Abstract
Background
Alloimmunization through blood transfusion, transplantation, or circulating fetal cells during pregnancy is a significant concern. Some exposed individuals make alloantibodies while others do not, implying variation in genetic risk factors.
Methods
We conducted a genome-wide association study (GWAS) of 9,427,497 Single Nucleotide Polymorphisms (SNPs) to identify genetic variants for HLA alloimmunization in previously pregnant blood donors with (N=752) and without (N=753) HLA Class I or II alloantibodies.
Results
A SNP in the Neurexophilin 2 (NXPH2) gene surpassed genome-wide significance (P=2.06×10−8), with multiple adjacent markers P<10−6, for women with anti-class I alloantibodies only. Little is currently known about the function of NXPH2, although gene family members have been shown to impact immunity. SNPs in the E2F7 gene, a transcription factor related to cell-cycle control and cellular proliferation, also approached genome-wide significance (P= 2.5 × 10−7).
Conclusion
Further work to extend the GWAS approach and to characterize variants in NXPH2 and E2F7 in the context of alloantibody formation is warranted.
Introduction
It has been appreciated for over 50 years that pregnancy can lead to alloimmunization, (1) and high titer HLA antibodies can persist for decades after pregnancy (2). Successive pregnancies carry further risk for antibody formation. Using sensitive assays for detection of HLA antibodies, it was shown that ~10% of women are alloimmunized after one pregnancy, and this rises to ~30% of women after four or more pregnancies (3). Alloimmunization is also a significant clinical problem in transfusion medicine (4–17). While this is usually not problematic for recipients who experience limited exposures to allogeneic blood products, alloimmunization is a concern for those who depend on regular platelet or RBC transfusions, including patients with hematological malignancies, myelodysplastic disease, or patients with sickle cell disease and other inherited hemoglobin disorders (RBC dependent). Currently, leukoreduction of transfusion products (3,5,16), prophylactic transfusion of antigen matched red blood cell units (5,18,19), and frequent laboratory monitoring for new antibody formation with subsequent use of cross-match compatible blood components (5,9,18), are the only tools available to manage transfusion therapy in patients with antibodies to red blood cell, HLA, or platelet antigens. Both prior pregnancy and blood transfusion were identified as major contributors to HLA alloimmunization for subjects awaiting kidney transplant (20). The presence of HLA antibodies is associated with increased risk for rejection and graft failure, both acutely (21) and over the lifetime of the graft (22).
Alloimmunization -- whether to RBC, platelet-specific, or HLA antigens, depending on the type of blood product received or other exposure -- occurs in roughly one-third of blood recipients in the absence of full or partial RBC antigen matching or leukoreduction (5–7,14,19,23). This consistency in the alloimmunization rates among individuals, regardless of the type of exposure (e.g., RBC, HLA or platelet-antigen antibodies following transfusions, or even HLA alloimmunization following pregnancy), together with indications of overlap among the subpopulations vulnerable to alloimmunization across these various exposures, suggests the existence of host susceptibility factors that are likely to be in part genetically determined (6,8,12,24). Similar conclusions were reached with a mathematical model based on a database of thousands of transfusion recipients in diverse patient populations (23,25,26). Data from the transplantation setting suggest that women who are not alloimmunized during pregnancy are less likely to develop donor-specific antibodies after transplantation (27).
Recent technological developments (i.e., high-throughput, genome-wide single nucleotide polymorphism (SNP) genotyping using commercial arrays) enable the comprehensive search for genetic variation associated with differing risks of alloimmunization (28). Here we report the results of a GWAS including 752 alloantibody positive and 753 alloantibody negative, previously pregnant female blood donors who had been enrolled in the Leukocyte Antibody Prevalence Study (LAPS). We found associations exceeding genome-wide significance for SNPs within the NXPH2 gene and close to genome-wide significance for the E2F7 gene.
