Abstract
Micropatterning is a widely used powerful tool to create highly ordered microstructures on material surfaces. However, due to technical limitations, the integration of micropatterned microstructures into bio-inspired three-dimensional (3D) scaffolds to successfully regenerate well-organized functional tissues has not been achieved. In this work, we report a unique maskless micropatterning technology to create 3D nanofibrous matrices with highly-organized tubular architecture for tissue regeneration. Our micropatterning method is a laser-guided, non-contact, high-precision, flexible computer programming of machining process that can create highly ordered tubules with the density ranged from 1,000–60,000/mm2 and the size varied from 300 nm - 30 μm in the bio-inspired 3D matrix. The tubular architecture presents pivotal biophysical cues to control dental pulp stem cell alignment, migration, polarization, and differentiation. More importantly, when using this 3D tubular hierarchical matrix as a scaffold, we have successfully regenerated functional tubular dentin that has the same well-organized microstructure as its natural counterpart. Our 3D maskless micropattern approach represents a powerful avenue not only for the exploration of cell-material interactions in 3D, but also for the regeneration of functional tissues with well-organized microstructures.
Keywords: micropattern, tubular, tissue regeneration, three-dimension, biomimetic
Graphical Abstract
A unique maskless micropatterning technology is developed to create 3D nanofibrous matrices with highly-organized tubular architecture for tissue regeneration. The tubular architecture presents crucial biophysical cues to dental pulp stem cell migration, polarization, and differentiation. When using this 3D tubular hierarchical matrix as a scaffold, functional tubular dentin is successfully regenerated with the same well-organized microstructure as its natural counterpart.
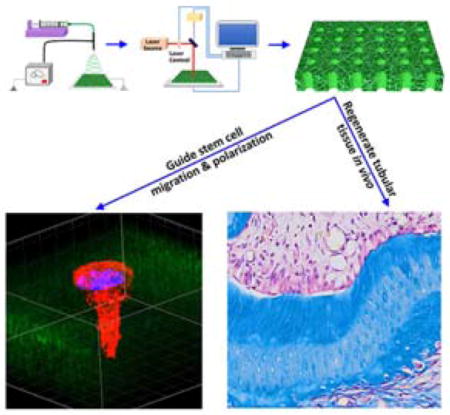
1. Introduction
Micropatterning is a widely used powerful tool to manipulate cell-material interactions.[1, 2] Since the late 1970s, a number of micropatterning technologies, such as photolithography, microcontact printing, microfluidic patterning and direct writing, have been developed to pattern the surfaces of materials to regulate cellular behaviors.[2, 3] Those efforts have greatly advanced our knowledge of how to control cell alignment, migration, proliferation, differentiation, and molecular signaling pathways and therefore, offer invaluable insight into scaffolding design for tissue engineering. The majority of the micropatterning technologies, however, focus on using non-biodegradable materials as substrates, such as silicon, poly(dimethylsiloxane) (PDMS), titanium, poly(ethylene glycol) (PEG), and polyacrylate,[4] which are not suitable to serve as scaffolding materials for tissue regeneration. While a few biodegradable polymers were used as substrates to produce micropatterns with ordered structures such as grooves, pillars, wells, and pits, most of those micropatterns were located on the surfaces of two-dimensional (2D) films or sheets.[5] To date, the integration of micropatterns into bio-inspired three-dimensional (3D) scaffolds to successfully regenerate well-organized functional tissues has not been achieved.
Many tissues in the body possess well-organized microstructures that are a prerequisite for their physiological functions. In particular, dentin is the major component of a tooth and is composed of millions of closely packed dentinal tubules that have a density of 15,000–48,000/mm2 and a diameter of 2–4 μm (Figure 1b–d).[6] The tubules are arranged in parallel and extend through the entire thickness of the dentin from the pulp chamber to the dentinoenamel junction (DEJ) (Figure 1b). The unique tubular microstructure of the dentin offers channels to transfer fluid and stimulus signals and provides strong mechanical support for the tooth.[7] Without the well-organized tubular structure, the tooth cannot perform its normal mechanical and biological functions. Therefore, when dentin is destroyed or damaged due to trauma or disease, regeneration of the tubular architecture of the dentin is crucial to fully recover its physiological functions. However, this effort has long been a challenge, and to date, only bone-like mineralized tissues or sporadic dentinal tubules were formed in the root canal chamber after in vivo tissue regeneration.[8] The main obstacle to regenerating such a well-ordered tubular microstructure is the lack of innovative technologies to prepare bio-inspired micropatterned scaffolds that closely mimic the natural dental extracellular matrix (ECM) and provide critical biophysical cues to guide dental stem cell growth and differentiation. Here, we report a unique maskless micropatterning technology to create a 3D nanofibrous tubular matrix with the same hierarchical architecture as natural dentin ECM. Using this bio-inspired 3D matrix as a scaffold, we are the first to guide dental pulp stem cell (DPSC) alignment, polariztion, and differentiation, and to successfully regenerate tubular dentin of the same structure as its natural counterpart.
