Abstract
Osteoarthritis (OA) is the most common joint disease and the leading cause of chronic disability in middle-aged and older populations worldwide. The development of disease modifying therapy for OA is in its infancy largely because the regulatory mechanisms for the molecular effectors of OA pathogenesis are poorly understood. Recent studies identified epigenetic events as a critical regulator of molecular players involved in the induction and development of OA. Epigenetic mechanisms include DNA methylation, non-coding RNA and histone modifications. The aim of this review is to briefly highlight the recent advances in the epigenetics of cartilage and potential of HDACs (Histone deacetylases) inhibitors in the therapeutic management of OA. We summarize the recent studies utilizing HDAC inhibitors as potential therapeutics for inhibiting disease progression and preventing the cartilage destruction in OA. HDACs control normal cartilage development and homeostasis and understanding the impact of HDACs inhibitors on the disease pathogenesis is of interest because of its importance in affecting overall cartilage health and homeostasis. These findings also shed new light on cartilage disease pathophysiology and provide substantial evidence that HDACs may be potential novel therapeutic targets in OA.
Keywords: Osteoarthritis, Epigenetics, HDACs, miRNA, lncRNA, DNA methylation
Graphical Abstract
1. Introduction
Osteoarthritis (OA), the most common form of joint disease, is associated with cartilage degradation, disability and poor quality of life and is a leading cause of chronic disability in middle-aged and older populations affecting more than 27 million Americans [1]. Aging is a major risk factor for OA, however other factors such as gender, obesity, joint injury, genetics and mechanical abnormalities have been shown to contribute to the development of OA [2]. There is no single specific cause that has been identified for OA till date, however, there is a growing body of evidence that suggest that OA is a result of the interactions between molecular events and mechanical issues in the affected joint [3]. Although OA is a disease of the whole joint and affect all the joint tissues, cartilage degradation is the main characteristic of OA [4]. The underlying molecular mechanisms of OA pathogenesis involve multiple components and include dysregulation of different layers of regulatory mechanisms.
Altered gene expression of matrix degrading proteases (MMPs), inflammatory mediators and extracellular matrix (ECM) related genes such as collagens and proteoglycans in articular chondrocytes isolated from OA cartilage has been documented [4–8]. However, the underlying regulatory mechanism for the expression of these genes in OA cartilage has not been fully understood. Epigenetics is an important layer of regulation of gene expression and is associated with the pathogenesis of a number of human diseases [9]. Epigenetics is defined as a stable change in gene expression between cell divisions, and sometimes generations, that involves no change in the underlying DNA sequence [10]. The “NIH Roadmap Epigenomics Project” defined epigenetics as heritable changes in gene activity and expression and stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable [11]. Here we reviewed the recent findings in this field with an emphasis on the potential of HDACs inhibitors for the management of OA. If we have missed a reference, it is inadvertent and not because the findings are not important.
2. Epigenetic regulation of OA pathogenesis
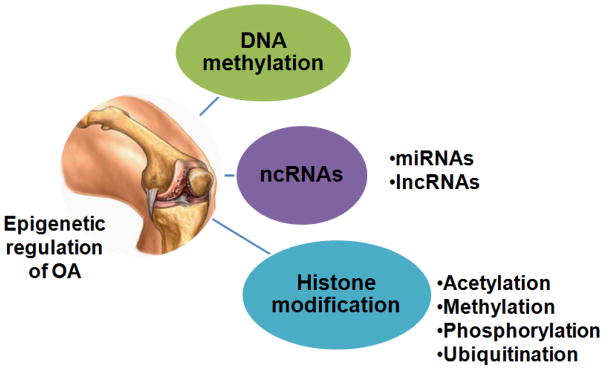
Although epigenome of each cell is unique but can undergo temporal and spatial changes in response to environmental stimuli such as diet, exercise, smoking and disease status. Aberrant epigenetic modifications due to environmental factors are associated with a number of pathological conditions and include DNA methylation, non-coding RNAs (ncRNAs) expression and histone modifications that regulate the gene expression at transcriptional and/or post-transcriptional levels (Figure 1). Here, we will briefly highlight the first two epigenetic mechanisms namely DNA methylation and ncRNA mediated regulation of gene expression in OA and will focus on HDACs mediated regulation of OA pathogenesis and explore the potential of HDACs inhibitors (HDACi) for the management of OA.
Figure 1. Epigenetic regulation of osteoarthritis.
Three different epigenetic regulation of molecular pathogenesis of OA. DNA methylation, non-coding RNAs (ncRNAs: miRNAs and lncRNAs) expression and histone modifications regulate the gene expression involved in the etiology of OA at transcription and/or post-transcription levels. Histone acetylation, histone methylation, phosphorylation and ubiquitination are major histone modifications involved in OA.
2.1 DNA methylation in OA
DNA methylation is the most widely studied epigenetic control mechanism and involves addition of a methyl group to the 5′ position of cytosine within a CpG dinucleotide to form 5-methylcytosine in presence of DNA methyltransferase (DNMT). Methylation within the gene promoter regions is associated with the suppression of gene expression, whereas methylation within gene bodies correlates with the enhanced gene expression [12, 13]. The pioneer findings of DNA methylation studies in OA lead to the discovery of hypomethylation of catabolic genes and hypermethylation of anabolic genes in chondrocytes [14]. Methylation changes of a few specific sites inside the promoter region or the gene itself can be crucial in determining transcription and expression of a gene in OA.
The candidate gene approach to examine the DNA methylation of matrix degrading proteases such as MMP3, MMP9, MMP13 and ADAMTS4 was the first study to describe the possible impact of DNA methylation in OA [15]. These studies demonstrated the hypomethylation in the promoter regions of selected catabolic genes in OA chondrocytes compared to control chondrocytes and found that this was associated with the increased expression of the gene [15]. The inflammatory cytokine IL-1β, which is recognized as a critical player in OA pathogenesis, is also epigenetically regulated and methylation levels within its proximal promoter region were found to be inversely correlated with gene transcription in OA chondrocytes [16]. Genome-wide DNA methylation studies in OA cartilage using Illumina Human Methylation 450K array allowing the quantitation of nearly 470,000 CpG sites covering all the coding genes identified 550 differentially methylated CpG sites among OA patients, with the RUNX1 (runt-related transcription factor) being most differentially methylated gene [17]. Further investigations are needed to clearly define the impact of methylation in the promoter region of a gene on the disease process in OA.
2.2 Non-coding RNAs in OA
Non-coding RNAs (ncRNA) are functional RNA molecules that regulate gene expression but do not translate into proteins. There are two types of ncRNAs- short ncRNAs which are <30 nucleotides and long ncRNAs (lncRNAs) which are >200 nucleotides in length [18, 19]. Further, short ncRNAs have been classified as mainly of three types- microRNAs (miRNAs), piwi-interacting RNAs (piRNAs) and short interfering RNAs (siRNAs) [20]. These ncRNAs regulate gene expression at transcription, splicing or translation levels. The miRNAs in general modify the protein expression thus acting mainly at the post-transcriptional level by binding to a specific complimentary sequence in the target mRNA and block its translation [21]. Recent advances in the field of ncRNAs have revealed their importance in the pathogenesis of many diseases including OA [22].
