Abstract
Background
While EoE is associated with certain gene variants, the rapidly increasing incidence of eosinophilic esophagitis (EoE) suggests that environmental factors contribute to disease development.
Objective
We tested for gene-environment interaction between EoE-predisposing polymorphisms (within TSLP, LOC283710/KLF13, CAPN14, CCL26, TGF-β) and implicated early life factors (antibiotic use in infancy, cesarean delivery, breastfeeding, neonatal intensive care unit (NICU) admission, and absence of pets in the home).
Methods
We conducted a case-control study using hospital-based cases (n=127) and controls representative of the hospital catchment area (n=121). We computed case-only interaction tests and, in secondary analyses, evaluated the combined and independent effects of genotype and environmental factors on risk of EoE.
Results
Case-only analyses identified interactions between rs6736278 (CAPN14) and breastfeeding (p=0.02) and rs17815905 (LOC283710/KLF13) and NICU admission (p=0.02), but not with any of the factors examined. Case-control analyses suggested that disease risk may be modifiable in individuals with certain gene variants. In particular, breastfeeding, in those with the susceptibility gene variant at rs6736278 (CAPN14), reduced the risk of developing EoE (aOR: 0.08; 95% CI: 0.01, 0.59). Admission to the NICU, in those without the susceptibility gene variant at rs17815905 (LOC283710/KLF13), significantly increased risk of developing disease (aOR 4.83; 95% CI: 1.49, 15.66).
Conclusions
The interplay of gene (CAPN14 and LOC283710/KLF13) and early life environment factors (breastfeeding, NICU admission) may contribute to EoE susceptibility.
Keywords: early life factors, eosinophilic esophagitis, gene-environment interaction, exposures
Introduction
Over the past two decades, eosinophilic esophagitis (EoE) has transformed from a case-reportable disease to a substantial cause of upper GI morbidity.1 The reasons for this increase are likely multifactorial including increased disease recognition and changing environmental factors, particularly those interacting with immunological pathways involved in EoE.2, 3 Numerous genetic susceptibility variants have been shown to contribute to EoE, including variants at 5q22 (TSLP/WDR36), 2p23 (CAPN14), and 11q13 (LRCC32), but the magnitude of association for disease susceptibility is modest (<2-fold), similar to the magnitude seen in other allergic and immunological diseases. Twin studies have established a role for genetics in disease susceptibility, but an even stronger role for environmental factors, particularly those encountered in early life, as shown by the marked increased concordance for EoE between dizygotic twins compared with non-twin siblings and environmental assessments in EoE case control studies.4, 5
Identification of gene-environment interactions for EoE offers an opportunity to improve understanding of the mechanisms for disease development. It also may offer a means for identifying possible modifiable risk factors for disease, specifically risk factors that may only be relevant in the presence of specific genetic susceptibility variants. Therefore, we conducted a preliminary, case-control study of gene-environment interaction between EoE genetic variants6–9 and early life exposures that have been associated with EoE and other atopic diseases (Supplementary Table 1).5, 10–12 Specifically, we examined interaction between 5 gene variants (rs6736278 within CAPN14 at 2p23, rs2302009 within CCL26 at 7q11, rs3806932 within TSLP at 5q22, rs17815905 within the LOC283710 and KLF13 region at 15q13, and rs1800469 within TGF-β at 19q13) and 6 maternally-reported early life exposures (cesarean delivery, preterm delivery, NICU admission, breastfeeding, antibiotics in infancy, and absence of a furry pet in infancy). We selected single nucleotide polymorphisms (SNPs) reported to be associated with EoE, or in high linkage disequilibrium with SNPs associated with EoE. These included SNPs from genes with demonstrated functionality in gene regulation and association with allergic sensitization. Specifically, these included SNPs from genes with demonstrated functionality in gene regulation and association with allergic sensitization (TSLP at 5q22 [rs3806932], the LOC283710 and LOC283710 region at 15q13 [rs4329885], CAPN14 [rs6736278], CCL26 [rs2302009], and TGF-β [rs1800469])).6–9 CAPN14 encodes an IL-13-induced calcium-activated cysteine protease involved in maintaining integrity of esophageal barrier function and is overexpressed in EoE.13, 14 The rs4329885 SNP, found on the 15q13 chromosome band between LOC283710 and KLF13, is in a region that is influential for gene regulation, may contribute to impaired esophageal epithelial cell development that has been observed to occur in EoE,15 and has been shown to be involved in Th2 cytokine production.16 TSLP, a gene that is critical to inducing allergen sensitization, is strongly associated with and overexpressed in EoE.9, 17 CCL26 encodes for eotaxin-3, the most upregulated gene in EoE, and acts as a chemoattractant for the activation and recruitment of eosinophils into esophageal tissue.8 Transforming growth factor (TGF) - β secretion, produced in part by eosinophils, is implicated in the fibrosis of esophageal tissue observed in EoE, as well as in immunomodulation.18 While the exact mechanism for a possible interaction is unknown, it may be that environmental factors have epigenetic effects that influence DNA expression.19, 20