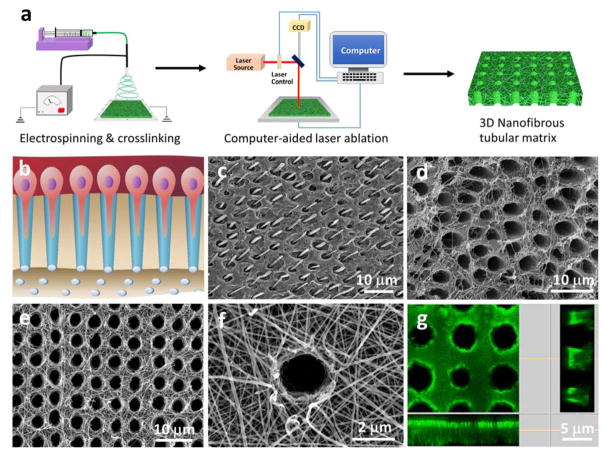
Figure 1.
Design, fabrication and characterization of micropatterned tubular 3D matrix. (a) Schematic of the electrospinning and laser-guided micropatterning processes to fabricate 3D nanofibrous tubular gelatin matrices. (b) Schematic of odontoblasts aligned on the interface between dentin and pulp and the extension of their long processes deep inside the dentinal tubules. (c, d) SEM images of human tubular dentin. In (c), the dentin tubules were filled with resin and acid-etched to retain the long process of odontoblasts in the tubules. After removal of the mineral and odontoblast processes, the collagen nanofibers surrounding the dentinal tubules were clearly seen in image (d). (e) A typical SEM image of the micropatterned tubular gelatin matrix, (f) High magnification of (e), showing the nanofibrous gelatin matrix. (g) Confocal image of the tubular gelatin matrix. The longitudinal section image of the matrix indicates that the tubular pores are completely open.
2. Results and Discussion
The challenge of fabricating synthetic dentin-like tubular matrix is twofold. First, the tubular micropattern, which has to be formed in a 3D matrix, cannot be fabricated by most of the current micropatterning methods, such as soft lithography and microcontact printing. While a fiber-templating method was reported to fabricate tubules, its minimal diameter (≥10 μm) was much larger than that of dentinal tubules (2–4 μm).[9] Moreover, the fiber-templating process required the polymerization of non-biodegradable acrylate, which is not suitable to serve as a scaffolding material. Second, dentin ECM is a nanofibrous structure at the nanoscale level; therefore, the synthetic tubular matrix should mimic this architecture to facilitate DPSC growth and differentiation. However, the incorporation of nanofibrous architecture into small tubules (1–10 μm) to form hierarchical structures is technically challenging and has not been reported via conventional micropatterning technologies. To overcome these challenges, we developed an innovative laser-guided micropatterning technology, which was further combined with an electrospinning process to prepare a 3D nanofibrous tubular matrix (Figure 1). Our micropatterning method is a non-contact, high-precision, flexible computer programming of machining process. While our method can be used for structuring a wide range of biomaterials, we chose gelatin as the scaffolding material to mimic the natural dentin ECM, because gelatin has almost the same chemical composition as the collagen that is the dominant organic component of dentin ECM. In addtion, gelatin is a denatured biopolymer and the selection of gelatin as a scaffolding material can circumvent the concerns of immunogenicity and pathogen transmission associated with collagen Figure 1a illustrates the fabrication process to prepare the 3D bio-inspired tubular matrix. First, the nanofibrous gelatin matrix was prepared using an electrospinning process and then crosslinked to stabilize its structure as we previously reported.[10, 11] This nanofibrous architecture has been proven to provide an excellent microenvironment for mesenchymal stem cell adhesion, proliferation, and differentiation.[12] Next, the micropattern was designed by using laser microdissection software (Leica Laser Microdissection V7.5.1). At room temperature and under the control of the micropatterning program, a Leica laser microdissection 7000 (Leica microsystem, Germany) was used to create the tubular micropattern within the nanofibrous gelatin matrix. The laser output power and aperture were used to control the size and the depth of the tubular pores, and the stage moving speed and laser pulse frequency were used to control the distance between the tubules. As shown in Figure 1e, the synthetic tubular matrix had the same size and pattern of tubules as those of natural dentin ECM (Figure 1d). A high-magnification image showed that the matrix was composed of gelatin nanofibers with an average diameter of approximately 200 nm (Figure 1f), which is at the same range of collagen fibers in dentin ECM.[11] To visualize the tubular architecture, we coupled the gelatin matrix with fluorescein isothiocyanate (FITC), and the confocal image clearly showed that each tubular pore of the gelatin matrix was completely open which is crucial for guiding DSPC alignment and differentiation to odontoblasts (Figure 1g).