2.2.1 miRNAs in OA
miRNAs are small, 20–23 base pair long cytoplasmic RNAs that regulate post-transcriptional gene expression through binding to target mRNAs via complementary base pairing between the miRNA and the “seed sequence” present in the 3′-UTR or the ORF of the target mRNA [23]. The interaction of miRNA with the target mRNA results in the degradation of mRNA leading to translational suppression [21]. The first studied miRNA in osteoarthritis was miR-140, which is specifically expressed in the cartilage and has been shown to play an important role in chondrogenesis [24–28]. The expression of miR-140 was reduced in human OA cartilage and its deletion lead to an accelerated OA phenotype in mouse [24–26]. Table 1 describes a brief summary and function of miRNAs differentially expressed in OA. Several miRNAs have been shown to affect the OA progression by mitigating the inflammation and influencing the anabolic function in cartilage.
Table 1.
Differentially expressed miRNAs and their molecular target(s) in OA
| miRNA | Differential expression in OA | Target gene(s) | Biological effects | References |
|---|---|---|---|---|
| miR-22 | Upregulated | PPARα, BMP7, ACAN | Lipid metabolism | [29] |
| miR-9 | Upregulated | MMP-13, MCPIP1 | Matrix degradation | [30] [31] |
| miR-25 | Upregulated | COX-2 | Inflammation | [30] [32] |
| miR-602 | Upregulated | SHH | Chondrocytes homeostasis | [33] |
| miR-608 | Upregulated | SHH | Chondrocytes homeostasis | [33] |
| miR-139 | Upregulated | MCPIP1 | Inflammation | [31] |
| miR-27a | Downregulated | MMP-13, IGFBP5 | Matrix degradation | [34] |
| miR-27b | Downregulated | MMP-13 | Matrix degradation | [35] |
| miR-140 | Downregulated | ADAMTS-5, MMP-13 | Matrix degradation | [36] |
| miR199a | Downregulated | COX-2 | Inflammation | [35] |
| miR-138 | Downregulated | Sp1, HIF2α | Chondrocytes differentiation | [37] [38] |
The first large scale screening of hundreds of miRNAs in OA cartilage was performed by Iliopoulos et al [29]. The screening of 365 differentially expressed miRNAs in articular cartilage established the cartilage specific profile of miRNA and identified 16 miRNAs (9 upregulated and 7 downregulated) and collaborative metabolic and inflammatory networks in OA pathogenesis [29, 39]. Later, Akhtar N et al were the first to demonstrate that miR-27b was differentially expressed in both normal and OA chondrocytes and involved in the regulation of MMP-13 expression in IL-1β stimulated human OA chondrocytes [38]. In other studies, the closely related miR-27a was also shown to exert anti-inflammatory effects by suppressing MMP13 and IGFBP-5 expression in cultured chondrocytes [34]. Yamasaki K et al (2009) showed that miR-146 is upregulated in IL-1β stimulated OA chondrocytes and regulate the expression of IRAK-1 (IL-1-receptor associated kinase 1) and TRAF-6 (TNF-receptor associated factor 6) by binding to the “seed sequence” present in the 3′-UTR of their mRNAs [40]. Several studies analyzed the miRNAs expression profile in the plasma and synovial fluid of OA patients and the data suggested that monitoring synovial fluid and plasma miRNAs have the potential as diagnostic biomarkers for OA and can be used for the analysis of the disease pathogenesis or outcome [41, 42]. These and other studies highlighted the importance of miRNA networks in the pathogenesis of OA and suggest that detection and targeting of specific miRNAs may be of value in early diagnosis and therapy of OA [31, 33, 35, 43, 44].
2.2.2 lncRNAs in OA
Long noncoding RNA (lncRNA) is defined as a regulatory ncRNA greater than 200 nucleotides in length and lacks an open reading frame of significant length and is not translated into a protein [20, 45]. LncRNAs have widespread biological functions and have been shown to exhibit 5′-cap, 3′-polyA tail and involved in splicing into multi-exons that exhibit transcriptional activity [19, 46]. Recent literature showed that lncRNAs are aberrantly expressed in OA cartilage and thus contributed to the degeneration of chondrocyte extracellular matrix [47, 48]. Table 2 provides a brief summary of differentially expressed lncRNAs in human OA cartilage tissue and potentially playing a role in OA.
Table 2.
Differentially expressed lncRNAs and their role in OA pathogenesis
| Target lncRNA | Expression in OA | Molecular effects in OA | References |
|---|---|---|---|
| HOTAIR | Upregulation | ECM degradation, Apoptosis, | [49] [50] |
| GAS5 | Upregulation | ECM degradation, Apoptosis | [49] |
| PCGEM1 | Upregulation | Apoptosis | [51] |
| HOTTIP | Upregulation | ECM degradation | [52] |
| LncRNA-CIR | Upregulation | ECM degradation | [48] |
| H19 | Downregulation | ECM degradation | [49] |
| MEG3 | Downregulation | Angiogenesis | [53] |
| CILinc01 | Downregulation | Inflammation | [54] |
| CILinc02 | Downregulation | Inflammation | [54] |
Xing D et al (2014) using lncRNA microarray analysis identified several differentially expressed lncRNAs (73 upregulated lncRNAs and 48 downregulated) in OA cartilage compared with normal cartilage [49]. These differentially expressed lncRNAs included the upregulated expression of HOTAIR, growth arrest-specific 5 (GAS5), PMS2L2, RP11-445H22.4, H19 and CTD-2574D22.4 [49]. Additionally, Fu and colleagues have demonstrated that there are 3007 upregulated lncRNAs and 1707 downregulated lncRNAs in OA cartilage when the expression was compared to the lncRNAs expression profile in normal cartilage samples [47]. Altogether, there are a number of differently expressed lncRNAs in human OA cartilage and functional investigation of these lncRNA is required to establish their role in OA pathogenesis.
Some recent studies have identified the role of lncRNAs in the regulation ECM degradation and the inflammatory response. Liu et al. (2014) identified a novel lncRNA termed as cartilage injury-related lncRNA (lncRNA-CIR) which was highly expressed in OA cartilage and OA chondrocytes and its knockdown was shown to inhibit the expression of MMP-13 and ADAMTS-5 and enhanced the expression of anabolic genes COL2A1 and ACAN in OA chondrocytes [48]. Additionally, Pearson et al. identified two novel chondrocyte lincRNAs (CILinc01 and CILinc02) having a role in the regulation of inflammation in OA
2.3 Histone modifications in OA
Histones are alkaline proteins found in the nucleus that envelope the DNA to form the nucleosomes. Histones undergo modifications that altered the chromatin conformation and influenced the binding of transcription factors with the promoter region. There are more than 150 histone modifications reported which have variable effects on gene transcription and such modification includes acetylation, methylation, phosphorylation and ubiquitination of histone residues [55]. A specific combination of histone marks is associated with silenced regions, promoters, enhancers, and transcribed regions of the genome. Acetylation/deacetylation and methylation/demethylation of histones are the primary modifications studied in OA.
