Abstract
Pesticide tolerance poses many challenges for pest control, particularly for destructive pests such as Bradysia odoriphaga. Imidacloprid has been used to control B. odoriphaga since 2013, however, imidacloprid resistance in B. odoriphaga has developed in recent years. Identifying actual and potential genes involved in detoxification metabolism of imidacloprid could offer solutions for controlling this insect. In this study, RNA-seq was used to explore differentially expressed genes in B. odoriphaga that respond to imidacloprid treatment. Differential expression data between imidacloprid treatment and the control revealed 281 transcripts (176 with annotations) showing upregulation and 394 transcripts (235 with annotations) showing downregulation. Among them, differential expression levels of seven P450 unigenes were associated with imidacloprid detoxification mechanism, with 4 unigenes that were upregulated and 3 unigenes that were downregulated. The qRT-PCR results of the seven differential expression P450 unigenes after imidacloprid treatment were consistent with RNA-Seq data. Furthermore, oral delivery mediated RNA interference of these four upregulated P450 unigenes followed by an insecticide bioassay significantly increased the mortality of imidacloprid-treated B. odoriphaga. This result indicated that the four upregulated P450s are involved in detoxification of imidacloprid. This study provides a genetic basis for further exploring P450 genes for imidacloprid detoxification in B. odoriphaga.
Introduction
The sciarid fly Bradysia odoriphaga is a serious crop pest with a wide range of more than 30 plant species from seven families1,2. The main host plant of B. odoriphaga is Chinese chive (Allium tuberosum Rottle ex Spreng). Chinese chive is a perennial vegetable with a high economic value and is grown over a vast geographic area from Asia through the Middle East, to Europe and North America, and is widely cultivated in China3–7. B. odoriphaga larvae usually gather in the roots, bulbs, and even in immature stems of Chinese chives, making the pest hard to control and allowing it to cause significant production losses of Chinese chives8. Failing to control B. odoriphaga could cause 50% yield reduction in Chinese chives at harvest time9,10.
Among the commonly used insecticides for control of B. odoriphaga larvae, neonicotinoids such as imidacloprid exhibit a good control efficacy11,12. Since 2013, insecticides containing imidacloprid as active ingredient has accounted for more than 60% of insecticides registered with the Ministry of Agriculture in China for B. odoriphaga control (www.chinapesticide.gov.cn). With the wide application of imidacloprid for B. odoriphaga control, increased imidacloprid resistance has recently developed in field populations of B. odoriphaga13–15. Therefore, in order to delay imidacloprid resistance development in B. odoriphaga, it is crucial to identify the imidacloprid detoxification metabolism related genes in B. odoriphaga, and then figure out the mechanism of imidacloprid detoxification.
The pathways by which imidacloprid is metabolized and detoxified in insects have been reported widely, highlighting the principal roles of cytochrome P450 enzyme and elevated gene expression levels of P450s in imidacloprid detoxification16,17. P450s are a superfamily of enzymes with many functions, including nutrition and xenobiotic detoxification by metabolizing a wide range of endogenous and exogenous compounds18,19. The over-expression of some P450 genes has been proven to be associated with increased metabolism of imidacloprid, including CYP6G1 (Drosophila melanogaster), CYP6D1 (Musca domestica), CYP6ER1, CYP6AY1 (Nilaparvata lugens), and CYP6CM1vQ (Bemisia tabaci)20–23. Therefore, the over-expression of P450s is a common mechanism to detoxify imidacloprid in insects. In addition, elevated P450s enzyme activity was found to be related with imidacloprid insensitivity in B. odoriphaga larvae in our previous study, which is currently the only report about the imidacloprid detoxification mechanism in B. odoriphaga14.
Although many ecological and physiological aspects of B. odoriphaga have been investigated in recent years1,24,25, the molecular mechanisms for insecticide metabolism in B. odoriphaga are still unexplored, especially those mechanisms related to metabolism of the commonly used insecticide imidacloprid. Recently, next-generation sequencing (NGS)-based RNA-Seq analysis has enhanced the efficiency and speed at which genes are discovered, especially for insects without a reference genome26. Moreover, transcriptome sequencing is an efficient way to uncover information about functional genes and differentially expressed genes in insects after insecticide treatment27,28.
In this study, we generated annotated transcriptome sequences and gene expression profiles that provide useful information for identifying genes involved in imidacloprid detoxification in B. odoriphaga. The assembled and annotated unigenes in the transcriptome were selected to explore key P450 genes related to imidacloprid detoxification, and to provide a molecular basis for exploring imidacloprid detoxification and functional analysis of these genes. In addition, the gene function of upregulated P450 transcripts were further explored through RNAi, which provided a better understanding of the role of these differentially expressed P450 genes in imidacloprid detoxification.
Results
Illumina sequencing and de novo assembly
According to the results of the bioassay, the LC50 value of imidacloprid against B. odoriphaga was 4.08 mg/L. Illumina sequencing and de novo assembly were performed by merging all samples of B. odoriphaga in the treatment and control groups, resulting in the generation of 43,453 total unigenes, a total length of 55,884,005 bp, an N50 length of 1,995 bp, and a mean length of 1,286.08 bp. The corresponding information of transcripts of B. odoriphaga is shown in Tables 1 and 2. The size distribution indicated that the lengths of 17,952 of the unigenes were more than 1000 bp (Fig. S1). The control 2 library produced the most data (52,347,280 clean reads), while the control 1 library produced the fewest clean reads (50,957,216). As a whole, all libraries exhibited good quality, with an average of 89.04% of the clean reads meeting base call quality at the Q30 standard. According to the alignments of sequences in the unigene library, the statistics of mapped reads in each sample are shown in Table 2. These clean reads were stored in the NCBI SRA database (http://www.ncbi.nlm.nih.gov/sra/) under the accession numbers SRX2939421.