Materials and Methods
Study Subjects
Subjects were drawn from the previously conducted Retrovirus Epidemiology Donor Study-II (Reds-II) LAPS cohort (3,17), a multi-center cross-sectional study designed to measure the prevalence and laboratory characteristics of HLA and neutrophil antibodies in blood donors with and without a history of known allogeneic exposures through blood transfusion or pregnancy. The LAPS protocol involved a questionnaire for enrolled donors that characterized donors for transfusion and pregnancy history, in addition to routine demographic variables such as age, race-ethnicity and gender. Donors gave specific consent to studies of genetic factors that may be involved in immune reactions to pregnancy and transfusion, including alloimmunization. Each subject in the LAPS repository has been tested for class I and II HLA allo-antibodies, and classified as positive if the signal on the detection assay was three standard deviations above the mean found in 1,138 non-transfused males (3). All subjects included in the present analysis were female and reported a history of pregnancy. Given that blood donors are enriched for subjects reporting white or European ancestry and the fact that population stratification decreases power due to differential linkage disequilibrium across ethnic groups, (29) the analysis was limited to subjects reporting white or European ancestry.. All 772 eligible subjects with detectable HLA alloantibodies were included as cases, and they were matched with an equal number of subjects without detectable HLA alloantibodies as controls. To maximize potential alloexposure in the non-immunized control population, women with the greatest number of reported pregnancies were selected (>2 prior pregnancies for all subjects).
A second set of samples from 373 transfused subjects from the Trial to Reduce Alloimmunization to Platelets (TRAP) (30) was examined to determine if the lead SNP implicated in the LAPS GWAS would associate with alloimmunization risk after transfusion. These samples were obtained from the NHLBI Biologic Specimen and Data Repository Information Coordinating Center (BioLINCC). Our group had previously tested these subjects for HLA antibody status pre- and serially post-transfusion (31).
Sample genotyping and data quality control
Two of the selected samples lacked sufficient DNA, and the remaining 1,542 subjects were genotyped with the Illumina Omni 1 array, containing probes for 1,008,624 SNPs. Subjects with <99% call-rate were eliminated (N=15), as well as planned and unexpected sample duplicates (identity by state or IBS > 90%) (N=11); cryptic 1st degree relatives (identity by descent or IBD > 10%) (N=11, 6 parent-offspring and 5 full siblings); and non-female gender on the basis of X-chromosome zygosity (N=0). Seven hundred fifty-two alloantibody positive and 753 alloantibody negative subjects passed these QC thresholds and were included in subsequent analyses. SNPs with call-rate < 99% (N=76,122), and Hardy-Weinberg disequilibrium in controls with P < 0.00001 (N=288) were discarded, leaving 932,214 SNPs for analysis.
Genotype imputation was performed using IMPUTE2 and the phase I integrated variant set release (v3) (32,33) and the 1,000 Genomes cosmopolitan reference haplotypes (34). This resulted in genotype calls for a total of 38,066,487 SNPs and insertion/deletion polymorphisms (indels), but only the 9,427,497 variants with a Minor Allele Frequencies (MAF) > 0.01 with and Rsq > 0.85 in the 1,000 Genomes European super population were analyzed further. In addition, imputation of 4-digit HLA alleles was performed with HLA*IMP:02 (35). Post-imputation QC followed the same criteria that were applied to the genotyped SNPs. No SNPs were eliminated on the basis of R-squared; rather R-squared and dosage were used to adjust the associations.
Genotype analyses
After imputing a common set of SNPs and combining cases and controls into a single dataset, EIGENSTRAT (36) was used to compute principal components based on the genotyped markers. No subjects were found to fall >2 standard deviations from the median of the centroid of the self-reported Caucasian samples in the first 3 dimensions of eigenvalue space; and thus no population outliers were excluded. An initial screen to test for association across the MHC and then the entire genome was performed by examining allele and genotype frequency distributions in the dataset of alloantibody positive and negative individuals. We generated allele frequencies for each SNP and performed association tests for allele counts in cases and controls using a Cochran-Armitage trend test (1 degree of freedom per SNP).