2.3.1 Histone acetylation in OA
Acetylation of specific lysine residues on the N-terminal tails of histones is mediated through enzymatic activity of histone acetyltransferases (HATs) which oxidize amine to amides and nullifies the positive charge of histones. Thus acetylation decreases the binding ability of histones with DNA, thereby preventing condensation of chromatin and therefore allows the binding of transcription factor leading to initiation of gene expression [56]. On the contrary, deacetylation is carried out by histone deacetylases (HDAC) that provide a positively charged histone tail, encouraging high affinity binding between the DNA backbone and histones, resulting in chromatin condensation, and thus preventing transcription leading to repression of gene expression. The HDAC proteins are classified into two functional groups based on DNA sequence similarity and activities. Classic HDAC proteins (class I, II, and IV) possess a zinc-dependent active site and these families have 11 known members named HDAC 1 to 11. HDAC class III proteins require the NAD+ cofactor for activity and are known as sirtuins that are further grouped into 7 subtypes named chronologically (SIRT 1 to 7) [57–59]. The balance between HAT and HDAC has a strong impact on the chondrocyte phenotype and was shown to be altered by biological treatments used in arthritis depicting the important role of histone modifications in the disease [60].
Recent studies demonstrated that altered activation of HDACs and their differential expression patterns contribute to the initiation and progression of OA. Expression levels of HDAC1 and HDAC2 were higher in OA chondrocytes and were shown to repress the expression of genes encoding COL2A1, ACAN, COMP, COL11A1 [61–64]. HDAC4 is known to be a major regulator of chondrocyte hypertrophy and abnormal expression of HDAC4 in OA cartilage suggests its involvement in promoting the catabolic activity of chondrocytes associated with OA pathogenesis [65]. Cao et al. further demonstrated that decreased HDAC4 expression in OA chondrocytes corresponds with increased expression of Runx2 and other MMPs suggesting that HDAC4 has a chondroprotective effect in OA cartilage [66]. Additionally, Higashiyama et al (2010) demonstrated the enhanced expression of HDAC7 in OA cartilage which correlated with the increased production of MMP-13 and ECM degradation [67]. These studies suggest that inhibiting or augmenting the expression of specific HDACs may be useful in the management of OA.
Class III HDAC family (Sirtuins family), which is composed of seven members, seems to play a pivotal role in chromatin regulation in cartilage. It has been shown that chondrocytes isolated from OA patients SIRT1 activity was essential for cartilage-specific expression of extracellular components ACAN and COL2A1 [68]. Other studies showed the in vivo relevance of SIRT1 in cartilage biology by demonstrating that the articular cartilage in Sirt1−/− mice had altered chondrocyte phenotype with high levels of MMP13 and increased chondrocyte apoptosis [69, 70]. Furthermore, over-expression of SIRT1 in chondrocytes under stimulation with IL-1β reduced the expression of matrix degrading proteases MMPs 1, 2, 9, 10, 11, 12, and 13 and ADAMTS-5 suggesting a protective role of SIRT1 in cartilage [71]. Recently, a known SIRT1 activator has been shown to inhibit the destruction of cartilage by increasing SIRT1 activity in mouse OA cartilage [72]. Collectively, these studies suggest that SIRT1 activators may be useful in the management of OA. In addition to SIRT1, SIRT6 has also been shown to play a protective role in OA. Wu Y et al (2015) in his recent study demonstrated that SIRT6 protein levels were significantly low in OA chondrocytes compared to normal human chondrocytes and over-expression of SIRT6 in mouse joints protected from cartilage degeneration by reducing the expression of NF-κB-dependent inflammatory genes [73].
2.3.2 Histone methylation in OA
Histones methylation is an important modification resulting in the formation of active and inactive genomic regions which is associated with both transcriptional activation and silencing [74]. Methylation of lysine or arginine residues on histone tails is catalysed by histone methyl transferases (HMTs) and protein arginine methyltransferases (PRMTs) whereas demethylation of histone is catalyzed by histone demethylases [75]. Histone methylation adds one or more methyl groups to regulate transcription. Depending on the residue involve in the histones methylation, various transcriptional consequences occur such as the addition of three methyl groups to lysine 27 of histone 3 (H3K27) which results in transcriptional repression whereas methylation of lysine 4 in histone 3 (H3K4) leads to transcriptional activation [76]. Few reports are available regarding histone methylation in chondrocytes. A recent study showed that IL-1 induces histone H3K4 di- and trimethylation around the COX2 and iNOS promoters but not that of MMP-13 and histone methyltransferase inhibition prevented the IL-1-induced COX-2 and iNOS expressions in chondrocytes [77]. Only limited number of studies investigated the role of histone methylation in OA and this field lack large studies of genome-wide histone modification patterns in OA, particularly integrated studies of histone methylation and gene expression patterns; this should be a focus of future research efforts.
2.4 Potential of HDAC inhibitors as therapeutics in OA
HDACs including HDAC1, HDAC2 and HDAC7 are upregulated in OA chondrocytes and their inhibition using specific HDAC inhibitor (HDACi) has been shown to confer protection and prevent the ECM degradation in human OA chondrocytes [14, 65, 78]. Inhibiting HDAC activity offers potential solutions to prevent or reverse the molecular events involved in OA pathogenesis. Although HDACi exerts chondroprotective effect and provide benefits to permanent and adult articular cartilage by decreasing production of catabolic cartilage matrix genes, there are few reports demonstrating that genetic and chemical inhibition of HDAC during development causes multiple skeletal deformities and negatively affects chondrocyte biology. HDACi includes a range of naturally occurring as well as synthetic compounds, which differ in terms of function and HDAC specificity. Pan-HDACi such as Trichostatin A and Vorinostat inhibit the activity of class I and II HDACs (HDAC 1 to 10), whereas others are class/isoform-selective inhibitors such as FK-228 inhibits HDAC 1 and 2.
Recent evidence demonstrates that HDAC inhibitors have chondroprotective effects in OA models. Wang X et al (2009) showed that broad-spectrum HDACi Trichostatin A prevents IL1β-mediated upregulation of MMPs in human OA chondrocyte [79]. Additionally, Culley et al. (2013) found that inhibition of Class I HDACs via two other agents (Valproic acid and MS275) was sufficient to prevent IL1β-induced upregulation of matrix degrading proteases-MMP-1 and MMP-13 in human OA chondrocytes [80]. These authors further demonstrated that systemic administration of Trichostatin A (TSA) prevents cartilage destruction in a model of surgically induced OA [80]. Similarly, other studies also demonstrated the chondroprotective potential of TSA in a rabbit experimental OA model where TSA prevented cartilage degradation and production of matrix degrading MMP1, MMP3, and MMP13; Cathepsins K, B, L, and S; and Cystatin C, as well as IL-1 [81]. Young et al. further demonstrated the chondroprotective effects of HDAC inhibition by demonstrating the TSA and sodium butyrate mediated inhibition of MMPs and ADAMTS5 expression in pro-inflammatory cytokine treated human chondrocytes [82].