Table 1.
Statistics for assembled unigenes.
| Total number | N50 length | Total Length | Max Length | Min Length | Average Length | |
|---|---|---|---|---|---|---|
| Unigene | 43453 | 1995 | 55884005 | 27155 | 301 | 1286.08 |
Table 2.
Aligning statistics of clean reads with assembled unigenes.
| Sample | Clean reads | Clean nucleotides (bp) | >Q30 (%) | (G + C) % | Mapped reads | Mapped ratio |
|---|---|---|---|---|---|---|
| Control 1 | 50957216 | 6369559907 | 87.93 | 41.50 | 45343685 | 88.98 |
| Control 2 | 52347280 | 6543269039 | 89.67 | 42.00 | 46485565 | 88.80 |
| Treatment group 1 | 51477692 | 6434711500 | 88.12 | 41.00 | 45542976 | 88.47 |
| Treatment group 2 | 52326200 | 6540775000 | 90.43 | 42.00 | 46503068 | 88.87 |
| All | 207108388 | 25888315446 | ||||
| mean | 89.04 | 41.63 | 45968824 | 88.78 |
Functional annotation of all unigenes
To classify the functions of predicted unigenes from the imidacloprid treatment and control groups, 21,705 of 43,453 total unigenes in B. odoriphaga were annotated using the NR (19442), SWISSPROT (16094), KOG (14723), KEGG (6737), and GO (15245) databases with a cut-off E-value of 10−5. For NR annotation, 44.74% of all unigenes provided a BLAST result, and the best-match results of the NR homologous species distribution are shown in Fig. S2. The sequences of B. odoriphaga showed 1820 matches with Aedes aegypti sequences followed by Vitrella brassicaformis (1394), Ae. albopictus (1147), and Culex quinquefasciatus (1143) (Fig. S2). Gene ontology (GO) analysis gives the representation of gene and gene product attributes across all libraries. In each of the three main categories (biological process, cellular component, and molecular function) of the GO classification, the terms “binding”, “cell part”, and “cellular process” were the most dominant, respectively (Fig. S3).
Expression variation of differential expression genes (DEG) related to imidacloprid detoxification
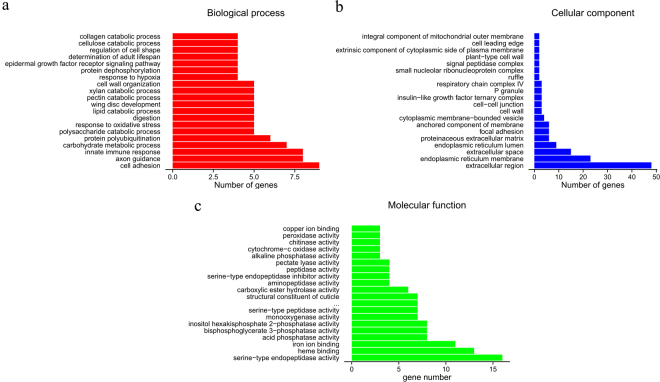
Based on the DEG analysis, out of 674 DEGs between imidacloprid treatment and control, 281 and 393 unigenes were upregulated and downregulated, respectively (Figs 1 and 2).
Figure 1.
The distribution of differential expression genes (DEGs) annotated in the Gene Ontology (GO) data library within the biological process (a), cellular component (b), and molecular function (c) categories. The x-axis indicates the number of unigenes sub-categories and the y-axis indicates the sub-categories.
Figure 2.
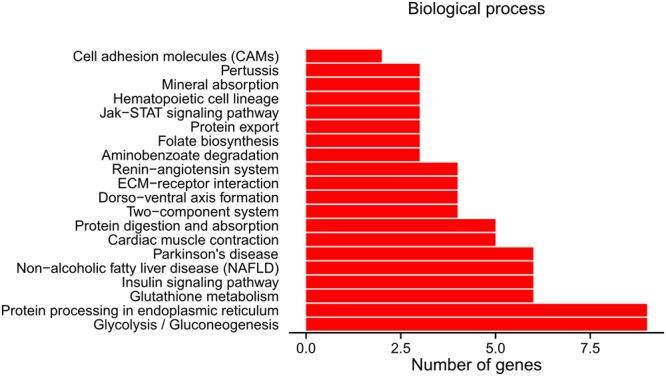
The distribution of pathways of differential expression genes (DEGs) annotated in the Kyoto Encyclopedia of Genes and Genomes (KEGG) data library. The x-axis indicates the number of unigenes sub-categories and the y-axis indicates the sub-categories.
In the comparison of imidacloprid treated and control larvae, 360 DEGs were annotated in the NR database. Among them, seven P450 unigenes, which are named with comp 30097, 30167, 34369, 36247, 38558, 40700 and 44013, were obtained according to the standard described above (Table 3). There are four P450 unigenes were upregulated (comp 30167, 38558, 40700 and 44013), while the other three P450 unigenes were downregulated (comp 30097, 34369 and 36247). Among the three main detoxifying enzymes in insect, namely P450s, carboxylesterase and GSTs, besides P450s, we also found a carboxylesterase unigene, but it showed down regulated. Thus, we selected these seven P450 unigenes as candidates to evaluate gene expression profiles.
