microscopy. Anal Biochem. 2011;419:250–259. doi: 10.1016/j.ab.2011.08.017. [DOI] [PubMed] [Google Scholar]
- Ferguson ML, Le Coq D, Jules M, Aymerich S, Radulescu O, Declerck N, Royer CA. Reconciling molecular regulatory mechanisms with noise patterns of bacterial metabolic promoters in induced and repressed states. Proc Natl Acad Sci USA. 2012;109:155–160. doi: 10.1073/pnas.1110541108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiche JB, Cattoni DI, Diekmann N, Langerak JM, Clerte C, Royer CA, Margeat E, Doan T, Nöllmann M (2013) Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by Superresolution microscopy. PLoS Biol 11(5): e1001557. 10.1371/journal.pbio.1001557 [DOI] [PMC free article] [PubMed]
- García-Marcos A, Sánchez SA, Parada P, Eid J, Jameson DM, Remacha M, Gratton E, Ballesta JPG. Yeast ribosomal stalk heterogeneity in vivo shown by two-photon FCS and molecular brightness analysis. Biophys J. 2008;94:2884–2890. doi: 10.1529/biophysj.107.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin AG, Costantino S, Lorenzo L-E, Swift JL, Sergeev M, Ribeiro-da-Silva A, De Koninck Y, Wiseman PW. Revealing protein oligomerization and densities in situ using spatial intensity distribution analysis. Proc Natl Acad Sci. 2011;108:7010–7015. doi: 10.1073/pnas.1018658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golebiewska U, Johnston JM, Devi L, Filizola M, Scarlata S. Differential response to morphine of the oligoemeric state of mu-opioid in the presence of delta-opioid receptors. Biochemistry. 2011;50:2829–2837. doi: 10.1021/bi101701x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiziou S, Sauveplane V, Chang H-J, Cler EC, Declerck N, Jules M, Bonnet J. A part toolbox to tune genetic expression in Bacillus subtilis. Nucleic Acids Res. 2016;44:7495–7508. doi: 10.1093/nar/gkw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haustein E, Schwille P. Fluorescence correlation spectroscopy: novel variations of an established technique. Annu Rev Biophys Biomol Struct. 2007;36:151–169. doi: 10.1146/annurev.biophys.36.040306.132612. [DOI] [PubMed] [Google Scholar]
- Hellriegel C, Caiolfa VR, Corti V, Sidenius N, Zamai M. Number and brightness image analysis reveals ATF-induced dimerization kinetics of uPAR in the cell membrane. FASEB J. 2011;25:2883–2897. doi: 10.1096/fj.11-181537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen J, Hur KH, Saunders CA, Luxton GWG, Mueller JD. Quantitative brightness analysis of protein oligomerization in the nuclear envelope. Biophys J. 2017;113:138–147. doi: 10.1016/j.bpj.2017.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrick-Davis K, Grinde E, Lindsley T, Cowan A, Mazurkiewicz JE. Oligomer size of the serotonin 5-hydroxytryptamine 2C (5-HT2C) receptor revealed by fluorescence correlation spectroscopy with photon counting histogram analysis: evidence for homodimers without monomers or tetramers. J Biol Chem. 2012;287:23604–23614. doi: 10.1074/jbc.M112.350249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinde E, Yokomori K, Gaus K, Hahn KM, Gratton E. Fluctuation-based imaging of nuclear Rac1 activation by protein oligomerisation. Sci Rep. 2014;4:4219. doi: 10.1038/srep04219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinde E, Pandžić E, Yang Z, Ng IHW, Jans DA, Bogoyevitch MA, Gratton E, Gaus K. Quantifying the dynamics of the oligomeric transcription factor STAT3 by pair correlation of molecular brightness. Nat Commun. 2016;7:11047. doi: 10.1038/ncomms11047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink MA, Shah K, Russinova E, de Vries SC, Visser AJWG. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and brassinosteroid insensitive 1 receptor oligomerization. Biophys J. 2008;94:1052–1062. doi: 10.1529/biophysj.107.112003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(80):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Huang S, Heikal AA, Webb WW. Two-photon fluorescence spectroscopy and microscopy of NAD(P)H and Flavoprotein. Biophys J. 2002;82:2811–2825. doi: 10.1016/S0006-3495(02)75621-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur K-H, Mueller JD (2015) Quantitative brightness analysis of fluorescence intensity fluctuations in E. coli. PLoS One 10:e0130063. 10.1371/journal.pone.0130063 [DOI] [PMC free article] [PubMed]