Cai et al. using Nrf2−/− mice explored the systemic histone deacetylase inhibition as a novel strategy to prevent OA and identify a role for Nrf2 in preventing the cartilage degeneration in preclinical model of OA [83]. These authors showed that loss of Nrf2 in mice causes severe cartilage damage in OA models and administration of TSA promotes the expression of Nrf2 in wild type mouse joint tissues and reduces cartilage destruction [83]. However, TSA did not offer significant protection from OA in Nrf2−/− mice, suggesting that Nrf2 is required for TSA-dependent protective benefits in OA [83]. Further an earlier study by Nasu et al. demonstrated the efficacy of systemic TSA administration in reducing the expression of various MMPs, as well as measures of cartilage damage in a collagen antibody-injection OA mouse model [84]. Chen et al demonstrated the alleviation of OA by TSA and found that elevated levels of MMPs and IL-1 were significantly reduced by TSA treatment in an experimental model of OA [85]. Further Saito et al (2013) has demonstrated that TSA suppressed the mechanical stress induced expression of RUNX-2 and ADAMTS-5 by inhibiting the activation of MAPK signaling (ERK1/2, p38MAPK, JNK) in human chondrocytes [86].
The United States Food and Drug Administration (FDA) approved several broad-acting HDACi for clinical use for various diseases. Vorinostat or SAHA, Zolinza; Romidepsin were approved for cutaneous T cell lymphoma, myeloma, other hematopoietic cancers and solid tumors, whereas Valproic acid, Valproate, Divalproex sodium, Depakote were approved for epilepsy and bipolar disorder. Other HDACis such as Panobinostat or LBH589; Entinostat or MS-275; Givinostat or ITI2357 are currently under evaluation in various clinical trials for treatments of other cancers and conditions such as inflammation, neurodegeneration, and diabetes [87]. There are 609 HDACi related human clinical trials completed/ongoing (https://clinicaltrials.gov; last accessed July 31, 2017); however, none of them are related to OA. One study has assessed the safety and efficacy of an oral HDACi Givinostat (ITF2357) in systemic-onset juvenile idiopathic arthritis and found significant therapeutic effects with regard to the arthritic component of the disease [88].
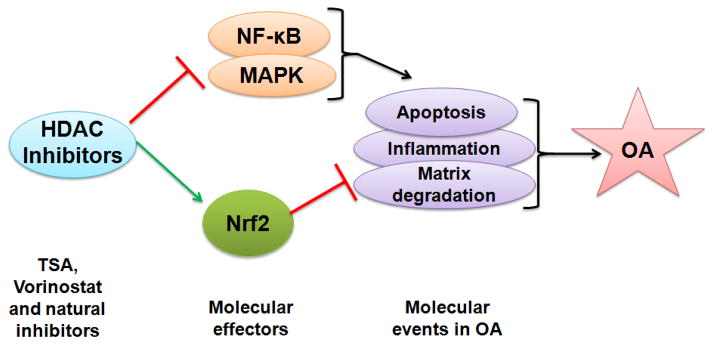
Vorinostat has emerged as a promising HDACi which is orally bioavailable and acts as a broad-spectrum inhibitor of class I and II HDACs. Vorinostat, chemically known as Suberoylanilide hydroxamic acid, and clinically as Zolinza, is clinically the most advanced HDACi [89]. Approximately half of all of the reported clinical trials on HDACi are with Vorinostat. Vorinostat was first approved in 2006 by the US Food and Drug Administration for the treatment of cutaneous manifestations in patients with cutaneous T-cell lymphoma in whom other treatments have failed. Zhong et al in his recent study used Vorinostat, a broad-spectrum HDACi and demonstrate that Vorinostat inhibits IL1β-induced upregulation of various MMPs as well as nitric oxide production in human chondrocytes in vitro, selectively acting through p38 and ERK1/2 inhibition [90]. Further Wang et al. demonstrated another indirect mechanism by which Vorinostat prevents cartilage degeneration in OA [91]. The HDACi- Vorinostat and Panobinostat has been shown to elevate miR-46, which is a negative regulator of inflammatory responses, in fibroblast-like synoviocytes from OA joints [91]. A recent report by Makki et al (2016) indicated that Vorinostat is an effective suppressor of IL-6 induced signaling events in OA [92]. This study opens a new avenue in the management of OA by demonstrating the Vorinostat-mediated suppression of MMP-13 through inhibition of IL-6 in human OA chondrocytes suggesting the potential of Vorinostat as a therapeutic agent for the management of OA. The molecular aspect of Vorinostat mediated protective effect demonstrate that it down-regulates iNOS (inducible nitric oxide synthase), and selectively inhibits NF-kB signaling and represses IL-1β induced p38-MAPK and ERK 1/2 activation without affecting JNK activation [90]. These studies indicate that the inhibition of HDACs could have a beneficial effect in the management of osteoarthritic conditions through selective inhibition of known molecular targets of OA (Figure 2).
Figure 2. Molecular effectors of HDAC inhibitors in osteoarthritis.
HDAC inhibitors (TSA, Vorinostat) inhibits the activation of NF-κB and MAPK (ERK1/2, JNK, p38MAPK) which are major players of OA pathogenesis known to be involved in the matrix degradation, apoptosis and inflammatory response in OA. HDAC inhibitor also activates transcription factor Nrf2 which is known to inhibit the molecular events of inflammation and matrix degradation in OA.
Interestingly class III HDACs (Sirtuins family) has also been shown to play a pivotal role in chromatin regulation in cartilage. SirT1 has recently been show as essential regulator of chondrocyte survival through activation of insulin-like growth factor signaling and inhibition p53 mediated apoptosis [93]. Additionally, SirT1 also promotes the expression of cartilage matrix gene such as ACAN, COL2A1, COL9A1 and COMP through deacetylation of SOX9 and activation of HIF-2α [68]. The inhibition of SIRT1 by siRNA induces significant downregulation of ACAN and upregulation of COL10A1 and ADAMTS-5 [94]. The inhibition of SirT1 leads to an increase in chondrocyte apoptosis while addition of resveratrol, a sirtuin activator, protects chondrocytes from cell death [95]. Thus, recent evidence suggests that HDACi have the potential to protect articular cartilage from destruction by preventing matrix degradation.
Although, many of HDACi are synthetic compounds but some HDACi are derived naturally from microbial metabolites (eg, TSA, isolated from Streptomyces hygroscopicus). The first molecule identified to affect histone acetylation status was allyl derivative isolated from garlic extract. Other prominent plant-based HDACi includes Sulforaphane (found in cruciferous vegetables) and Quercetin (found in a variety of fruits and beverage) [57]. Because HDACi seems to possess therapeutic potential against a variety of diseases, there is increasing interest in the potential of dietary compounds that possess HDAC inhibitory activities. However, some of the natural agents appear to have dual effects on HDACs. For example, Resveratrol is a reported activator of class III HDAC SIRT1 but also has been shown to inhibit the other 3 class of HDACs (class I, II, and IV) [96]. Other examples are of Curcumin and Genistein, which exhibit both HDAC inhibitory and histone acetylation activator activities [57]. It appears that further research is required to determine whether the naturally occurring HDAC modulators may be useful in the management of OA.