Table 3.
The imidacloprid detoxification related P450s genes detected in the Bradysia odoriphaga unigenes dataset.
| Unigene ID | Accession ID | NR annotation | RNA-seq | |
|---|---|---|---|---|
| Log2 FC | P value | |||
| comp30167 | gi|170049288 | cytochrome P450 9b2 | 1.179 | 0.016 |
| comp38558 | gi|568255592 | cytochrome P450 | 0.398 | 0.033 |
| comp40700 | gi|157119361 | cytochrome P450 49a1 | 0.769 | 0.022 |
| comp44013 | gi|170049356 | cytochrome P450 12b1 | 0.772 | 0.004 |
| comp30097 | gi|916344537 | cytochrome P450 6BQ37 | −0.852 | 0.038 |
| comp34369 | gi|109603635 | cytochrome P450 9e2-like | −1.506 | 0.003 |
| comp36247 | gi|906471773 | Cytochrome P450 18a1 | −0.917 | 0.006 |
Quantitative real-time PCR validation of P450s
qRT-PCR was conducted to verify the expression level of seven P450 DEGs (4 upregulated and 3 downregulated) between the treatment group and the control group after imidacloprid treatment. The number of unigenes in each gene expression profile based on qRT-PCR (Fig. 3a,b) was similar to the RNA-Seq DEG gene expression data (Fig. S4).
Figure 3.
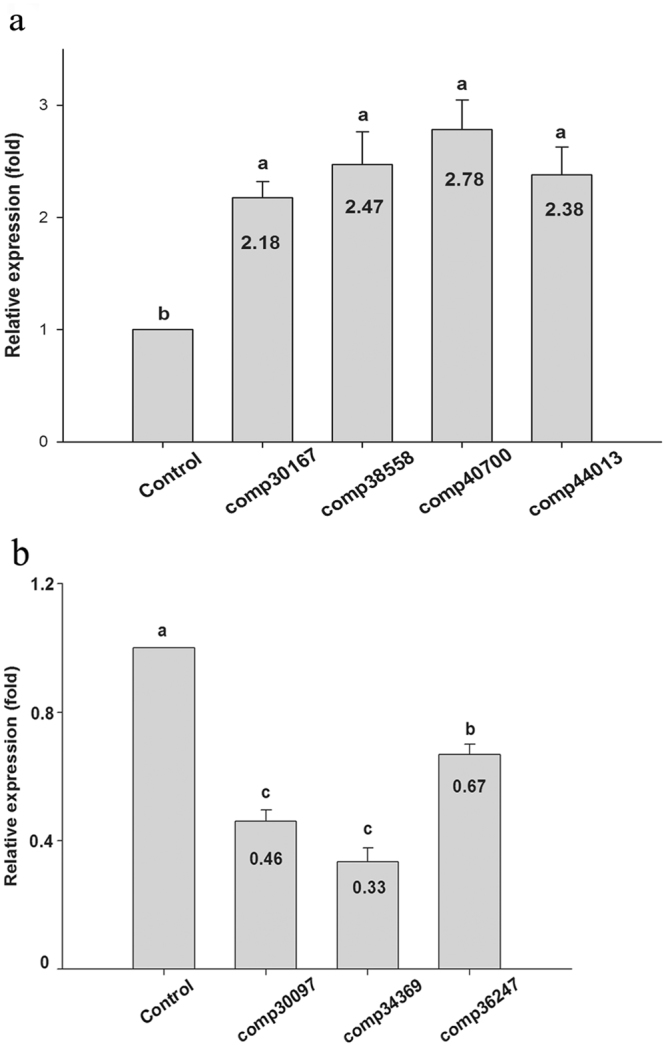
Relative expression levels of seven P450 unigenes in imidacloprid-treated B. odoriphaga. (a) Four upregulated P450s unigenes; (b) Three downregulated P450 unigenes. The transcription levels of the seven P450s unigenes were determined by quantitative real time PCR. Each bar indicates the mean of three biological replicates (±SE). 18 s was used as a reference gene. The different letters on the bars represent significant differences between treatment and control group based on Duncan’s multiple comparison test (P < 0.05). Numbers in each bar represent fold change compared with the control.
Functional analysis of P450s by RNAi
After feeding B. odoriphaga larvae individual P450 dsRNAs, the relative expression levels of mRNA were examined to investigate the knockdown efficiency of P450 gene expression. After feeding larvae the specific P450 dsRNAs, the expression of the target P450s gene decreased significantly compared to the control (Fig. 4a), indicating an effective silencing of P450s by RNAi in B. odoriphaga. Furthermore, after feeding larvae P450 dsRNAs, the mortality of B. odoriphaga larvae caused by imidacloprid increased significantly, with increases ranging from 18.93 to 35.78%, when treated with an LC50 dose of imidacloprid, Fig. 4b.
Figure 4.