The micropatterned gelatin matrix (a tubule density of 20,000/mm2 and a pore size of 2–5 μm) had a tensile strength of 7.67±0.88 MPa and 2.79±0.13 MPa under dry and wet conditions, respectively. Furthermore, the tubular matrix had the maximal elongation ratio of 4.2±0.7% under dry status and 55.0±2.5% under wet condition (Support Information, Figure S1), indicating that the micropatterned gelatin matrix was a ductile biomaterial with appropriate mechanical strength. In addition, the tubular gelatin matrix was biodegradable, and its degradation rate was modulated by the crosslinking density, ranging from several days to a few months (Supporting Information, Figure S2 and S3).
Laser-capture microdissection is usually applied to procure specific single cells or an entire area of tissues under direct microscopic visualization. It is typically used in functional genomics and proteomic studies. Here we are the first to use the laser-capture microdissection system as a tool for tubular structure fabrication with precise control of the micropattern size and scaffolding architecture. The Leica Laser Microdissection 7000 system has high energy per pulse and ultra-short pulse length (<4 ns), which allows high cutting resolution. Energy-dispersive X-ray (EDX) spectroscopy showed that the composition of the matrix adjacent to the tubules remained intact, indicating that the ultra-short laser pulses of this system had minimal thermal stress and collateral damage to the gelatin matrix (data now shown). This is a flexible technique that is performed at room temperature in a mild atmosphere without requiring expensive clean-room facilities. In addition, this mothed does not need a mask as a template and can be used to micropattern a wide range of biomaterials.
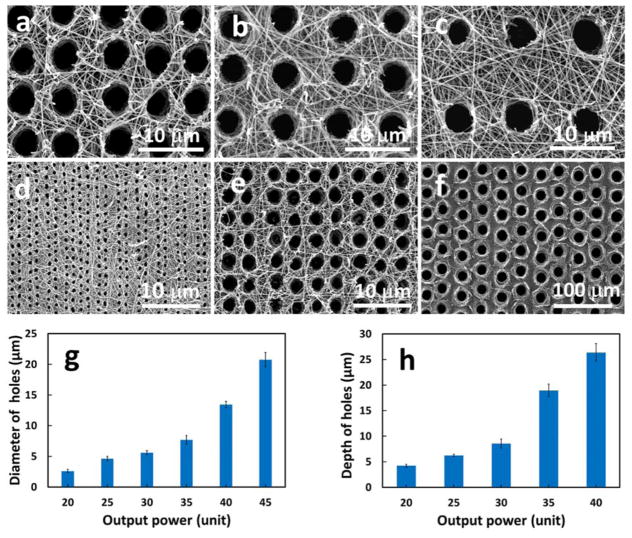
The micropattern (e.g., tubular density and size) of the matrix was precisely controlled by the laser pulse frequency, laser aperture, writing speed, and laser power. As shown in Figure 2, the tubule density was set from 1,000–60,000/mm2 by laser writing speed (Figure 2a–c), and the size of the tubule varied from less than 300 nm to more than 30 μm based on the laser output power (Figure 2d–f). When the output power increased from 20 to 45 units, the size of the tubules increased from 2.6±0.3 μm to 20.8±1.2 μm (Figure 2g). As shown in Figure 2h, the depth of the tubules in the matrix created by a single laser pulse also increased with the laser power. In addition, the depth of the tubules increased with multiple pulses on the same pore. Since we mimicked human dentin matrix, the synthetic tubular gelatin matrix in this study had a tubule density of 20,000/mm2 and a pore size of 2–5 μm. The speed of the micropattering was fast and it took approximately 8.3 minutes to complete every square millimeter. Next, we used this micropatterned 3D matrix for tubular dentin regeneration.
Figure 2.
The SEM tubular matrix images with different densities and sizes of the tubules controlled by laser frequency (a) 40Hz, (b) 30 Hz, and (c) 20Hz; and by laser power (d) 20 Unit, (e) 30 Unit, and 45 Unit. (g) The diameter of tubular holes increased with the laser output power. (h) The depth of the tubular holes increased with the laser output power.
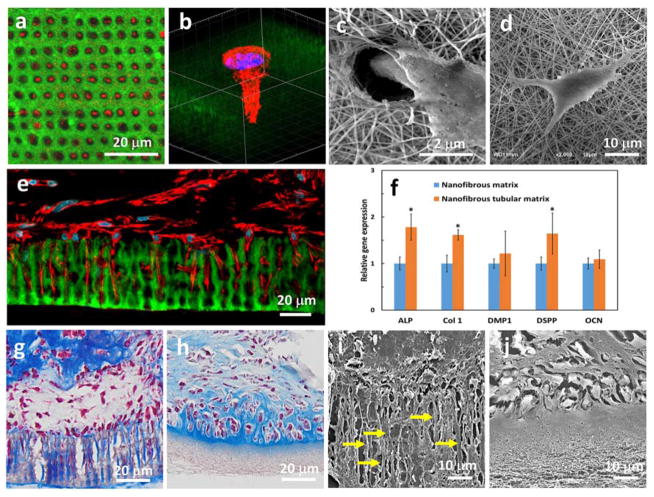
DPSC is one of the most promising candidates for dental tissue regeneration. It is highly proliferative, multipotent, and capable of forming a dentin-like complex in vivo.[13] Compared to other cell sources, DPSC can be easily obtained from extracted teeth that are generally discarded in the clinic. Therefore, we chose DPSCs for tubular dentin regeneration in this work. We first examined the adhesion of the DPSCs on the micropatterned 3D matrix. Forty-eight hours after cell seeding, a layer of DPSCs had aggregated on the top of the matrix. More importantly, the DPSCs that directly contacted the matrix surface polarized and elongated with long extensions (red) invading the tubular matrix (green) (Figure 3a). Over 80% micro- patterns were occupied by the cells (n>300). A high-magnification confocal image shows that the DPSC extended its process into the tubular structure of the matrix, while the nuclei of the DPSC stayed on the surface of the matrix (Figure 3b). A scanning electron microscopy (SEM) image further confirmed the long process of the DPSC directionally migrated into a tubule of the matrix (Figure 3c).