2.5 Conclusions
There is an increasing interest in exploiting the therapeutic use of epigenetics and HDACi in the treatment of a variety of disorders linked to HDAC dysfunction. HDACs post-translationally alter the histone and non-histone proteins, which affect the final gene expression outcome. Expression of certain HDACs has been shown to be deregulated in OA. In recent years, the roles of HDACs in cartilage development and homeostasis, as well as in the advancement of OA pathogenesis, have become better clarified; however, this area of research is still in its infancy and detailed investigations are needed to understand the functional and therapeutic significance of specific HDACs inhibition in OA. There is still much to be discovered about their mechanisms of action. Most studies investigating the roles of HDACs in cartilage have studied in the context of chondrocytes from young animals, but there are limited mechanistic studies in articular cartilage of adults or aged animals. Determining which HDAC is to be targeted and whether a selective HDACi or a pan-HDACi would be useful in the therapeutic management of OA is an important area of future research. A recent report from our lab is an important effort in this direction that provides substantial evidence for the potential use of HDACi-Vorinostat in the management of OA. Although use of HDACi as a therapeutic intervention in OA is promising, use of newer generations of HDACi is expected to be useful for the selective and effective inhibition of molecular events involved in OA pathogenesis.
Acknowledgments
This work was supported by National Institute of Health grants (RO1-AT-005520; RO1-AT-007373; RO1-AR- 067056; R21-AR- 064890) and funds from the Northeast Ohio Medical University to TMH.
Footnotes
Conflict of Interest Disclosure
Authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Murphy L, Helmick CG. The impact of osteoarthritis in the United States: a population-health perspective. The American journal of nursing. 2012;112(3 Suppl 1):S13–9. doi: 10.1097/01.NAJ.0000412646.80054.21. [DOI] [PubMed] [Google Scholar]
- 2.Blagojevic M, Jinks C, Jeffery A, Jordan KP. Risk factors for onset of osteoarthritis of the knee in older adults: a systematic review and meta-analysis. Osteoarthritis and cartilage. 2010;18(1):24–33. doi: 10.1016/j.joca.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 3.Chen C, Tambe DT, Deng L, Yang L. Biomechanical properties and mechanobiology of the articular chondrocyte. American journal of physiology Cell physiology. 2013;305(12):C1202–8. doi: 10.1152/ajpcell.00242.2013. [DOI] [PubMed] [Google Scholar]
- 4.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis and rheumatism. 2012;64(6):1697–707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldring MB, Berenbaum F. Emerging targets in osteoarthritis therapy. Curr Opin Pharmacol. 22:51–63. doi: 10.1016/j.coph.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldring SR, Goldring MB. Changes in the osteochondral unit during osteoarthritis: structure, function and cartilage-bone crosstalk. Nat Rev Rheumatol. 2016;12(11):632–644. doi: 10.1038/nrrheum.2016.148. [DOI] [PubMed] [Google Scholar]
- 7.Houard X, Goldring MB, Berenbaum F. Homeostatic mechanisms in articular cartilage and role of inflammation in osteoarthritis. Curr Rheumatol Rep. 2013;15(11):375. doi: 10.1007/s11926-013-0375-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khan NM, Haseeb A, Ansari MY, Devarapalli P, Haynie S, Haqqi TM. Wogonin, a plant derived small molecule, exerts potent anti-inflammatory and chondroprotective effects through the activation of ROS/ERK/Nrf2 signaling pathways in human Osteoarthritis chondrocytes. Free radical biology & medicine. 2017;106:288–301. doi: 10.1016/j.freeradbiomed.2017.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bui C, Barter MJ, Scott JL, Xu Y, Galler M, Reynard LN, Rowan AD, Young DA. cAMP response element-binding (CREB) recruitment following a specific CpG demethylation leads to the elevated expression of the matrix metalloproteinase 13 in human articular chondrocytes and osteoarthritis. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2012;26(7):3000–11. doi: 10.1096/fj.12-206367. [DOI] [PubMed] [Google Scholar]
- 10.Bird A. Perceptions of epigenetics. Nature. 2007;447(7143):396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 11.Romanoski CE, Glass CK, Stunnenberg HG, Wilson L, Almouzni G. Epigenomics: Roadmap for regulation. Nature. 2015;518(7539):314–6. doi: 10.1038/518314a. [DOI] [PubMed] [Google Scholar]
- 12.Lev Maor G, Yearim A, Ast G. The alternative role of DNA methylation in splicing regulation. Trends in genetics: TIG. 2015;31(5):274–80. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466(7303):253–7. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im GI, Choi YJ. Epigenetics in osteoarthritis and its implication for future therapeutics. Expert opinion on biological therapy. 2013;13(5):713–21. doi: 10.1517/14712598.2013.764410. [DOI] [PubMed] [Google Scholar]
- 15.Roach HI, Yamada N, Cheung KS, Tilley S, Clarke NM, Oreffo RO, Kokubun S, Bronner F. Association between the abnormal expression of matrix-degrading enzymes by human osteoarthritic chondrocytes and demethylation of specific CpG sites in the promoter regions. Arthritis and rheumatism. 2005;52(10):3110–24. doi: 10.1002/art.21300. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez K, de Andres MC, Takahashi A, Oreffo RO. Effects of hypoxia on anabolic and catabolic gene expression and DNA methylation in OA chondrocytes. BMC Musculoskelet Disord. 15:431. doi: 10.1186/1471-2474-15-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeffries MA, Donica M, Baker LW, Stevenson ME, Annan AC, Humphrey MB, James JA, Sawalha AH. Genome-wide DNA methylation study identifies significant epigenomic changes in osteoarthritic cartilage. Arthritis & rheumatology (Hoboken NJ) 2014;66(10):2804–15. doi: 10.1002/art.38762. [DOI] [PubMed] [Google Scholar]
- 18.Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Seminars in cell & developmental biology. 2014;34:9–14. doi: 10.1016/j.semcdb.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma L, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA biology. 2013;10(6):925–33. doi: 10.4161/rna.24604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15(Spec No 1):R17–29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Barter MJ, Young DA. Epigenetic mechanisms and non-coding RNAs in osteoarthritis. Curr Rheumatol Rep. 2013;15(9):353. doi: 10.1007/s11926-013-0353-z. [DOI] [PubMed] [Google Scholar]