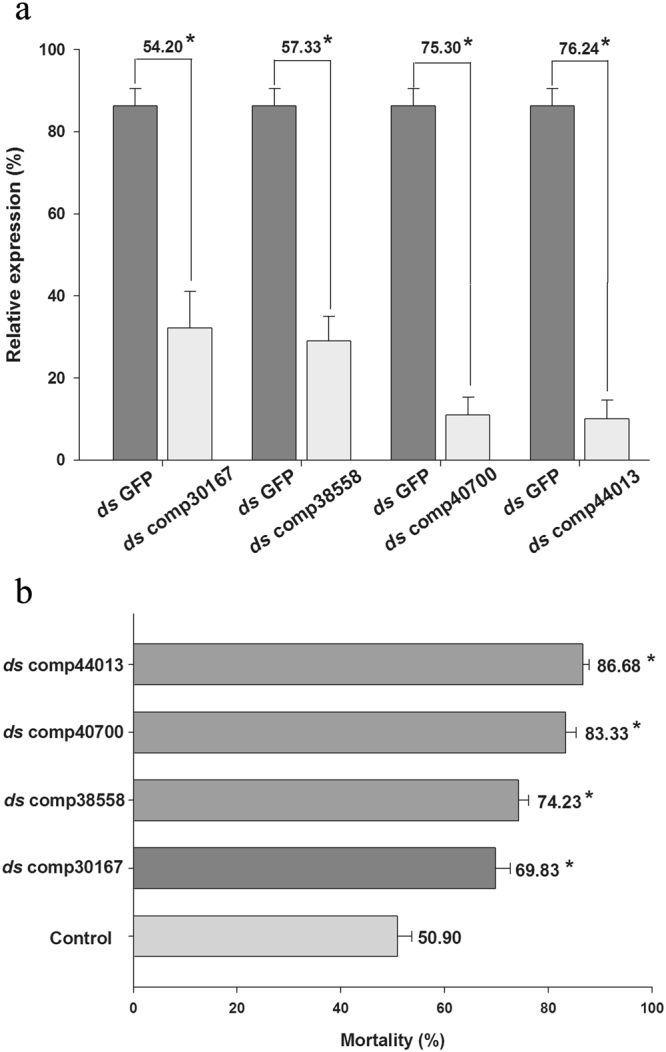
The dsRNA-mediated suppression of P450s transcript expression in B. odoriphaga fed on the artificial diet with dsRNA. The control group was fed with dsGFP. (a) Repressions of the transcripts of four P450s genes after larvae were fed an artificial diet with dsRNA for 48 h. The transcript levels of four P450s genes were examined using qRT-PCR, and 18 s was selected as a reference gene. (b) Insecticide bioassays were conducted 48 h after the uptake of P450 dsRNA by the standard contact and stomach bioassay method. Mortality was assessed 48 h after the insecticide treatments. Results are mean ± SE of three biological replications (n = 3). The asterisks on the bars indicate significant differences between the control and treatment group after P450s dsRNA uptake (Student’s t-test, P < 0.05).
Discussion
To discover and annotate the genes involved in imidacloprid detoxification, we used de novo assembly and analyzed differentially expressed genes in B. odoriphaga after imidacloprid treatment. This approach was necessary because B. odoriphaga does not have a reference genome. In this study, an Illumina platform was used to sequence the B. odoriphaga transcriptome. We identified 43,453 unigenes, and 19,442 of these unigenes were found to be above the assigned cutoff (e < 1e-5) after BLAST analysis using the NR database. The results were similar to previous RNA-seq studies on B. odoriphaga by Gao26, in which 47,578 unigenes were assembled using de novo sequencing. This subgroup of unigenes was further annotated using the GO and KEGG databases. These unigenes from transcriptome sequencing after imidacloprid treatment could provide useful information at the genomic level for controlling B. odoriphaga and other dipteran insects.
To discover the genes related to imidacloprid detoxification in B. odoriphaga, the functions of DEGs were annotated from the GO and KEGG databases according to the annotated information of all unigenes of B. odoriphaga. By searching the GenBank database, the four up-regulated unigene comp30167, 38558, 40700 and 44013 were annotated similar with CYP9b2 in Culex quinquefasciatus, P450s gene in Anopheles darlingi genome, CYP49a1 in Aedes aegypti and CYP12b1 in Culex quinquefasciatus, respectively. While it had been reported that CYP9b2 in Drosophila melanogaster was related to toxin resistance29, CYP49a1 expressed in Bombyx morito was involved in phoxim metabolism30 and CYP12b1 as a mitochondrial P450 may be related to biological process in Drosophila acanthoptera31. So far, no any imidaclodprid detoxification related annotation could be found for the four up-regulated P450 unigenes. Thus, in order to give an insight into the gene functions, we conducted qRT-PCR to verify the response of these DEGs after imidacloprid treatment. Moreover, RNAi was used for characterization of the roles of upregultaed P450 unigenes in detoxification of imidacloprid.
In addition, among the B. odoriphaga transcripts related to imidacloprid detoxification, four P450 differentially expressed transcripts were upregulated and three P450 transcripts were downregulated. In insects, more than half of all P450 genes belong to only two subfamilies, CYP 4 and CYP 632. In our study, one differentially expressed P450s transcripts belonged to CYP 6 and two belonged to the CYP 9 clade. Genes in the CYP 6 subfamily are involved in imidacloprid detoxification metabolism in Bemisia tabaci, Myzus persicae, Nilaparvata lugens, and Musca domestica33–36, while the CYP 9 subfamily genes are related to the oxidative metabolism of insecticides18.
To evaluate the validity of Illumina analysis after imidacloprid treatment, a total of seven differentially expressed unigenes, including four upregulated and three downregulated P450 transcripts, were selected to test expression changes by qRT-PCR between the imidacloprid treatment group and the control group. The variation in expression of these unigenes is shown in Fig. 3a.b. The qRT-PCR results of these seven P450 DEGs related to imidacloprid detoxification were identical to those obtained by DEG expression profiling.