Determining HLA discordance between mother and child
Alloantibody formation is more likely if the HLA alleles of the mother and those of her fetus differ. While the tremendous allelic diversity of the HLA loci makes it unlikely that a mother and fetus will be HLA identical, we addressed this possibility in a sub-study including ~250 mothers from LAPS. For these subjects, the father and at least one child were also enrolled and directly typed for DRB1 alleles, with SNP-based imputation available for the remaining class I and class II loci. This enabled us to determine the extent of HLA matching between mother-child pairs, and to correlate that with the risk of alloimmunization in the ~1/3 of mothers exhibiting HLA alloantibodies.
Results
Genome wide association with alloimmunization
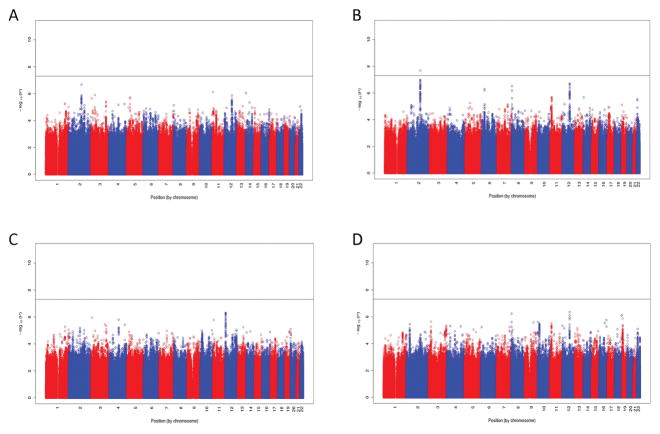
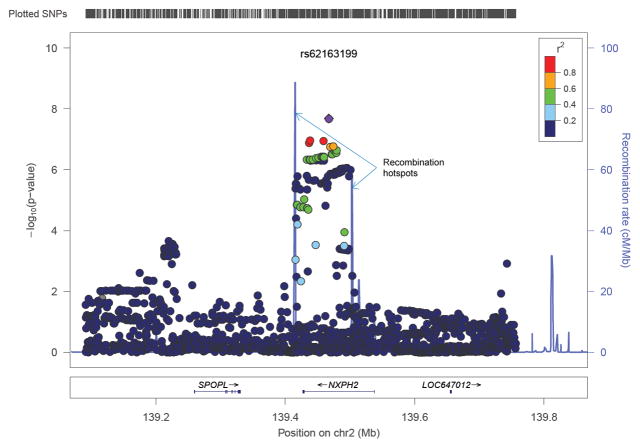
Principal components analysis (PCA) of cases and controls was consistent with the self-reported European ancestry of all study subjects (Supplemental Figure 1). There was no evidence of inflated p-values from population stratification across the four main association analyses, after adjustment for the first two principal components. Statistics of association among cases (those with alloantibodies) and controls were calculated for each of four different alloantibody scenarios, and plotted as negative log p-values by chromosome (Figure 1). Of the 772 women with detectable HLA antibodies, 440 possessed HLA class I antibodies and 525 possessed HLA class II antibodies. The analysis of individuals with class I alloantibodies versus controls was the only analysis in which SNPs on chromosome (chr) 2 exceeded the threshold of genome-wide significance defined as P ≤ 5 × 10−8 (Figure 1B). The lead SNP was rs62163199 (P = 2.4 × 10−8; OR = 0.51[0.41–0.65]). A detailed plot of the SNPs at this locus is shown in Figure 2A, which illustrates that the SNPs exhibiting association are coincident with the entire span of the NXPH2 gene. Recombination hotspots at both ends of the NXPH2 gene define the precise limits of the associated SNPs, which strongly implicates variants in this gene alone as responsible for the signal of association. The second most significant locus for class I alloantibodies is on chr12, just 5′ of the E2F7 gene (Figure 2B), although these SNPs do not exceed genome-wide significance (rs7487338; P= 2.4 × 10−7; OR = 0.41[0.29–0.57]), and this result will require additional replication. None of the other analyses of alloantibodies against class I, class II, or both class I and class II antigens resulted in association statistics that reached genome-wide thresholds of significance.