- 23.Sondag GR, Haqqi TM. The Role of MicroRNAs and Their Targets in Osteoarthritis. Curr Rheumatol Rep. 2016;18(8):56. doi: 10.1007/s11926-016-0604-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyaki S, Asahara H. Macro view of microRNA function in osteoarthritis. Nature reviews Rheumatology. 2012;8(9):543–52. doi: 10.1038/nrrheum.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyaki S, Nakasa T, Otsuki S, Grogan SP, Higashiyama R, Inoue A, Kato Y, Sato T, Lotz MK, Asahara H. MicroRNA-140 is expressed in differentiated human articular chondrocytes and modulates interleukin-1 responses. Arthritis and rheumatism. 2009;60(9):2723–30. doi: 10.1002/art.24745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araldi E, Schipani E. MicroRNA-140 and the silencing of osteoarthritis. Genes & development. 2010;24(11):1075–80. doi: 10.1101/gad.1939310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barter MJ, Tselepi M, Gomez R, Woods S, Hui W, Smith GR, Shanley DP, Clark IM, Young DA. Genome-Wide MicroRNA and Gene Analysis of Mesenchymal Stem Cell Chondrogenesis Identifies an Essential Role and Multiple Targets for miR-140-5p. Stem cells (Dayton Ohio) 2015;33(11):3266–80. doi: 10.1002/stem.2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Min Z, Zhang R, Yao J, Jiang C, Guo Y, Cong F, Wang W, Tian J, Zhong N, Sun J, Ma J, Lu S. MicroRNAs associated with osteoarthritis differently expressed in bone matrix gelatin (BMG) rat model. International journal of clinical and experimental medicine. 2015;8(1):1009–17. [PMC free article] [PubMed] [Google Scholar]
- 29.Iliopoulos D, Malizos KN, Oikonomou P, Tsezou A. Integrative microRNA and proteomic approaches identify novel osteoarthritis genes and their collaborative metabolic and inflammatory networks. PloS one. 2008;3(11):e3740. doi: 10.1371/journal.pone.0003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones SW, Watkins G, Le Good N, Roberts S, Murphy CL, Brockbank SM, Needham MR, Read SJ, Newham P. The identification of differentially expressed microRNA in osteoarthritic tissue that modulate the production of TNF-alpha and MMP13. Osteoarthritis and cartilage. 2009;17(4):464–72. doi: 10.1016/j.joca.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Makki MS, Haseeb A, Haqqi TM. MicroRNA-9 promotion of interleukin-6 expression by inhibiting monocyte chemoattractant protein-induced protein 1 expression in interleukin-1beta-stimulated human chondrocytes. Arthritis & rheumatology (Hoboken NJ) 2015;67(8):2117–28. doi: 10.1002/art.39173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YJ, Maizel A, Chen X. Traffic into silence: endomembranes and post-transcriptional RNA silencing. The EMBO journal. 2014;33(9):968–80. doi: 10.1002/embj.201387262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akhtar N, Makki MS, Haqqi TM. MicroRNA-602 and microRNA-608 regulate sonic hedgehog expression via target sites in the coding region in human chondrocytes. Arthritis & rheumatology (Hoboken NJ) 2015;67(2):423–34. doi: 10.1002/art.38952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tardif G, Hum D, Pelletier JP, Duval N, Martel-Pelletier J. Regulation of the IGFBP-5 and MMP-13 genes by the microRNAs miR-140 and miR-27a in human osteoarthritic chondrocytes. BMC musculoskeletal disorders. 2009;10:148. doi: 10.1186/1471-2474-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Akhtar N, Haqqi TM. MicroRNA-199a* regulates the expression of cyclooxygenase-2 in human chondrocytes. Annals of the rheumatic diseases. 2012;71(6):1073–80. doi: 10.1136/annrheumdis-2011-200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li ZC, Han N, Li X, Li G, Liu YZ, Sun GX, Wang Y, Chen GT, Li GF. Decreased expression of microRNA-130a correlates with TNF-alpha in the development of osteoarthritis. International journal of clinical and experimental pathology. 2015;8(3):2555–64. [PMC free article] [PubMed] [Google Scholar]
- 37.Seidl CI, Martinez-Sanchez A, Murphy CL. Derepression of MicroRNA-138 Contributes to Loss of the Human Articular Chondrocyte Phenotype. Arthritis & rheumatology (Hoboken NJ) 2016;68(2):398–409. doi: 10.1002/art.39428. [DOI] [PubMed] [Google Scholar]
- 38.Akhtar N, Rasheed Z, Ramamurthy S, Anbazhagan AN, Voss FR, Haqqi TM. MicroRNA-27b regulates the expression of matrix metalloproteinase 13 in human osteoarthritis chondrocytes. Arthritis and rheumatism. 2010;62(5):1361–71. doi: 10.1002/art.27329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diaz-Prado S, Cicione C, Muinos-Lopez E, Hermida-Gomez T, Oreiro N, Fernandez-Lopez C, Blanco FJ. Characterization of microRNA expression profiles in normal and osteoarthritic human chondrocytes. BMC musculoskeletal disorders. 2012;13:144. doi: 10.1186/1471-2474-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamasaki K, Nakasa T, Miyaki S, Ishikawa M, Deie M, Adachi N, Yasunaga Y, Asahara H, Ochi M. Expression of MicroRNA-146a in osteoarthritis cartilage. Arthritis and rheumatism. 2009;60(4):1035–41. doi: 10.1002/art.24404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borgonio Cuadra VM, Gonzalez-Huerta NC, Romero-Cordoba S, Hidalgo-Miranda A, Miranda-Duarte A. Altered expression of circulating microRNA in plasma of patients with primary osteoarthritis and in silico analysis of their pathways. PLoS One. 2014;9(6):e97690. doi: 10.1371/journal.pone.0097690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H, Nakamura T. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12(3):R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin EA, Kong L, Bai XH, Luan Y, Liu CJ. miR-199a, a bone morphogenic protein 2-responsive MicroRNA, regulates chondrogenesis via direct targeting to Smad1. The Journal of biological chemistry. 2009;284(17):11326–35. doi: 10.1074/jbc.M807709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sumiyoshi K, Kubota S, Ohgawara T, Kawata K, Nishida T, Shimo T, Yamashiro T, Takigawa M. Identification of miR-1 as a micro RNA that supports late-stage differentiation of growth cartilage cells. Biochemical and biophysical research communications. 2010;402(2):286–90. doi: 10.1016/j.bbrc.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 45.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nature reviews Genetics. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 46.Fitzgerald KA, Caffrey DR. Long noncoding RNAs in innate and adaptive immunity. Current opinion in immunology. 2014;26:140–6. doi: 10.1016/j.coi.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, Yu B, Meng F. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis and cartilage. 2015;23(3):423–32. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 48.Liu Q, Zhang X, Dai L, Hu X, Zhu J, Li L, Zhou C, Ao Y. Long noncoding RNA related to cartilage injury promotes chondrocyte extracellular matrix degradation in osteoarthritis. Arthritis & rheumatology (Hoboken NJ) 2014;66(4):969–78. doi: 10.1002/art.38309. [DOI] [PubMed] [Google Scholar]