Post-transcriptional gene silencing by RNA interference (RNAi) is a useful tool for examining the functions of individual genes37. As the reported imidacloprid metabolism resistance was mainly related with up-regulate of P450s gene38. The functions of the 4 up-regulated P450s unigenes in imidacloprid metabolism resistance were further studied by RNAi, while the roles of down-regulated genes need to be examined in other work. Oral delivery mediated RNAi method has been applied to the study of gene function, including in insecticide detoxification metabolism genes39–41. Then, by using a feeding based RNAi technique, we were able to reduce the four P450s transcripts levels by 54.20 to 76.24% in larvae fed on a diet containing P450s dsRNA, compared to larvae fed on a diet containing GFP dsRNA. Suppression of the P450 transcripts in the larvae fed on P450s dsRNA resulted in a significant increase of imidacloprid sensitivity in B. odoriphaga larvae. This indicated that these four up-regulated P450 unigenes are involved in imidacloprid detoxification in B. odoriphaga. In addition, this finding also enriched the growing body of literature showing that the RNAi technique can be applied in B. odoriphaga. The low level of detoxification enzyme gene expression and elevated insecticide sensitivity has been widely found in many other kinds of insects after RNAi42–45. Overall, oral delivery mediated RNAi not only suggested that ingestion of dsRNA through an artificial diet could be exploited for functional genomic studies in B. odoriphaga, but also indicated that P450s can be considered a major control target for delaying resistance to neonicotinoid insecticides in B. odoriphaga. The full length and expression character of these upregulated P450 transcripts need to be further studied.
In conclusion, RNA-Seq techniques have revolutionized our understanding of molecular genetics in insecticide resistance. In this study, RNA-Seq was performed to obtain several differentially expressed P450 unigenes related to imidacloprid detoxification in B. odoriphaga after imidacloprid treatment. As expected, four upregulated and three downregulated P450 unigenes were found. qRT-PCR expression results were consistent with RNA-Seq data. Furthermore, gene silencing followed by an insecticide bioassay was applied to understand the role of the four upregulated P450s transcripts in imidacloprid detoxification, which indicated that these upregulated P450 transcripts played an important role in the detoxification of imidacloprid in B. odoriphaga. Information from the transcriptome, DEG analysis, and gene function experiment provide a genetic basis for investigating imidacloprid detoxification P450 genes in B. odoriphaga.
Materials and Methods
Insect material
The population of B. odoriphaga used in this study was obtained from the Key Laboratory of Pesticide Toxicology & Application Technique of Shandong Province, College of Plant Protection, Shandong Agricultural University. This strain has been maintained in the laboratory without exposure to any insecticides since 2013. They were maintained at 25 ± 1 °C and 60–70% relative humidity with a photoperiod of 16:8 h (L:D).
Bioassay
Bioassays were conducted on newly emerged 4th instar larvae of B. odoriphaga using a standard contact and stomach bioassay method46. Briefly, three fresh Chinese chive pseudo stems and B. odoriphaga larvae were dipped into insecticide or control solutions for 15 s with gentle agitation. Then, the larvae and stems were transferred to a 9-well tissue-culture plate containing 2% agar covered with filter paper. Three replicates were conducted with at least 20 larvae for each replicate. Distilled water containing 0.1% (v/v) Triton X-100 and 1% acetone was used as a control. Mortality was assessed after 24 h. Larvae were considered dead if they were unable to move when touched by a moist brush. Probit analysis was used to calculate the median lethal concentration (LC50) with 95% confidence intervals (SPSS 19.0). The treatment group was newly emerged 4th instar larvae exposed to an LC50 concentration of imidacloprid for 24 h, and the control group was treated with the control solution. Two replicates were used in the treatment group and in the control group, and four samples in total were used for transcriptomic analysis.
RNA isolation and quality controls
Total RNA was extracted from 20 larvae in each sample using TRIzol reagent (Ambion, USA) according to the manufacturer’s protocol. The quality, quantity, and integrity of each RNA sample were assessed using two spectrophotometers: the Nanodrop (Thermo Scientific, USA) and the Aglient 2100 (Life Tech, USA). RNA samples were only used if they had a 260:230 ratio from 2.0 to 2.5, a 260:280 ratio from 1.9 to 2.1, and an RNA integrity number (RIN) higher than 8.0.
Illumina sequencing and de novo assembly
According to the Illumina manufacturer’s instructions, mRNA was enriched from DNase I-treated total RNA using the oligo (dT) magnetic beads, and cDNA was reverse-transcribed using the random hexamer primer. After purification with magnetic beads, cDNA was ligated at the 39-end with adenine and sequencing adaptors, followed by PCR amplification to create a cDNA library.
The cDNA library was sequenced on an Illumina sequencing platform (Hiseq. 2500, Illumina, USA). After removal of low-quality reads containing primer/adaptor sequences and trimming of read lengths using NGS QC Toolkit (v2.3.3), high-quality reads considered as clean data with an identity value of 95% and a coverage length of 125 bp were assembled de novo using Trinity (vesion:trinityrnaseq_r20131110) software and clustered using the TGICL to generate unigenes (Shanghai Oebiotech Co., Ltd., China)47–49.
Sequence clustering and functional categorization of unigenes
Unigene annotation of B. odoriphaga in the treatment and control groups was performed by searching the GenBank database with the BLASTX algorithm (http://www.ncbi.nlm.nih.gov/). NR (ftp://ftp.ncbi.nih.gov/blast/db/), GO (http://www.geneontology.org/), SWISSPROT (http://www.uniprot.org/downloads), KOG (http://www.ncbi.nlm.nih.gov/COG/), and KEGG (http://www.genome.jp/kegg/) annotations of the unigenes were determined using BLAST software.