Figure 3.
(a) Confocal image of DPSCs seeded on the micropatterned matrix with their processes (red) invading the tubular matrix (green). (b) High-magnification confocal image showing a DPSC extending its process into a tubule, while the nuclei are located on the surface of the matrix. (c) An SEM image showing that the DPSC spread their extension into a tubule of the micropatterned gelatin matrix. (d) An SEM image showing that a DPSC presented several randomly-oriented protrusions attached to the nanofibers on the surface of the non-patterned gelatin matrix. (e) A confocal image taken after the DPSC/matrix construct was cultured in vitro for 7 days. The DPSCs were polarized and their cellular processes (red) extended into the tubular matrix (green). (f) Comparison of gene expression of DPSCs on the micropatterned and non-patterned gelatin matrix. (g, h) Trichrome staining after the cell/matrix constructs were cultured in vitro in different media for 2 weeks. In (g), the odontoblasts (red) aligned on the pore surfaces and secreted a large number of collagen fibers (blue) that were arranged along the tubules of the micropatterned matrix. In (h), the cells seeded on the non-micropatterned matrix did not present processes but formed lacuna-like cell clusters. (i, j) SEM images of cell/matrix constructs after being cultured in vitro in differentiation media for 2 weeks. The long processes of cells were only observed in the micropatterned matrix, but not in the non-patterned counterpart.
A cell shape circularity index (CI) =4πA/L2 was used to describe the polarization of a cell, where A is the area of the cell, and L is the perimeter of the cell.[14] The value of CI was no more than 1.0 under any circumstance, and a lower value of the CI is characteristic of a migratory polarized cell. The value of the CI was calculated to be 0.25±0.05, indicating that the DPSC was in a high polarized morphology on the tubular matrix. In contrast, the DPSC on the non-tubular matrix presented branched morphology and had several randomly-oriented protrusions attached to the nanofibers on the surface of the matrix (Figure 3d). No major processes were formed, and the cell shape CI of the DPSC on the non-tubular matrix was 0.51±0.10, which was much higher than that of the DPSC on the tubular matrix.
After culturing in proliferation media for 7 days, almost all the tubules of the gelatin matrix were occupied by the processes of the DPSCs (Figure 3e). Furthermore, while the nuclei of the columnar DPSCs were still located on the outside of the tubules, the processes of the elongated DPSCs were more than 20 μm long and extended deep into the entire tubules. These polarized DPSCs had cell morphology very similar to odontoblasts, which are the most distinctive cells in dental pulp. In a natural tooth, the odontoblasts form a layer aligning on the interface between dentin and pulp and extend their long processes deep inside the dentin tubules. This result indicated that the tubular matrix presented pivotal biophysical cues to guide DPSC polarization and long process formation.
Fourteen days after culturing in differentiation media, we examined the gene expression of DPSCs in the tubular matrix and found the differential markers (ALP, Col I, and DSPP) on the tubular matrix were significantly higher than those on the non-tubular matrix (Figure 3f), indicating that the tubular architecture accelerated the differentiation of the DPSCs to the odontoblasts. It was reported that micropattern cues on silicon wafers can be sensed by cells via integrin-mediated focal adhesion signaling that subsequently activates mitogen-activated protein kinase/extracellular signaling-regulated kinase (MAPK/ERK) cascades to enhance adult neural stem cell differentiation.[15] Also, the phosphorylation levels of the MAPKs, ERK1/2 and p38 were up-regulated when the DPSCs were cultured in a demineralized dentin matrix and their inhibitors significantly inhibited the odontogenic differentiation of the DPSCs.[16] Since our synthetic tubular matrix had the same architecture as that of the demineralized dentin matrix, we speculate that our topography-induced differentiation to odontoblasts on the tubular matrix may follow the MAPK/ERK1/2 signaling pathway. However, the detailed studies of the underlying mechanism are beyond the scope of this work and will be examined in a separate study.