- 49.Xing D, Liang JQ, Li Y, Lu J, Jia HB, Xu LY, Ma XL. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthopaedic surgery. 2014;6(4):288–93. doi: 10.1111/os.12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang C, Wang P, Jiang P, Lv Y, Dong C, Dai X, Tan L, Wang Z. Upregulation of lncRNA HOTAIR contributes to IL-1beta-induced MMP overexpression and chondrocytes apoptosis in temporomandibular joint osteoarthritis. Gene. 2016;586(2):248–53. doi: 10.1016/j.gene.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Kang Y, Song J, Kim D, Ahn C, Park S, Chun CH, Jin EJ. PCGEM1 stimulates proliferation of osteoarthritic synoviocytes by acting as a sponge for miR-770. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2016;34(3):412–8. doi: 10.1002/jor.23046. [DOI] [PubMed] [Google Scholar]
- 52.Kim D, Song J, Han J, Kim Y, Chun CH, Jin EJ. Two non-coding RNAs, MicroRNA-101 and HOTTIP contribute cartilage integrity by epigenetic and homeotic regulation of integrin-alpha1. Cellular signalling. 2013;25(12):2878–87. doi: 10.1016/j.cellsig.2013.08.034. [DOI] [PubMed] [Google Scholar]
- 53.Su W, Xie W, Shang Q, Su B. The Long Noncoding RNA MEG3 Is Downregulated and Inversely Associated with VEGF Levels in Osteoarthritis. BioMed research international. 2015;2015:356893. doi: 10.1155/2015/356893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson MJ, Philp AM, Heward JA, Roux BT, Walsh DA, Davis ET, Lindsay MA, Jones SW. Long Intergenic Noncoding RNAs Mediate the Human Chondrocyte Inflammatory Response and Are Differentially Expressed in Osteoarthritis Cartilage. Arthritis & rheumatology (Hoboken NJ) 2016;68(4):845–56. doi: 10.1002/art.39520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 56.Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Molecular cell. 2006;23(3):289–96. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 57.Bassett SA, Barnett MP. The role of dietary histone deacetylases (HDACs) inhibitors in health and disease. Nutrients. 2014;6(10):4273–301. doi: 10.3390/nu6104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. Journal of molecular biology. 2004;338(1):17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Inoue T, Hiratsuka M, Osaki M, Oshimura M. The molecular biology of mammalian SIRT proteins: SIRT2 in cell cycle regulation. Cell cycle (Georgetown Tex) 2007;6(9):1011–8. doi: 10.4161/cc.6.9.4219. [DOI] [PubMed] [Google Scholar]
- 60.Toussirot E, Wendling D, Herbein G. Biological treatments given in patients with rheumatoid arthritis or ankylosing spondylitis modify HAT/HDAC (histone acetyltransferase/histone deacetylase) balance. Joint, bone spine: revue du rhumatisme. 2014;81(6):544–5. doi: 10.1016/j.jbspin.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 61.Bradley EW, Carpio LR, van Wijnen AJ, McGee-Lawrence ME, Westendorf JJ. Histone Deacetylases in Bone Development and Skeletal Disorders. Physiological reviews. 2015;95(4):1359–81. doi: 10.1152/physrev.00004.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong S, Derfoul A, Pereira-Mouries L, Hall DJ. A novel domain in histone deacetylase 1 and 2 mediates repression of cartilage-specific genes in human chondrocytes. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23(10):3539–52. doi: 10.1096/fj.09-133215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huh YH, Ryu JH, Chun JS. Regulation of type II collagen expression by histone deacetylase in articular chondrocytes. The Journal of biological chemistry. 2007;282(23):17123–31. doi: 10.1074/jbc.M700599200. [DOI] [PubMed] [Google Scholar]
- 64.Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. The Journal of biological chemistry. 2004;279(45):47081–91. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 65.Lu J, Sun Y, Ge Q, Teng H, Jiang Q. Histone deacetylase 4 alters cartilage homeostasis in human osteoarthritis. BMC musculoskeletal disorders. 2014;15:438. doi: 10.1186/1471-2474-15-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao K, Wei L, Zhang Z, Guo L, Zhang C, Li Y, Sun C, Sun X, Wang S, Li P, Wei X. Decreased histone deacetylase 4 is associated with human osteoarthritis cartilage degeneration by releasing histone deacetylase 4 inhibition of runt-related transcription factor-2 and increasing osteoarthritis-related genes: a novel mechanism of human osteoarthritis cartilage degeneration. Arthritis research & therapy. 2014;16(6):491. doi: 10.1186/s13075-014-0491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higashiyama R, Miyaki S, Yamashita S, Yoshitaka T, Lindman G, Ito Y, Sasho T, Takahashi K, Lotz M, Asahara H. Correlation between MMP-13 and HDAC7 expression in human knee osteoarthritis. Modern rheumatology. 2010;20(1):11–7. doi: 10.1007/s10165-009-0224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dvir-Ginzberg M, Gagarina V, Lee EJ, Hall DJ. Regulation of cartilage-specific gene expression in human chondrocytes by SirT1 and nicotinamide phosphoribosyltransferase. The Journal of biological chemistry. 2008;283(52):36300–10. doi: 10.1074/jbc.M803196200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gabay O, Zaal KJ, Sanchez C, Dvir-Ginzberg M, Gagarina V, Song Y, He XH, McBurney MW. Sirt1-deficient mice exhibit an altered cartilage phenotype. Joint, bone spine: revue du rhumatisme. 2013;80(6):613–20. doi: 10.1016/j.jbspin.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gabay O, Sanchez C, Dvir-Ginzberg M, Gagarina V, Zaal KJ, Song Y, He XH, McBurney MW. Sirtuin 1 enzymatic activity is required for cartilage homeostasis in vivo in a mouse model. Arthritis and rheumatism. 2013;65(1):159–66. doi: 10.1002/art.37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsushita T, Sasaki H, Takayama K, Ishida K, Matsumoto T, Kubo S, Matsuzaki T, Nishida K, Kurosaka M, Kuroda R. The overexpression of SIRT1 inhibited osteoarthritic gene expression changes induced by interleukin-1beta in human chondrocytes. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2013;31(4):531–7. doi: 10.1002/jor.22268. [DOI] [PubMed] [Google Scholar]
- 72.Li W, Cai L, Zhang Y, Cui L, Shen G. Intra-articular resveratrol injection prevents osteoarthritis progression in a mouse model by activating SIRT1 and thereby silencing HIF-2alpha. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2015;33(7):1061–70. doi: 10.1002/jor.22859. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y, Chen L, Wang Y, Li W, Lin Y, Yu D, Zhang L, Li F, Pan Z. Overexpression of Sirtuin 6 suppresses cellular senescence and NF-kappaB mediated inflammatory responses in osteoarthritis development. Scientific reports. 2015;5:17602. doi: 10.1038/srep17602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee DY, Teyssier C, Strahl BD, Stallcup MR. Role of protein methylation in regulation of transcription. Endocrine reviews. 2005;26(2):147–70. doi: 10.1210/er.2004-0008. [DOI] [PubMed] [Google Scholar]