Methods
Cases were recruited from the Cincinnati Center for Eosinophilic Disorders (CCED) at Cincinnati Children’s Hospital Medical Center (CCHMC) and met criteria for EoE diagnosis as defined by current consensus guidelines.21 Controls, used to assess violation of the assumption of independence between exposure and the genotype in the non-cases, were recruited from the CCHMC Genomic Control Cohort, a population-based control cohort representative of the Greater Cincinnati population.22 Cases and controls were restricted to those less than age 18 at the time of enrollment and all were self-reported Caucasian race.
Early life, environmental factors assessment
Early life environmental factors for cases and controls were assessed through an on-line self-administered questionnaire provided to mothers. The questionnaire, described previously in detail,10 was developed to study early life exposures, through an interactive process that included cognitive interviewing and testing. Survey domains included prenatal, intrapartum and pediatric exposures, and the exposures of interest for this study were limited to those previously associated with EoE or demonstrating main effect associations in the current study population,5 including caesarean delivery, preterm birth, admission to the neonatal intensive care unit (NICU), formula feeding and antibiotic use (Supplementary Table 1).5, 10–12, 23
SNP genotyping and quality control
DNA from cases and controls was obtained from peripheral blood samples. Cases and controls were genotyped using the genome-wide OMNI5 BeadChip array (Illumina, Inc.).22 Examination of call rates identified <0.1% missing genotypes. We identified no evidence of departure from Hardy Weinberg equilibrium in cases, controls or cases and controls combined (Supplementary Table 2). Prior to analyses, we performed principal components analysis using EIGENSOFT24 on 639 ancestry informative markers from the OMNI5 Beadchip (Illumina, Inc.).22
Statistical analyses
In our primary analysis we conducted a case-only gene-environment interaction analysis. We used a generalized linear model (logit link, binomial distribution) to assess the association between each environmental factor and genotype in cases. We modeled the risk allele using a recessive mode of inheritance if the risk allele was the major allele (e.g. GG or GA vs GG) and used a dominant mode of inheritance if the risk allele was the minor allele (e.g. AA vs AG or GG). If there was marginal evidence of an interactive effect (p<0.10) in the case-only analysis, we tested the assumption of independence between the exposure and genotype in the source population by modelling the association between exposure and genotype in the controls. Lack of independence was identified by evidence of a similar direction and magnitude of association between exposure and genotype in cases and controls.
In secondary analyses, to facilitate interpretation of any interactive effect, we used cases and controls to estimate the odds of disease in those with the susceptibility genotype only, those with the environmental factor only, and those with both the susceptibility genotype and the environmental factor, relative to those with neither the susceptibility genotype nor the environmental factor. Additionally, we modeled disease risk within those with the susceptibility genotype, as a means for assessing the contribution of each early life factor in those with the susceptibility genotype. For all models, we estimated both crude and adjusted (adjusted for maternal education and two principal components for ancestry) associations. The presence of interaction was determined by indication of a case-only interaction (p<0.05) and indication of stratum-specific differences in the magnitude of association in the case-control assessment. Finally, we calculated the relative excess risk due to interaction (RERI) to assess for interaction on the additive scale.25, 26 We generated both crude and adjusted estimates for both primary and secondary analyses. Adjusted models included adjustment for maternal education, as a proxy for socioeconomic status, and population stratification. The study was approved by the Cincinnati Children’s Hospital Medical Center IRB.