We further used primary dental pulp cells from first molar tooth buds of mice to test the feasibility of forming a tubular structure on the synthetic tubular matrix in vitro. After culturing in differentiation media for two weeks, a layer of odontoblasts (red) was aligned on the surface of the tubular matrix, and an abundant amount of Type I collagen (blue) was deposited along the long tubules of the matrix (Figure 3g). In contrast, the cells seeded on the non-micropatterned nanofibrous matrix did not generate processes (Figure 3h). Instead, they formed lacuna-like cell clusters and were encapsulated by the Type I collagen secreted by themselves. SEM images showed the same results (Figure 3i&j), confirming the pivotal role of the tubular architecture of the synthetic matrix in guiding cell polarization and differentiation.
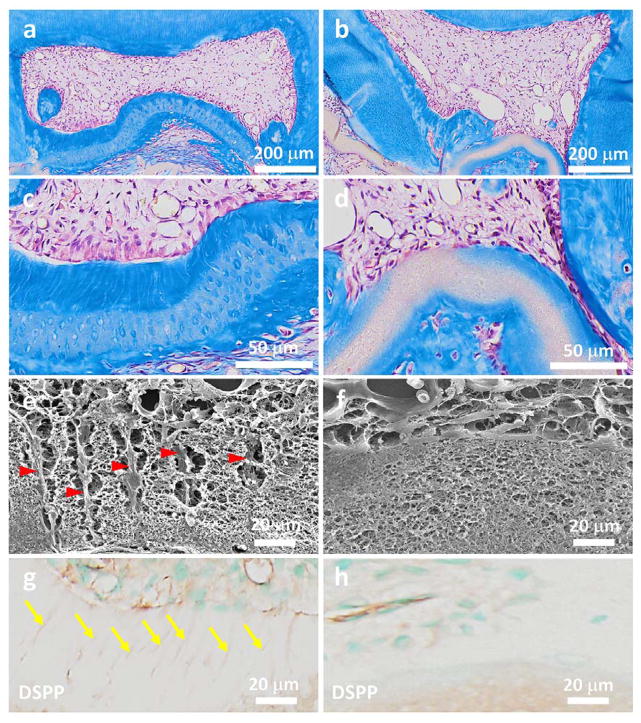
While various micropatterning matrices have been fabricated to control stem cell fate, none of them was truly designed to regenerate tissues. Here we show the first example of regenerating functional tubular dentin using our synthetic tubular 3D matrix as a scaffold. We subcutaneously implanted cell/nanofibrous tubular gelatin matrix constructs in immunocompromised nude mice (NU/NU, 5 weeks, Charles Rivers) to examine tubular dentin regeneration. For comparison, the nanofibrous gelatin scaffold without tubular architecture was included as a control. The animals were sacrificed, and samples were retrieved after 4 weeks of transplantation. Masson’s trichrome staining shows that a thick layer of well-organized tubular dentin with an average length of 45 μm was successfully regenerated in the tubular matrix group (Figures 4a&c). A palisading odontoblast layer was aligned along the surface of the newly formed dentin (Figure 4c). The SEM image further indicated that the odontoblastic processes were embedded in the mineralized tissue and reached the entire layer of the new dentin matrix (Figure 4e). Furthermore, a number of newly formed blood vessels were observed in the pulp tissue area. Immunohistochemical staining shows dentin sialophosphoprotein (DSPP), which is considered to be the dentin-specific marker of odontoblasts, was strongly expressed in the newly formed matrix (Figure 4g). These results indicated the success of regenerating the tubular and vascularized pulpodentin complex using our tubular gelatin scaffold. In contrast, no tubular dentin structures were formed in the non-patterned control group (Figures 4b&d). There were no odontoblasts aligned along the surface of the non-patterned gelatin matrix (Figure 4d). In addition, little collagen was deposited and no DSPP was detected in the non-patterned gelatin matrix (Figures 4d&h). Taken together, these results demonstrated the importance of the physical architecture of the tubular scaffold in odontoblastic differentiation and proved that the bio-inspired 3D scaffolds with dentinal tubule-like architecture were critical for tubular dentin regeneration.
Figure 4.
Regeneration of tubular tissues in vivo using a 3D micropatterned matrix. The cell/matrix constructs were implanted in nude mice for 4 weeks. (a–d) Masson’s trichrome staining shows a thick layer of well-organized tubular dentin that was regenerated in the tubular matrix group (a, c); while no tubular dentin structure was formed in the non-patterned control group (b, d). In addition, a palisading odontoblast layer was aligned along the surface of the newly formed tubular dentin. (b) and (d) are the high magnification images of (a) and (b), respectively. (e, f) SEM images of cell/matrix constructs after being implanted in vivo for 4 weeks. The long processes of cells embeded in the fibrous matrix were clearly observed in the micropatterned group (yellow arrowheads in e), but not in the non-patterned group (f). (g, h) Immunohistochemical staining of DSPP in both the tubular micropatterned and non-micropatterned groups. DSPP, which is considered the dentin-specific marker of odontoblasts, was highly expressed in the tubular micropatterned group (yellow arrows in g), but not in the non-micropatterned control group, indicating the pivotal role of the tubular architecture in odontoblastic differentiation and tubular dentin regeneration.