- 75.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125(2):213–7. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 76.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, Gold DL, Sekido Y, Huang TH, Issa JP. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nature genetics. 2008;40(6):741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 77.El Mansouri FE, Chabane N, Zayed N, Kapoor M, Benderdour M, Martel-Pelletier J, Pelletier JP, Duval N, Fahmi H. Contribution of H3K4 methylation by SET-1A to interleukin-1-induced cyclooxygenase 2 and inducible nitric oxide synthase expression in human osteoarthritis chondrocytes. Arthritis and rheumatism. 2011;63(1):168–79. doi: 10.1002/art.27762. [DOI] [PubMed] [Google Scholar]
- 78.Carpio LR, Westendorf JJ. Histone Deacetylases in Cartilage Homeostasis and Osteoarthritis. Current rheumatology reports. 2016;18(8):52. doi: 10.1007/s11926-016-0602-z. [DOI] [PubMed] [Google Scholar]
- 79.Wang X, Song Y, Jacobi JL, Tuan RS. Inhibition of histone deacetylases antagonized FGF2 and IL-1beta effects on MMP expression in human articular chondrocytes. Growth factors (Chur, Switzerland) 2009;27(1):40–9. doi: 10.1080/08977190802625179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Culley KL, Hui W, Barter MJ, Davidson RK, Swingler TE, Destrument AP, Scott JL, Donell ST, Fenwick S, Rowan AD, Young DA, Clark IM. Class I histone deacetylase inhibition modulates metalloproteinase expression and blocks cytokine-induced cartilage degradation. Arthritis and rheumatism. 2013;65(7):1822–30. doi: 10.1002/art.37965. [DOI] [PubMed] [Google Scholar]
- 81.Chen WP, Bao JP, Tang JL, Hu PF, Wu LD. Trichostatin A inhibits expression of cathepsins in experimental osteoarthritis. Rheumatology international. 2011;31(10):1325–31. doi: 10.1007/s00296-010-1481-7. [DOI] [PubMed] [Google Scholar]
- 82.Young DA, Lakey RL, Pennington CJ, Jones D, Kevorkian L, Edwards DR, Cawston TE, Clark IM. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis research & therapy. 2005;7(3):R503–12. doi: 10.1186/ar1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai D, Yin S, Yang J, Jiang Q, Cao W. Histone deacetylase inhibition activates Nrf2 and protects against osteoarthritis. Arthritis research & therapy. 2015;17:269. doi: 10.1186/s13075-015-0774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nasu Y, Nishida K, Miyazawa S, Komiyama T, Kadota Y, Abe N, Yoshida A, Hirohata S, Ohtsuka A, Ozaki T. Trichostatin A, a histone deacetylase inhibitor, suppresses synovial inflammation and subsequent cartilage destruction in a collagen antibody-induced arthritis mouse model. Osteoarthritis and cartilage. 2008;16(6):723–32. doi: 10.1016/j.joca.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 85.Chen WP, Bao JP, Hu PF, Feng J, Wu LD. Alleviation of osteoarthritis by Trichostatin A, a histone deacetylase inhibitor, in experimental osteoarthritis. Molecular biology reports. 2010;37(8):3967–72. doi: 10.1007/s11033-010-0055-9. [DOI] [PubMed] [Google Scholar]
- 86.Saito T, Nishida K, Furumatsu T, Yoshida A, Ozawa M, Ozaki T. Histone deacetylase inhibitors suppress mechanical stress-induced expression of RUNX-2 and ADAMTS-5 through the inhibition of the MAPK signaling pathway in cultured human chondrocytes. Osteoarthritis and cartilage. 2013;21(1):165–74. doi: 10.1016/j.joca.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 87.Dinarello CA, Fossati G, Mascagni P. Histone deacetylase inhibitors for treating a spectrum of diseases not related to cancer. Molecular medicine (Cambridge Mass) 2011;17(5–6):333–52. doi: 10.2119/molmed.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vojinovic J, Damjanov N, D’Urzo C, Furlan A, Susic G, Pasic S, Iagaru N, Stefan M, Dinarello CA. Safety and efficacy of an oral histone deacetylase inhibitor in systemic-onset juvenile idiopathic arthritis. Arthritis and rheumatism. 2011;63(5):1452–8. doi: 10.1002/art.30238. [DOI] [PubMed] [Google Scholar]
- 89.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nature biotechnology. 2007;25(1):84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 90.Zhong HM, Ding QH, Chen WP, Luo RB. Vorinostat, a HDAC inhibitor, showed anti-osteoarthritic activities through inhibition of iNOS and MMP expression, p38 and ERK phosphorylation and blocking NF-kappaB nuclear translocation. International immunopharmacology. 2013;17(2):329–35. doi: 10.1016/j.intimp.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 91.Wang JH, Shih KS, Wu YW, Wang AW, Yang CR. Histone deacetylase inhibitors increase microRNA-146a expression and enhance negative regulation of interleukin-1beta signaling in osteoarthritis fibroblast-like synoviocytes. Osteoarthritis and cartilage. 2013;21(12):1987–96. doi: 10.1016/j.joca.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 92.Makki MS, Haqqi TM. Histone Deacetylase Inhibitor Vorinostat (SAHA) Suppresses IL-1beta-Induced Matrix Metallopeptidase-13 Expression by Inhibiting IL-6 in Osteoarthritis Chondrocyte. The American journal of pathology. 2016;186(10):2701–8. doi: 10.1016/j.ajpath.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gagarina V, Gabay O, Dvir-Ginzberg M, Lee EJ, Brady JK, Quon MJ, Hall DJ. SirT1 enhances survival of human osteoarthritic chondrocytes by repressing protein tyrosine phosphatase 1B and activating the insulin-like growth factor receptor pathway. Arthritis and rheumatism. 2010;62(5):1383–92. doi: 10.1002/art.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujita N, Matsushita T, Ishida K, Kubo S, Matsumoto T, Takayama K, Kurosaka M, Kuroda R. Potential involvement of SIRT1 in the pathogenesis of osteoarthritis through the modulation of chondrocyte gene expressions. Journal of orthopaedic research: official publication of the Orthopaedic Research Society. 2011;29(4):511–5. doi: 10.1002/jor.21284. [DOI] [PubMed] [Google Scholar]
- 95.Takayama K, Ishida K, Matsushita T, Fujita N, Hayashi S, Sasaki K, Tei K, Kubo S, Matsumoto T, Fujioka H, Kurosaka M, Kuroda R. SIRT1 regulation of apoptosis of human chondrocytes. Arthritis and rheumatism. 2009;60(9):2731–40. doi: 10.1002/art.24864. [DOI] [PubMed] [Google Scholar]
- 96.Venturelli S, Berger A, Bocker A, Busch C, Weiland T, Noor S, Leischner C, Schleicher S, Mayer M, Weiss TS, Bischoff SC, Lauer UM, Bitzer M. Resveratrol as a pan-HDAC inhibitor alters the acetylation status of histone [corrected] proteins in human-derived hepatoblastoma cells. PloS one. 2013;8(8):e73097. doi: 10.1371/journal.pone.0073097. [DOI] [PMC free article] [PubMed] [Google Scholar]