Differentially expressed genes after imidacloprid treatment in B. odoriphaga
Reads sequenced from each sample of B. odoriphaga were aligned with the unigene library by Bowtie (http://bowtie-bio.sourceforge.net/index.shtml). To obtain relative expression levels in each sample, fragments per kilobase of transcript per million mapped reads (FPKM) in each sample were counted. The differential gene expression analysis was then performed using DESeq (http://www.bioconductor.org/packages/release/bioc/html/DESeq.html) for the treatment group and the control group. A gene was considered to be differentially expressed when results from the above tests were all significant at a level of P ≤ 0.05. The GO and KEGG annotations were performed using BLAST software for the differential expression of genes.
Reverse transcription (RT-PCR) and quantitative real-time PCR (qRT-PCR) validation
Approximately 20 B. odoriphaga larvae were collected in an Eppendorf tube. Total RNA was extracted using RNA-Solv@ Reagent (E.Z.N.A., USA) according to the manufacturer’s instructions. First-strand cDNA was synthesized from 1 μg of the total RNA using a PrimeScript RT-reagent Kit with gDNA Eraser (TaKaRa Biotechnology, China). Three independent RNA preparations representing three biological replicates were used for cDNA synthesis. The expression levels of P450 genes were quantified by qRT-PCR using a Biosystems 7500 Real-time PCR System (Applied Biosystems Inc, Foster City, USA) following the manufacturer’s protocols with the SYBR® Premix Ex Taq™ II kit (TaKaRa). Each 20 μL reaction contained 10 μL SYBR Green, 0.4 μL of each primer, 0.4 μL ROX, 1 μL cDNA template, and double distilled water. The cycling parameters consisted of an initial step at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. The relative expression of each P450 gene was calculated according to the 2−ΔΔCT method50. Difference analysis was performed by using Duncan’s multiple comparison test with SPSS software in transcriptome verification experiment. Each qRT-PCR experiment consisted of three independent biological replicates, each with three technical replicates.
RNAi and bioassays with imidacloprid after RNAi
Upregulated P450s transcripts were selected as candidates from differentially expressed gene (DEG) analysis. Then, dsRNAs were synthesized from purified PCR products using the Transcript Aid T7 High Yield Transcription Kit (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s instructions. PCR was performed using gene-specific primers containing T7 polymerase sites. All primer sets are listed in Table S1. PCR was performed with cycling parameters starting with 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s and 72 °C for 45 s, with a final extension at 72 °C for 10 min. Template DNA and single-stranded RNA were removed from the transcription reaction with DNase and RNase treatments, respectively. The dsRNA was purified using the phenol (pH 4.7): chloroform extraction and ethanol precipitation methods and eluted in diethyl pyrocarbonate (DEPC)-treated nuclease-free water. The dsRNA concentrations were measured using a Nanodrop spectrophotometer. DEPC-treated nuclease-free water was used as the negative control. Difference analysis was performed by using student t tests with SPSS software. A value of P < 0.05 was considered significant. The same for the following experiment.
To evaluate the functional role of specific P450s genes, B. odoriphaga 4th instar larvae fed with dsRNA-P450s at a concentration of 30 μg/g artificial diet and with an interval of 48 h post-feeding were used for qRT-PCR and bioassay. Primers used in this study are shown in Supplementary Table S1.
The LC50 dose of imidacloprid was applied for bioassays of the B. odoriphaga larvae after RNAi. The 4th instar larvae of B. odoriphaga were treated with different P450 dsRNA for 48 h. The control group was fed with an artificial diet mixed with GFP dsRNA. Bioassays were conducted as described in bioassay section.
Electronic supplementary material
Acknowledgements
The authors wish to thank the insect keeper who participated in this work. This study was funded by the Special Fund for Agro-scientific Research in the Public Interest from the Ministry of Agriculture of China (grant number: 201303027) and the National Key Research and Development Program of China (Project No. 2017YFD0201204).