Results
Of the 237 patients successfully contacted, 169 (71%) consented to participation. Of these, 136 (80%) completed the questionnaire. Similarly, for controls, of the 208 potential subjects successfully contacted, 147 (71%) consented to participation and, of these, 125 (85%) completed the questionnaire. Our final analytic sample, after exclusion of observations with missing data, included 127 cases and 121 controls. The demographic features of the cases and controls were similar, with the exception of atopic disease comorbidities (e.g. 83.3% cases versus 12.4% controls reporting food allergies) and sex (80.3% males in cases versus 52.1% in controls) (Table 1).
Table 1.
Characteristics of study population
| Characteristic | Controls n=121 % or mean; std | Cases n=127 % or mean |
|---|---|---|
| Age at enrollment (mean yrs.; std) | 13.8; 2.8 | 10.6; 3.8 |
| Sex (% male) | 52.1 | 80.3 |
| Atopic illnesses (%) | ||
| Food allergies | 12.4 | 83.3 |
| Environmental allergies | 42.2 | 69.3 |
| Antibiotic allergies | 14.5 | 25.8 |
| Eczema | 21.5 | 59.1 |
| Asthma | 18.2 | 44.9 |
| Maternal marital status (%) | ||
| Married or civil union | 93.4 | 94.4 |
| Single | 6.6 | 5.6 |
| Maternal education (%) | ||
| <HS* | 0 | 2.4 |
| HS or GED** | 17.4 | 11.8 |
| Technical or associates | 21.5 | 18.9 |
| Bachelor’s | 40.5 | 37.0 |
| Graduate or professional | 20.7 | 29.9 |
HS=High School
GED=General Education Development test certification
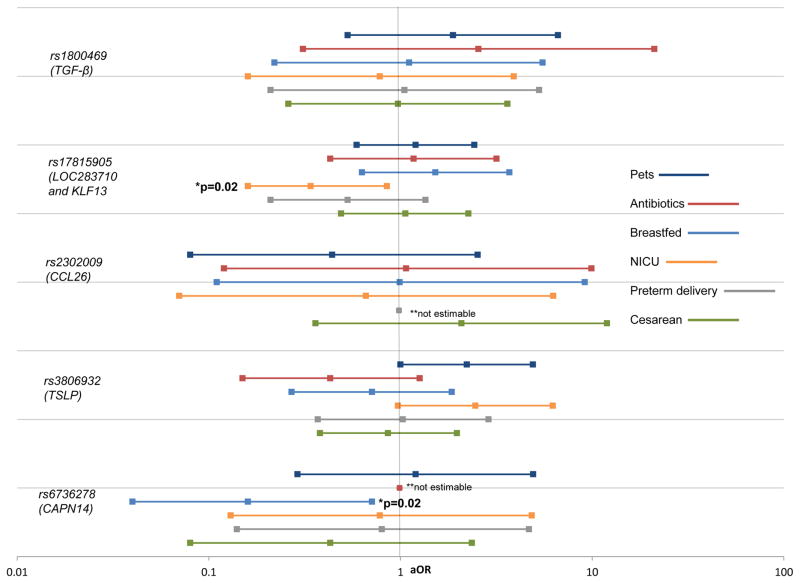
In our primary case-only analyses, we observed interaction between rs6736278 within CAPN14 and breastfeeding (p=0.02) (Figure 1). We also observed interaction between rs17815905 in the LOC283710 and KLF13 region and NICU admission (p=0.02). There was no evidence of statistically significant interaction between the SNPs in CCL26, TSLP or TGF-β with any of the early life factors examined (Supplementary Table 3). Where there was evidence of interaction, or marginal evidence of interaction (TSLP and NICU admission and TSLP and no furred pets in infancy), we found no evidence of violation of independence (Supplementary Table 4). Secondary analyses confirmed the case-only interactions observed (Figure 2, Tables 2,3) and stratum-specific assessments identified breastfeeding as a potentially modifiable factor in developing EoE among children with a certain genotype (Table 2). While the exact mechanism of how breastfeeding and CAPN14 may interact and become protective for disease is unknown, the protective effect of breastfeeding was demonstrated in a study of interaction between breastfeeding and the cluster of differentiation 14 (CD14) genotype (CD14C-159T) in relation to atopic sensitization.27 CAPN14 is integral to maintaining barrier function in the esophagus.13, 14 Potentially, with impaired barrier function, breastfeeding may confer a higher protective effect than that which is observed when barrier function is preserved, however this must be investigated in follow-up mechanistic studies. Notably, we observed a 92% reduction in the odds of developing EoE among those who were breastfed within the subgroup of participants harbouring the susceptibility genotype (Table 2). Examination of interaction on the additive scale, RERI, identified a strong departure from additivity for CAPN14 and breastfeeding (RERI: −6.27 [95% CI: −17.27, 4.72])) (Table 2) and the LOC283710 and KLF13 region and NICU admission (RERI: −4.53 [95% CI: −10.81, 1.76]), however neither of these reached statistical significance (Table 3).