3. Conclusions
In summary, we have presented a unique micropatterning technology to fabricate 3D micropatterned tubular matrices that truly mimic natural dentin ECM using synthetic biomaterials in this work. Our study shows that this bio-inspired scaffold provides crucial biophysical cues to guide dental stem cell migration, polarization, and differentiation, and regenerate tubular dentin. It should be noted that our 3D maskless micropatterning technique can be readily employed for regenerating other tissues with tubular structures such as nephrons and blood vessels. In addition, the micropatterned synthetic matrix is composed of a biocompatible material with a clear background; therefore, it is an excellent platform for exploring cell-material and cell-cell interactions (e.g., epithelial-mesenchymal interactions). Combining our technology with other processes, more complex micropatterns and other cell types can be implemented to achieve a broader degree of 3D cell patterning for functional tissue regeneration. Our 3D maskless micropattern approach can be readily used on a broad range of both natural and synthetic biomaterials; therefore, represents a powerful avenue not only for the exploration of a range of fundamental biological questions, but also for the regeneration of functional tissues with well-organized structures.
4. Experimental Section
Preparation of 3D tubular matrices
Gelatin (Type B, 225g Bloom) was dissolved in hexafluoroisopropanol at a concentration of 5% (wt/v) and used for electrospinning. The electrospinning process was performed in an electrospinning system (SPRAYBASE® PLATFORM, Ireland) with a voltage of 12 kV (Gamma High Voltage, USA) and a feed rate of at 0.5 ml/h using a digital controlled infusion pump (Cole Palmer Inc, USA). The distance between the drum collector and the spray tip was 10 cm. The collected gelatin nanofibers were crosslinked in the solution containing 50 mM 2-ethanesulfonic acid (MES), 10 mM N-Ethyl-N-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), and 10 mM N-Hydroxysuccinimide (NHS) at 4°C for 24h. The crosslinked gelatin matrix was incubated in 50 mM glycine solution to neutralize the unreacted EDC, washed with distilled water, dehydrated in absolute ethanol, and dried in a vacuum oven.
The laser-guided micropatterning process was carried out using a Leica LMD 7000 system. This system has a laser wavelength of 349 nm, a pulse length of less than 4 ns, and maximal pulse energy of 120 μJ. The operation parameters included real pulse energy (laser power and aperture), pulse frequency, speed of the moving stage, and drill times. The power and aperture were used to control the size and depth of the micropattern. The pulse frequency was used to adjust the times of the laser pulse in every time unit. When the speed of the moving stage was constant, a lower pulse frequency increased the distance between each laser pulse. The parameters for fabricating the tubular matrix in Figure 1e were as follows: laser aperture=29, laser pulse energy=32, laser speed=19, and laser pulse frequency=40 Hz. Using these parameters, more than 1.4×105 tubular pores were created per hour, which could be further increased with the pulse frequency. The resulting nanofibrous micropatterned matrix had a tubular pore density of 20,000/mm2 and a tubular diameter of less than 5 μm, similar to that of a natural dentinal matrix.
Cell seeding onto micropatterned 3D matrices
The human DPSCs were a gift from Dr. Songtao Shi, of the University of Pennsylvania School of Dental Medicine. The thawed DPSCs (passage 2) were cultured in α-modified essential medium (α-MEM, Gibco, Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and penicillin-streptomycin (Invitrogen) in a humidified incubator with 5% CO2 at 37°C. DPSCs of passages 3–5 were used for all the cellular studies. The tubular matrix was soaked in 70% ethanol for 30 min, washed three times with PBS for and then washed with culture medium. After the DPSCs were seeded onto the matrix, the cell/matrix construct was cultured in the odontogenic differentiation medium (containing 50 μg/mg ascorbic acid, 5mM β-glycerophosphate, and 10 nM dexmethasone) with 5% CO2 at 37°C. The culture medium was changed every day.
Reverse transcription polymerase chain reaction (RT-PCR)
The DPSCs on the matrix were harvested for the RT-PCR test 14 days after cell culturing. The total RNA was extracted using the RNeasy plus MiniKit (Qiagen, USA), and the first-strand cDNA was reverse-transcribed with QuantiTect Rev. Transcription Kit (Qiagen, USA). Human-specific primers were designed and synthesized as follows: ALP (sense5′-CCACGTCTTCACATTTGGTG-3′; antisense 5′-AGACTGCGCCTGGTAGTTGT-3′), Col 1 (sense 5′-AAAAGGAAGCTTGGTCCACT-3′; antisense 5′-GTGTGGAGAAAGGAGCAGAA-3′), DMP1 (sense 5′-TGGGGATTATCCTGTGCTCT-3′; antisense 5′-TACTTCTGGGGTCACTGTCG-3′); OCN (sense 5′-ACTGTGACGAGTTGGCTGAC-3′; antisense 5′-CAAGGGCAAGAGGAAAGAAG-3′) and DSPP (sense 5′-TTAAATGCCAGTGGAACCAT-3′, antisense 5′-ATTCCCTTCTCCCTTGTGAC-3′). GAPDH (sense 5′-AGCCGCATCTTCTTTTGCGTC-3′; antisense 5′-TCATATTTGGCAGGTTTTTCT-3′) was used as the internal control. RT-PCR was carried out using a Bio-Rad C1000™ Thermal Cycler. The conditions for the RT-PCR were as follows: denaturation at 95°C (10 min), 40 cycles at 95°C (15s) with a final 1 min extension at 60°C. The results were calculated from three independent experiments.