Author Contributions
C.Y.C., X.Y.S. and X.W.G. conceived the experiments, C.Y.C. and C.C.W. conducted the experiments, C.Y.C. and Y.L. analysed the results. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-20981-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li WX, et al. Effects of temperature on the age-stage, two-sex life table of Bradysia odoriphaga (Diptera: Sciaridae) J. Econ. Entomol. 2015;108:126–134. doi: 10.1093/jee/tou011. [DOI] [PubMed] [Google Scholar]
- 2.Yang Y, et al. Development of Bradysiaodoriphaga (Diptera: Sciaridae) as affected by humidity: an age–stage, two-sex, life-table study. Appl. Entomol. Zool. 2015;50:3–10. doi: 10.1007/s13355-014-0295-6. [DOI] [Google Scholar]
- 3.Mau JL, Chen CP, Hsieh PC. Antimicrobial effect of extracts from Chinese chive, cinnamon, and corni fructus. J. Agric. Food Chem. 2001;49:183–188. doi: 10.1021/jf000263c. [DOI] [PubMed] [Google Scholar]
- 4.Imahori Y, et al. Physiological and quality responses of Chinese chive leaves to low oxygen atmosphere. Postharvest Biol. Technol. 2004;31:295–303. doi: 10.1016/j.postharvbio.2003.09.004. [DOI] [Google Scholar]
- 5.Yabuki Y, et al. Characterisation of volatile sulphur-containing compounds generated in crushed leaves of Chinese chive (Allium tuberosum Rottler) Food Chem. 2010;120:343–348. doi: 10.1016/j.foodchem.2009.11.028. [DOI] [Google Scholar]
- 6.Misawa T, Kuninaga S. First report of white leaf rot on Chinese chives caused by Rhizoctonia solani AG-2-1. J. Gen. Plant Pathol. 2013;79:280–283. doi: 10.1007/s10327-013-0455-5. [DOI] [Google Scholar]
- 7.Zhang H, Mallik A, Zeng RS. Control of Panama disease of banana by rotating and intercropping with Chinese chive (Allium tuberosum Rottler): role of plant volatiles. J. Chem. Ecol. 2013;39:243–252. doi: 10.1007/s10886-013-0243-x. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Lin L, Xie M, Zhang G, Su W. De novo sequencing and characterization of the Bradysia odoriphaga (Diptera: Sciaridae) larval transcriptome. Comp. Biochem. Physiol., Part D: Genomics Proteomics. 2015;16:20–27. doi: 10.1016/j.cbpa.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 9.Dang ZH, et al. Biology and injury of Bradysia odoriphaga on leek in different types of cultivation. J Agric Univ Hebei. 2001;24:65–68. [Google Scholar]
- 10.Li HJ, He XK, Zeng AJ, Liu YJ, Jiang SR. Bradysia odoriphaga copulatory behavior and evidence of a female sex pheromone. J. Agric. Urban Entomol. 2007;24:27–34. doi: 10.3954/1523-5475-24.1.27. [DOI] [Google Scholar]
- 11.Li XX, Ma XD, Xue M, Li ZX. Toxicity of insecticides to Bradysia odoriphaga at different temperature and their control effect in the fields. North Hortic. 2014;9:125–128. [Google Scholar]
- 12.Zhang P, Chen CY, Li H, Liu F, Mu W. Selective toxicity of seven neonicotinoid insecticides to fungus gnat Bradysia odoriphaga and earthworm Eisenia foetida. Acta Phytophylacica Sin. 2014;41:79–86. [Google Scholar]
- 13.Qi SM, et al. Detection of insecticide resistance of Bradysia odoriphaga in Shandong province. Plant Prot. 2016;42:179–183. [Google Scholar]
- 14.Chen CY, et al. Neonicotinoid insecticide resistance in Bradysia odoriphaga Yang et Zhang. Chin J Appl. Entomol. 2016;53:1250–1254. [Google Scholar]
- 15.Ding Q, et al. Monitoring the resistance of field populations of Bradysia odoriphaga Yang et Zhang in the main leek producing areas in China. Chin J Appl. Entomol. 2016;53:1242–1249. [Google Scholar]
- 16.Casida JE. Neonicotinoid metabolism: compounds, substituents, pathways, enzymes, organisms, and relevance. J. Agric. Food Chem. 2011;59:2923–2931. doi: 10.1021/jf102438c. [DOI] [PubMed] [Google Scholar]
- 17.Feyereisen R. Arthropod CYPomes illustrate the tempo and mode in P450 evolution. Biochim. Biophys. Acta Protein Proteomics. 2011;1814:19–28. doi: 10.1016/j.bbapap.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Feyereisen R. Insect P450 enzymes. Annu. Rev. Entomol. 1999;44:507–533. doi: 10.1146/annurev.ento.44.1.507. [DOI] [PubMed] [Google Scholar]
- 19.Clements J, et al. RNA interference of three up-regulated transcripts associated with insecticide resistance in an imidacloprid resistant population of Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2017;135:35–40. doi: 10.1016/j.pestbp.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Karunker I, et al. Structural model and functional characterization of the Bemisia tabaci CYP6CM1vQ, a cytochrome P450 associated with high levels of imidacloprid resistance. Insect Biochem. Mol. Biol. 2009;39:697–706. doi: 10.1016/j.ibmb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Markussen MD, Kristensen M. Cytochrome P450 monooxygenase-mediated neonicotinoid resistance in the house fly Musca domestica L. Pestic. Biochem. Physiol. 2010;98:50–58. doi: 10.1016/j.pestbp.2010.04.012. [DOI] [Google Scholar]
- 22.Ding Z, et al. Biochemical mechanisms of imidacloprid resistance in Nilaparvata lugens: over-expression of cytochrome P450 CYP6AY1. Insect Biochem. Mol. Biol. 2013;43:1021–1027. doi: 10.1016/j.ibmb.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Yang Y, Sun H, Liu Z. Metabolic imidacloprid resistance in the brown planthopper, Nilaparvata lugens, relies on multiple P450 enzymes. Insect Biochem. Mol. Biol. 2016;79:50–56. doi: 10.1016/j.ibmb.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Zhao, Y. et al. Proteomic profile of the Bradysia odoriphaga in response to the microbial secondary metabolite benzothiazole. Sci. Rep. 6, 10.1038/srep37730 (2016). [DOI] [PMC free article] [PubMed]
- 25.Cheng J, et al. Effects of Heat Shock on the Bradysia odoriphaga (Diptera: Sciaridae) J. Econ. Entomol. 2017;0:1–9. doi: 10.1093/jee/tox118. [DOI] [PubMed] [Google Scholar]