Figure 1.
Case-only assessment of gene-environment interaction in EoE
Illustration of the association between exposure and genotype in cases only to assess for gene-environment interaction, adjusted for maternal education.
*Suggestive associations examined in case-control data assessment of interaction
**Data too sparse for estimation of association
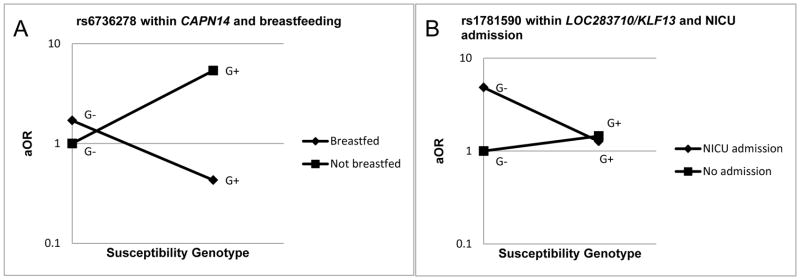
Figure 2.
Case-control analysis interaction plots for early life factors and susceptibility genotype
Plots denote interaction between selected exposures and genotype variants in the case-control analyses. Interaction models include adjustment for maternal education. G+=exposed to susceptibility genotype variant; G−=unexposed to susceptibility genotype variant. Figure A illustrates the difference in risk for those breastfed and not breastfed, among those with the susceptibility genotype variant. Figure B illustrates the increase in risk for NICU admission and those without the susceptibility variant.
Table 2.
CAPN14 (rs6736278) and breastfeeding
| Cases n | Controls n | OR (95% CI) | aOR (95% CI) | |
|---|---|---|---|---|
| Interaction | ||||
|
| ||||
| G−E− | 26 | 11 | referent | referent |
| G+E− | 2 | 5 | 5.91 (0.99, 35.21) | 5.38 (0.89, 32.71) |
| G−E+ | 75 | 60 | 1.89 (0.86, 4.14) | 1.70 (0.76, 3.77) |
| G+E+ | 18 | 4 | 0.53 (0.14, 1.91) | 0.43 (0.12, 1.62) |
|
| ||||
| Case only p for interaction | 0.01 | 0.02 | ||
| RERI (95% CI) | −6.27 (−17.27, 4.72) | |||
|
| ||||
| Effect of exposure among those with susceptibility genotype | ||||
|
| ||||
| G+E− | 2 | 5 | referent | referent |
| G+E+ | 18 | 4 | 0.09 (0.01, 0.63) | 0.08 (0.01, 0.59) |
Table 3.