Tubular dentin regeneration in vivo
The animal surgical procedure was approved by the University Committee on the Use and Care of Animals (UCUCA) of the Texas A&M College of Dentistry. Tooth molar crowns were obtained from the mandibular first molars of Sprague-Dawley rats. Each molar was carefully cleaned with a fine needle (30 G) and washed with PBS (2% Penicillin-Streptomycin). The rat crowns were dissected along with the tooth cervix using a saw. The pulp cells were seeded on the matrices (tubular and non-tubular) and then transferred to the surface of the rat crown. The scaffold/crown constructs were fixed using 6-0 silk suture and subcutaneously implanted on the backs of nude mice (NU/NU, 5 weeks, Charles Rivers). Four weeks after transplantation, the samples were retrieved, fixed in 4% paraformaldehyde (PFA), decalcified and processed for histological and immunohistochemical staining and SEM analysis.
Histological and immunohistochemical staining
After dehydration, the specimens were treated in xylene and embedded in paraffin. The specimens were cut in a lateral direction into alternating 5 μm-thick sections and processed for histological observation using Masson’s trichrome staining. For immunohistochemical analysis, the sections were dewaxed in xylene and ethanol and allowed to react with primary antibody of dentin sialophosphoprotein (DSPP, mouse monoclonal to DSPP gifted by Dr. Chunlin Qin, Texas A&M University College of Dentistry) (1:200). All the immunohistochemical experiments were detected using a 3, 3-diaminobenzidine kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturer’s instructions.
Characterization of the tubular matrix and histological sections
For SEM observation, the tubular matrices were coated with gold for 120 s using a sputter coater (SPI-module Sputter Coater Unit, SPI Supplies/Structure Probe, Inc.). During the gold-coating process, the gas pressure was 50m Torr, and the current was 20 mA. The surface morphology of the scaffolds was examined using SEM (JEOL JSM-6010LA) with an accelerating voltage of 10 kV. The average diameter and density of the tubular matrix was calculated from 20 images using ImageJ software (http://imagej.nih.gov/ij/download.html). For the confocal observation, the samples were stained with Alexa Fluor@phalloidin 633 (Invitrogen) for 1 h at 37°C and mounted with ProLong Gold Antifade Mountant with DAPI (Thermo) according to the manufacturer’s protocol. The images were taken with a confocal laser microscope (TCS SP5, Leica, Buffalo, USA) and analyzed (six samples in each group) with ImageJ software.
Statistical analysis
Quantitative results are presented as mean ± standard deviation (SD). To test the significance of the observed differences between two groups, an unpaired Student’s t-test was applied and a value of P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by NIH/NIDCR grant R01DE024979 (X. L.). We would like to thank Dr. Songtao Shi (UPenn) for generously providing us with the DPSCs, and we thank Jeanne Santa Cruz for her assistance with the editing of this article.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]; Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Biotechnol Prog. 1998;14:356. doi: 10.1021/bp980031m. [DOI] [PubMed] [Google Scholar]
- 2.Nikkhah M, Edalat F, Manoucheri S, Khademhosseini A. Biomaterials. 2012;33:5230. doi: 10.1016/j.biomaterials.2012.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nie ZH, Kumacheva E. Nat Mater. 2008;7:277. doi: 10.1038/nmat2109. [DOI] [PubMed] [Google Scholar]; Thery M. J Cell Sci. 2010;123:4201. doi: 10.1242/jcs.075150. [DOI] [PubMed] [Google Scholar]; Cooper A, Munden HR, Brown GL. Exp Cell Res. 1976;103:435. doi: 10.1016/0014-4827(76)90283-4. [DOI] [PubMed] [Google Scholar]
- 4.Fu JP, Wang YK, Yang MT, Desai RA, Yu XA, Liu ZJ, Chen CS. Nat Methods. 2010;7:733. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhang FM, Li GC, Yang P, Qin W, Li CH, Huang N. Colloids Surf B-Biointerfaces. 2013;102:457. doi: 10.1016/j.colsurfb.2012.02.018. [DOI] [PubMed] [Google Scholar]; Hahn MS, Miller JS, West JL. Adv Mater. 2006;18:2679. [Google Scholar]