- 26.Gao HH, et al. Transcriptome analysis and discovery of genes relevant to development in Bradysia odoriphaga at three developmental stages. PloS one. 2016;11:e0146812. doi: 10.1371/journal.pone.0146812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X, Sun HH, Zhang YX, Liu CJ, Liu ZW. Transcriptional changes in nAChRs, interactive proteins and P450s in Locusta migratoria manilensis (Orthoptera: Acrididae) CNS in response to high and low oral doses of imidacloprid. J. Insect Sci. 2015;15:102. doi: 10.1093/jisesa/iev080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu JC, et al. Discovery of organophosphate resistance-related genes associated with well-known resistance mechanisms of Plutella xylostella (L.) (Lepidoptera: Plutellidae) by RNA-Seq. J. Econ. Entomol. 2016;109:1378–1386. doi: 10.1093/jee/tow070. [DOI] [PubMed] [Google Scholar]
- 29.Najarro MA, Hackett JL, Macdonald SJ. Loci contributing to boric acid toxicity in two reference populations of Drosophila melanogaster. G3: Genes, Genomes, Genet. 2017;7:1631–1641. doi: 10.1534/g3.117.041418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, et al. Expression profile analysis of silkworm P450 family genes after phoxim induction. Pestic. Biochem. Physiol. 2015;122:103–109. doi: 10.1016/j.pestbp.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 31.Danielson PB, Fogleman JC. Isolation and sequence analysis of cytochrome P450 12B1: the first mitochondrial insect P450 with homology to lα, 25 dihydroxy-D3 24-hydroxylase. Insect Biochem. Mol. Biol. 1997;27:595–604. doi: 10.1016/S0965-1748(97)00035-0. [DOI] [PubMed] [Google Scholar]
- 32.Tijet N, Helvig C, Feyereisen R. The cytochrome P450 gene superfamily in Drosophila melanogaster: annotation, intron-exon organization and phylogeny. Gene. 2001;262:189–198. doi: 10.1016/S0378-1119(00)00533-3. [DOI] [PubMed] [Google Scholar]
- 33.Bass C, et al. Overexpression of a cytochrome P450 monooxygenase, CYP6ER1, is associated with resistance to imidacloprid in the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 2011;20:763–773. doi: 10.1111/j.1365-2583.2011.01105.x. [DOI] [PubMed] [Google Scholar]
- 34.Bass C, et al. Gene amplification and microsatellite polymorphism underlie a recent insect host shift. Proc. Natl. Acad. Sci. 2013;110:19460–19465. doi: 10.1073/pnas.1314122110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nauen R, Vontas J, Kaussmann M, Wölfel K. Pymetrozine is hydroxylated by CYP6CM1, a cytochrome P450 conferring neonicotinoid resistance in Bemisia tabaci. Pest Manage. Sci. 2013;69:457–461. doi: 10.1002/ps.3460. [DOI] [PubMed] [Google Scholar]
- 36.Højland DH, Vagn Jensen KM, Kristensen M. A comparative study of P450 gene expression in field and laboratory Musca domestica L. strains. Pest Manage. Sci. 2014;70:1237–1242. doi: 10.1002/ps.3681. [DOI] [PubMed] [Google Scholar]
- 37.Azorsa DO, Mousses S, Caplen NJ. Gene silencing through RNA interference: Potential for therapeutics and functional genomics. Lett. Pept. Sci. 2003;10:361–372. doi: 10.1007/s10989-004-4900-3. [DOI] [Google Scholar]
- 38.Bass C, Denholm I, Williamson MS, Nauen R. The global status of insect resistance to neonicotinoid insecticides. Pestic Biochem Physiol. 2015;121:78–87. doi: 10.1016/j.pestbp.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 39.El Halim HMA, et al. RNAi-mediated knockdown of the voltage gated sodium ion channel TcNav causes mortality in Tribolium castaneum. Sci. Rep. 2016;6:29301. doi: 10.1038/srep29301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vyas M, et al. Knock down of Whitefly Gut Gene Expression and Mortality by Orally Delivered Gut GeneSpecific dsRNAs. PLoS One. 2017;12:e.0168921. doi: 10.1371/journal.pone.0168921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang XY, et al. RNAi in the striped stem borer, Chilo suppressalis, establishes a functional role for aminopeptidase N in Cry1Ab intoxication. J. Invertebr. Pathol. 2017;143:1–10. doi: 10.1016/j.jip.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 42.Gong YH, Yu XR, Shang QL, Shi XY, Gao XW. Oral delivery mediated RNA interference of a carboxylesterase gene results in reduced resistance to organophosphorus insecticides in the cotton aphid, Aphis gossypii Glover. PloS one. 2014;9:e102823. doi: 10.1371/journal.pone.0102823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gellatly KJ, et al. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic. Biochem. Physiol. 2015;121:107–115. doi: 10.1016/j.pestbp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J, et al. Two homologous carboxylesterase genes from Locusta migratoria with different tissue expression patterns and roles in insecticide detoxification. J. Insect Physiol. 2015;77:1–8. doi: 10.1016/j.jinsphys.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Shen XM, et al. Involvement of Three Esterase Genes from Panonychus citri (McGregor) in Fenpropathrin Resistance. Int. J. Mol. Sci. 2016;17:1361. doi: 10.3390/ijms17081361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mu W, Liu F, Jia ZM, He MH, Xiang GQ. A simple and convenient rearing technique for Bradysia odoriphaga. Entomol J East China. 2003;12:87–89. [Google Scholar]
- 47.Pertea G, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinf. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 48.Grabherr MG, et al. Nat. Biotechnol. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data; pp. 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PloS one. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.