LOC283710 and KLF13 region (rs17815905) and NICU admission
| Cases n | Controls n | OR (95% CI) | aOR (95% CI) | |
|---|---|---|---|---|
| Interaction | ||||
|
| ||||
| G−E− | 38 | 51 | referent | referent |
| G+E− | 63 | 56 | 1.47 (0.85, 2.55) | 1.45 (0.83, 2.53) |
| G−E+ | 16 | 4 | 5.23 (1.62, 16.89) | 4.83 (1.49, 15.66) |
| G+E+ | 9 | 10 | 1.18 (0.44, 3.17) | 1.27 (0.46, 3.53) |
|
| ||||
| Case only p for interaction | 0.02 | 0.02 | ||
| RERI (95% CI) | −4.53 (−10.81, 1.76) | |||
|
| ||||
| Effect of exposure among those with susceptibility genotype | ||||
|
| ||||
| G+E− | 63 | 56 | referent | referent |
| G+E+ | 9 | 10 | 0.80 (0.30, 2.11) | 0.88 (0.32, 2.38) |
Discussion
In this study we examined interaction between 5 gene variants (rs6736278 within CAPN14 at 2p23, rs2302009 within CCL26 at 7q11, rs3806932 within TSLP at 5q22, rs17815905 within the LOC283710 and KLF13 region at 15q13, and rs1800469 within TGF-β at 19q13) and 6 maternally-reported early life exposures (cesarean delivery, preterm delivery, NICU admission, breastfeeding, antibiotics in infancy, and absence of a furry pet in infancy). We conducted case-only tests of interaction (with subsequent confirmation of results with the case-control analysis) and observed evidence of statistical interaction between rs6736278 (CAPN14) and breastfeeding and rs17815905 (LOC283710 and KLF13 region) and NICU admission. Case-control analyses of these associations provided additional evidence to support the presence of an interaction. The combined effect of susceptibility genotype and environmental exposure was less than expected (e.g. antagonistic) after examining the independent effect of genotype and environmental exposure. For example, the combined effect for breastfeeding and genotype was significantly less (aOR: 0.43; 95% CI: 0.12, 1.62) than the effect of genotype (aoR: 5.38; 95% CI: 0.89, 32.71) or breastfeeding alone (aOR: 1.70; 95% CI: 0.76, 3.77). Indeed, breastfeeding had a strong protective effect (aOR 0.08; 95% CI: 0.01, 0.59) in those with the susceptibility genotype (CAPN14 in rs6736278). While the exact mechanism of how breastfeeding and CAPN14 may interact and become protective for disease is unknown, the protective effect of breastfeeding was demonstrated in a study of interaction between breastfeeding and the cluster of differentiation 14 (CD14) genotype (CD14C-159T) in relation to atopic sensitization.27 CAPN14 is integral to maintaining barrier function in the esophagus.13 It is tempting to speculate that with impaired barrier function, breastfeeding may confer a higher protective effect than that which is observed when barrier function is preserved, but this would need to be investigated in mechanistic studies.
The early life factors examined in this study have been associated with dysbiosis in gut colonization and early life.28–36 Indeed, the esophagus harbors its own microbiome that is dysregulated in EoE patients.37 It is interesting to speculate that the interacting SNPs might mediate their effects by modifying esophageal microbiota content and/or their interaction with EoE-related immunological pathways, even before the onset of EoE.28, 38–40 While this study was not designed to directly assess the influence of these exposures on the microbiome, the results provide the first potential evidence of gene-environment interactions in EoE, and support future work examining the microbiome and EoE.
Our results must be interpreted with caution as this preliminary study had a relatively small sample size and we have not sought to replicate these findings in an independent sample. Further, the associations reported required only nominal significance (p< 0.05) and did not account for multiple testing. To try to minimize the risk of false positives, we limited the analyses and environmental effects which had been associated with EoE risk in prior studies. Given many of the exposures are correlated (e.g. preterm delivery and NICU admission), each exposure-genotype interaction assessment did not represent a completely independent test and the exposures and SNPs selected were selected a priori, based on hypotheses of an interactive effect, as opposed to an agnostic study of gene-environment interaction where multiple correction testing may be more relevant.41 While we observed interaction for several of the environmental exposures examined, these exposures may be a proxy for some other factor(s) that are interacting with the identified genes. Additionally, while early life factors such as cesarean delivery, breastfeeding, NICU admission, and having pets in the home are likely not hampered by recall bias, recall of antibiotic use and, possibly, preterm delivery, could be biased.
In conclusion, we have preliminary evidence that the etiology of EoE may involve the interplay of gene and environmental factors. Further understanding of these interactions provides an opportunity to potentially alter the development of EoE, as environmental factors can be modified in individuals harbouring specific genetic variants.
Supplementary Material
Clinical Implications.
EoE-predisposing polymorphisms interact with early life factors to modify risk of EoE disease development. Risk for EoE disease may be modifiable in individuals with certain environmental exposures and gene variants.