- 5.Kenar H, Kose GT, Hasirci V. Biomaterials. 2006;27:885. doi: 10.1016/j.biomaterials.2005.07.001. [DOI] [PubMed] [Google Scholar]; Wang K, Cai L, Zhang L, Dong JY, Wang SF. Adv Healthc Mater. 2012;1:292. doi: 10.1002/adhm.201200030. [DOI] [PubMed] [Google Scholar]; Zorlutuna P, Elsheikh A, Hasirci V. Biomacromolecules. 2009;10:814. doi: 10.1021/bm801307y. [DOI] [PubMed] [Google Scholar]; Lim Y, Johnson J, Fei Z, Wu Y, Farson D, Lannutti J, Choi H, Lee L. Biotechnol Bioeng. 2011;108:116. doi: 10.1002/bit.22914. [DOI] [PubMed] [Google Scholar]
- 6.Garberoglio R, Brannstrom M. Arch Oral Biol. 1976;21:355. doi: 10.1016/s0003-9969(76)80003-9. [DOI] [PubMed] [Google Scholar]; Lenzi TL, Guglielmi CDB, Arana-Chavez VE, Raggio DP. Microsc Microanal. 2013;19:1445. doi: 10.1017/S1431927613012725. [DOI] [PubMed] [Google Scholar]
- 7.Pashley DH. J Endodont. 1986;12:465. doi: 10.1016/S0099-2399(86)80201-1. [DOI] [PubMed] [Google Scholar]
- 8.Guo WH, He Y, Zhang XJ, Lu W, Wang CM, Yu H, Liu Y, Li Y, Zhou YL, Zhou J, Zhang MJ, Deng ZH, Jin Y. Biomaterials. 2009;30:6708. doi: 10.1016/j.biomaterials.2009.08.034. [DOI] [PubMed] [Google Scholar]; Ishimatsu H, Kitamura C, Morotomi T, Tahata Y, Nishihara T, Chen KK, Terashita M. J Endodont. 2009;35:858. doi: 10.1016/j.joen.2009.03.049. [DOI] [PubMed] [Google Scholar]; Galler KM, Hartgerink JD, Cavender AC, Schmalz G, D’Souza RN. Tiss Eng Part A. 2012;18:176. doi: 10.1089/ten.tea.2011.0222. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chang B, Ahuja N, Ma C, Liu XH. Mater Sci Eng R-Rep. 2017;111:1. doi: 10.1016/j.mser.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; Qu TJ, Jing JJ, Jiang Y, Taylor RJ, Feng JQ, Geiger B, Liu XH. Tiss Eng Part A. 2014;20:2422. doi: 10.1089/ten.tea.2013.0741. [DOI] [PMC free article] [PubMed] [Google Scholar]; Qu TJ, Jing JJ, Ren YS, Ma C, Feng JQ, Yu Q, Liu XH. Acta Biomaterialia. 2015;16:60. doi: 10.1016/j.actbio.2015.01.029. [DOI] [PubMed] [Google Scholar]; Qu TJ, Liu XH. J Mater Chem B. 2013;1:4764. doi: 10.1039/C3TB21002B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lluch AV, Fernandez AC, Ferrer GG, Pradas MM. J Biomed Mater Res Part B. 2009;90B:182. doi: 10.1002/jbm.b.31272. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Smith LA, Hu J, Ma PX. Biomaterials. 2009;30:2252. doi: 10.1016/j.biomaterials.2008.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu XH, Ma PX. Biomaterials. 2009;30:4094. doi: 10.1016/j.biomaterials.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sachar A, Strom TA, San Miguel S, Serrano MJ, Svoboda KKH, Liu XH. J Tiss Eng Reg Med. 2014;8:862. doi: 10.1002/term.1588. [DOI] [PubMed] [Google Scholar]; Sachar A, Strom TA, Serrano MJ, Benson MD, Opperman LA, Svoboda KKH, Liu XH. J Biomed Mater Res Part A. 2012;100A:3029. doi: 10.1002/jbm.a.34253. [DOI] [PMC free article] [PubMed] [Google Scholar]; Li Z, Qu TJ, Ding C, Ma C, Sun HC, Li SR, Liu XH. Acta Biomaterialia. 2015;13:88. doi: 10.1016/j.actbio.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ma C, Jing Y, Sun HC, Liu XH. Adv Healthc Mater. 2015;4:2699. doi: 10.1002/adhm.201500531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Proc Natl Acad Sci USA. 2000;97:13625. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lomakin AJ, Lee KC, Han SJ, Bui DA, Davidson M, Mogilner A, Danuser G. Nat Cell Bio. 2015;17:1435. doi: 10.1038/ncb3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L, Li N, Huang R, Song Q, Wang L, Zhang Q, Su RG, Kong T, Tang ML, Cheng GS. Plos One. 2013;8 doi: 10.1371/journal.pone.0059022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang HM, Liu SY, Zhou YL, Tan JL, Che HL, Ning F, Zhang XM, Xun WX, Huo N, Tang L, Deng ZH, Jin Y. Tissue Eng Part A. 2012;18:677. doi: 10.1089/ten.TEA.2011.0269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.