- James NG, Digman MA, Gratton E, Barylko B, Ding X, Albanesi JP, Goldberg MS, Jameson DM. Number and brightness analysis of LRRK2 oligomerization in live cells. Biophys J. 2012;102:L41–L43. doi: 10.1016/j.bpj.2012.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James NG, Digman MA, Ross JA, Barylko B, Wang L, Li J, Chen Y, Mueller JD, Gratton E, Albanesi JP, Jameson DM. A mutation associated with centronuclear myopathy enhances the size and stability of dynamin 2 complexes in cells. Biochim Biophys Acta. 2014;1840:315–321. doi: 10.1016/j.bbagen.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kask P, Palo K, Ullmann D, Gall K. Fluorescence-intensity distribution analysis and its application in biomolecular detection technology. Proc Natl Acad Sci USA. 1999;96:13756–13761. doi: 10.1073/pnas.96.24.13756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Barylko B, Johnson J, Mueller JD, Albanesi JP, Chen Y. Molecular brightness analysis reveals phosphatidylinositol 4-kinase IIβ association with clathrin-coated vesicles in living cells. Biophys J. 2012;103:1657–1665. doi: 10.1016/j.bpj.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Y, Li M, Carpenter MA, McDougle RM, Luengas EM, Macdonald PJ, Harris RS, Mueller JD. APOBEC3 multimerization correlates with HIV-1 packaging and restriction activity in living cells. J Mol Biol. 2014;426:1296–1307. doi: 10.1016/j.jmb.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald P, Johnson J, Smith E, Chen Y, Mueller JD (2013) Brightness analysis. Methods Enzymol 518:71–98 [DOI] [PubMed]
- Macdonald PJ, Chen Y, Wang X, Chen Y, Mueller JD. Brightness analysis by Z-scan fluorescence fluctuation spectroscopy for the study of protein interactions within living cells. Biophys J. 2010;99:979–988. doi: 10.1016/j.bpj.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D, Elson E, Webb WW. Fluorescence correlation spectroscopy. 2. Experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- Medina MA, Schwille P. Fluorescence correlation spectroscopy for the detection and study of single molecules in biology. BioEssays. 2002;24:758–764. doi: 10.1002/bies.10118. [DOI] [PubMed] [Google Scholar]
- Ming AYK, Yoo E, Vorontsov EN, Altamentova SM, Kilkenny DM, Rocheleau JV. Dynamics and distribution of Klothoβ (KLB) and fibroblast growth factor receptor-1 (FGFR1) in living cells reveal the fibroblast growth factor-21 (FGF21)-induced receptor complex. J Biol Chem. 2012;287:19997–20006. doi: 10.1074/jbc.M111.325670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Claus J, Jovin TM, Arndt-Jovin DJ. Distribution of resting and ligand-bound ErbB1 and ErbB2 receptor tyrosine kinases in living cells using number and brightness analysis. Proc Natl Acad Sci USA. 2010;107:16524–16529. doi: 10.1073/pnas.1002642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P, Hayles J. The cell in an era of systems biology. Cell. 2011;144:850–854. doi: 10.1016/j.cell.2011.02.045. [DOI] [PubMed] [Google Scholar]