Acknowledgments
Grant support: American College of Gastroenterology – Clinical Research Award. This work was also supported by National Institutes of Health R01 DK101856 (ESD), R37 AI045898, R01 AI124355; the Campaign Urging Research for Eosinophilic Disease (CURED); the Buckeye Foundation; and the Sunshine Charitable Foundation and its supporters, Denise A. Bunning and David G. Bunning.
Abbreviations
- EoE
eosinophilic esophagitis
- RERI
relative excess risk due to interaction
- aOR
adjusted odds ratio
- NICU
neonatal intensive care unit
- SNP
single nucleotide polymorphism
Footnotes
Disclosures: M.E.R. is a consultant for Immune Pharmaceuticals, NKT Therapeutics, Pulm One, Spoon Guru, Celgene, Shire, Astra Zeneca and Novartis and has an equity interest in the first four companies listed, and royalties from reslizumab (Teva Pharmaceuticals). M.E.R. is an inventor of several patents owned by Cincinnati Children’s. None of the other authors have any potential conflicts of interest relevant to the manuscript. This research was supported in part by the Cincinnati Children’s Research Foundation and its Cincinnati Genomic Control Cohort.
Author contributions: Jensen, Dellon and Rothenberg contributed to the development of the study concept and design. Rothenberg, Kuhl, and Martin contributed to the data acquisition, Jensen and Langefeld contributed to gene-environment analyses. All authors contributed to the interpretation of the data. Jensen and Dellon drafted the manuscript and all other authors reviewed and contributed intellectual content to the manuscript. Jensen and Dellon secured funding for conduct of the study. Kuhl provided research coordination support to the study.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dellon ES, Gonsalves N, Hirano I, Furuta GT, Liacouras CA, Katzka DA. ACG Clinical Guideline: Evidenced Based Approach to the Diagnosis and Management of Esophageal Eosinophilia and Eosinophilic Esophagitis (EoE) The American journal of gastroenterology. 2013;108:679–92. doi: 10.1038/ajg.2013.71. [DOI] [PubMed] [Google Scholar]
- 2.Hruz P, Bussmann C, Heer P, Simon HU, Zwahlen M, Beglinger C, et al. Escalating Epidemiology of Eosinophilic Esophagitis: 21 Years of Prospective Population-Based Documentation in Olten County. Gastroenterology. 2011;140(Suppl 1):S238–9. [Google Scholar]
- 3.Dellon ES, Erichsen R, Baron JA, Shaheen NJ, Vyberg M, Sorensen HT, et al. The increasing incidence and prevalence of eosinophilic oesophagitis outpaces changes in endoscopic and biopsy practice: national population-based estimates from Denmark. Aliment Pharmacol Ther. 2015;41:662–70. doi: 10.1111/apt.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander ES, Martin LJ, Collins MH, Kottyan LC, Sucharew H, He H, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134:1084–92. e1. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jensen ET, Kuhl JT, Martin LJ, Rothenberg ME, Dellon ES. Prenatal, intrapartum, and postnatal factors are associated with pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sherrill JD, Rothenberg ME. Genetic dissection of eosinophilic esophagitis provides insight into disease pathogenesis and treatment strategies. J Allergy Clin Immunol. 2011;128:23–32. doi: 10.1016/j.jaci.2011.03.046. quiz 3–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins MH, Blanchard C, Abonia JP, Kirby C, Akers R, Wang N, et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin Gastroenterol Hepatol. 2008;6:621–9. doi: 10.1016/j.cgh.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116:536–47. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42:289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen ET, Kappelman MD, Kim HP, Ringel-Kulka T, Dellon ES. Early life exposures as risk factors for pediatric eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2013;57:67–71. doi: 10.1097/MPG.0b013e318290d15a. [DOI] [PubMed] [Google Scholar]
- 11.Radano MC, Yuan Q, Katz A, Fleming JT, Kubala S, Shreffler W, et al. Cesarean section and antibiotic use found to be associated with eosinophilic esophagitis. J Allergy Clin Immunol Pract. 2014;2:475–7. e1. doi: 10.1016/j.jaip.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Slae M, Persad R, Leung A-T, Gabr R, Brocks D, Huynh H. Role of Environmental Factors in the Development of Pediatric Eosinophilic Esophagitis. Digestive Diseases and Sciences. 2015:1–9. doi: 10.1007/s10620-015-3740-7. [DOI] [PubMed] [Google Scholar]
- 13.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1:e86355. doi: 10.1172/jci.insight.86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litosh VA, Rochman M, Rymer JK, Porollo A, Kottyan LC, Rothenberg ME. Calpain-14 and its association with eosinophilic esophagitis. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The Human Genome Browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126:160–5. e3. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rieder F, Nonevski I, Ma J, Ouyang Z, West G, Protheroe C, et al. T-helper 2 cytokines, transforming growth factor beta1, and eosinophil products induce fibrogenesis and alter muscle motility in patients with eosinophilic esophagitis. Gastroenterology. 2014;146:1266–77. e1–9. doi: 10.1053/j.gastro.2014.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J Allergy Clin Immunol. 2011;128:3–20. e6. doi: 10.1016/j.jaci.2011.02.040. [DOI] [PubMed] [Google Scholar]
- 22.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46:895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, Blumberg RS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol. 2014;109:646–57. doi: 10.1038/ajg.2014.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Knol MJ, VanderWeele TJ. Recommendations for presenting analyses of effect modification and interaction. Int J Epidemiol. 2012;41:514–20. doi: 10.1093/ije/dyr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knol MJ, VanderWeele TJ, Groenwold RH, Klungel OH, Rovers MM, Grobbee DE. Estimating measures of interaction on an additive scale for preventive exposures. Eur J Epidemiol. 2011;26:433–8. doi: 10.1007/s10654-011-9554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SY, Kang MJ, Kwon JW, Park KS, Hong SJ. Breastfeeding Might Have Protective Effects on Atopy in Children With the CD14C-159T CT/CC Genotype. Allergy Asthma Immunol Res. 2013;5:239–41. doi: 10.4168/aair.2013.5.4.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huurre A, Kalliomaki M, Rautava S, Rinne M, Salminen S, Isolauri E. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology. 2008;93:236–40. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 29.Mulder IE, Schmidt B, Lewis M, Delday M, Stokes CR, Bailey M, et al. Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity. PLoS One. 2011;6:e28279. doi: 10.1371/journal.pone.0028279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel AL, Mutlu EA, Sun Y, Koenig L, Green S, Jakubowicz A, et al. Longitudinal Survey of Microbiota in Hospitalized Preterm Very Low Birth Weight Infants. J Pediatr Gastroenterol Nutr. 2015 doi: 10.1097/MPG.0000000000000913. [DOI] [PMC free article] [PubMed]
- 31.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13:440–7. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taft DH, Ambalavanan N, Schibler KR, Yu Z, Newburg DS, Ward DV, et al. Intestinal microbiota of preterm infants differ over time and between hospitals. Microbiome. 2014;2:36. doi: 10.1186/2049-2618-2-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tormo-Badia N, Hakansson A, Vasudevan K, Molin G, Ahrne S, Cilio CM. Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol. 2014;80:250–60. doi: 10.1111/sji.12205. [DOI] [PubMed] [Google Scholar]
- 34.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128:948–55. e1–3. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 35.Walker WA, Iyengar RS. Breast milk, microbiota, and intestinal immune homeostasis. Pediatr Res. 2015;77:220–8. doi: 10.1038/pr.2014.160. [DOI] [PubMed] [Google Scholar]
- 36.Westerbeek EA, van den Berg A, Lafeber HN, Knol J, Fetter WP, van Elburg RM. The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin Nutr. 2006;25:361–8. doi: 10.1016/j.clnu.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Harris JK, Fang R, Wagner BD, Choe HN, Kelly CJ, Schroeder S, et al. Esophageal microbiome in eosinophilic esophagitis. PLoS One. 2015;10:e0128346. doi: 10.1371/journal.pone.0128346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012;336:489–93. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker A. Intestinal colonization and programming of the intestinal immune response. J Clin Gastroenterol. 2014;48(Suppl 1):S8–11. doi: 10.1097/MCG.0000000000000230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rothman KJ. No adjustments are needed for multiple comparisons. Epidemiology. 1990;1:43–6. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.