- Patel RC, Kumar U, Lamb DC, Eid JS, Rocheville M, Grant M, Rani A, Hazlett T, Patel SC, Gratton E, et al. Ligand binding to somatostatin receptors induces receptor-specific oligomer formation in live cells. Proc Natl Acad Sci USA. 2002;99:3294–3299. doi: 10.1073/pnas.042705099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen NO. Scanning fluorescence correlation spectroscopy I. Theory and simulation of aggregation measurements. Biophys J. 1986;49:809–815. doi: 10.1016/S0006-3495(86)83709-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presman DM, Levi V, Pignataro OP, Pecci A. Melatonin inhibits glucocorticoid-dependent GR-TIF2 interaction in newborn hamster kidney (BHK) cells. Mol Cell Endocrinol. 2012;349:214–221. doi: 10.1016/j.mce.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Qian H, Elson EL. On the analysis of high order moments of fluorescence fluctuations. Biophys J. 1990;57:375–380. doi: 10.1016/S0006-3495(90)82539-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales T, Georget V, Malide D, Smirnov A, Xu J, Combs C, Knutson JR, Nicolas J-C, Royer CA. Quantitative detection of the ligand-dependent interaction between the androgen receptor and the co-activator, Tif2, in live cells using two color, two photon fluorescence cross-correlation spectroscopy. Eur Biophys J. 2007;36:153–161. doi: 10.1007/s00249-006-0095-1. [DOI] [PubMed] [Google Scholar]
- Ross JA, Digman MA, Wang L, Gratton E, Albanesi JP, Jameson DM. Oligomerization state of dynamin 2 in cell membranes using TIRF and number and brightness analysis. Biophys J. 2011;100:L15–L17. doi: 10.1016/j.bpj.2010.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Chen Y, Müller J, Barylko B, Wang L, Banks HB, Albanesi JP, Jameson DM. Dimeric endophilin A2 stimulates assembly and GTPase activity of dynamin 2. Biophys J. 2011;100:729–737. doi: 10.1016/j.bpj.2010.12.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Andres A, Chen Y, Müller JD. Molecular brightness determined from a generalized form of Mandel’s Q-parameter. Biophys J. 2005;89:3531–3547. doi: 10.1529/biophysj.105.067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savatier J, Jalaguier S, Ferguson ML, Cavaillès V, Royer CA. Estrogen receptor interactions and dynamics monitored in live cells by fluorescence cross-correlation spectroscopy. Biochemistry. 2010;49:772–781. doi: 10.1021/bi9013006. [DOI] [PubMed] [Google Scholar]
- Schwille P, Rigler R (1997) Dual-color fluorescence cross-correlation spectroscopy for 72 [DOI] [PMC free article] [PubMed]
- Schwille P, Haupts U, Maiti S, Webb WW. Molecular dynamics in living cells observed by fluorescence correlation spectroscopy with one- and two-photon excitation. Biophys J. 1999;77:2251–2265. doi: 10.1016/S0006-3495(99)77065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeev M, Swift JL, Godin AG, Wiseman PW. Ligand-induced clustering of EGF receptors: a quantitative study by fluorescence image moment analysis. Biophys Chem. 2012;161:50–53. doi: 10.1016/j.bpc.2011.11.003. [DOI] [PubMed] [Google Scholar]
- Sergeev M, Godin AG, Kao L, Abuladze N, Wiseman PW, Kurtz I. Determination of membrane protein transporter oligomerization in native tissue using spatial fluorescence intensity fluctuation analysis. PLoS One. 2012;7:1–11. doi: 10.1371/journal.pone.0036215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc Natl Acad Sci USA. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter BD, Huff JM, Wiegraebe W, Schwartz JW, Li R. SAM domain-based protein oligomerization observed by live-cell fluorescence fluctuation spectroscopy. PLoS One. 2008;3:e1931. doi: 10.1371/journal.pone.0001931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift JL, Godin AG, Doré K, Freland L, Bouchard N, Nimmo C, Sergeev M, De Koninck Y, Wiseman PW, Beaulieu J-M. Quantification of receptor tyrosine kinase transactivation through direct dimerization and surface density measurements in single cells. Proc Natl Acad Sci USA. 2011;108:7016–7021. doi: 10.1073/pnas.1018280108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetri V, Ossato G, Militello V, Digman MA, Leone M, Gratton E. Fluctuation methods to study protein aggregation in live cells: concanavalin a oligomers formation. Biophys J. 2011;100:774–783. doi: 10.1016/j.bpj.2010.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]