Figure 1.
Manhattan plots showing negative log p-values of associations, plotted by chromosome and position. (A) HLA class I or II alloantibody positive; (B) HLA class I alloantibody positive only, (C) HLA class II alloantibody positive, and (D) both HLA class I and class II alloantibody positive. Controls in all cases are neither HLA class I nor II positive.
Figure 2.
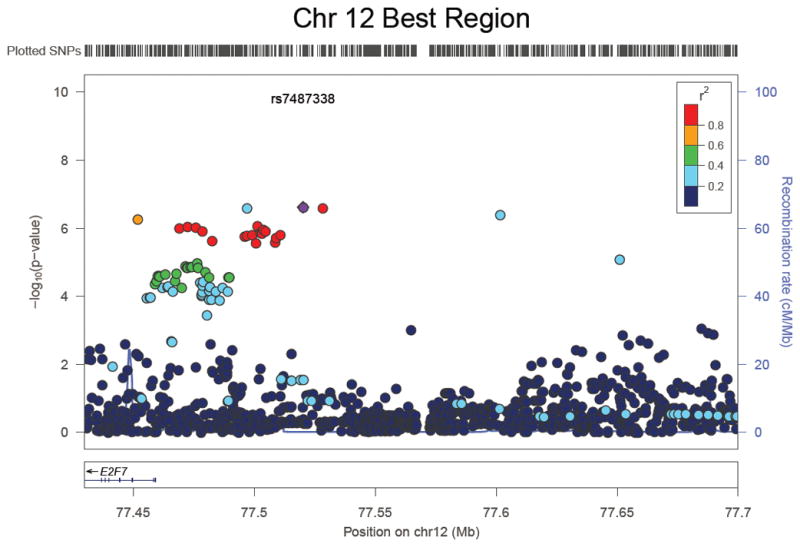
Expanded view of SNP associations with HLA class I alloimmunization. (A) Detailed view of SNP associations for the genome-wide significant hit on chromosome 2 for the analysis of HLA class I alloantibody formation. The dots represent the –log10 of the p-values for each SNP. The colored dots show the linkage disequilibrium between the most significant SNP and other SNPs. The purple graph in the background shows the recombination rate and indicates two 2 recombination hot spots flanking the significant result group of SNPs that are in high LD with one another. (B) Detailed view of SNP associations for the second most significant hit near the E2F7 gene on chromosome 12.
Examination of NXPH2 in a cohort of transfusion recipients
As part of LAPS, information on prior transfusion but not transplantation was collected. We examined if the rate of prior transfusion differed between women who did or did not form HLA class I antibodies and found no difference in the rate of transfusion between those with or without HLA class I antibodies (8.8% vs. 9.5%, p=0.8). To determine if the association between the NXPH2 minor allele variant and alloimmunization was limited to exposure via pregnancy, we genotyped a cohort of 373 well-characterized samples from subjects in the TRAP trial for whom samples were available for genotyping and determination of HLA alloimmunization. TRAP participants were subjects with acute myelogenous leukemia who received transfusion and did or did not get alloimmunized (30). Using the lead implicated SNP rs62163199, we were able to determine that 9 subjects were homozygous for the AA allele, 99 were heterogeneous, and 265 were homogeneous for the GG allele (Table 1). The proportion of alloimmunized AA homozygous subjects was not significantly different compared to the proportion of alloimmunized AG and GG subjects (p=0.4).
Table 1.
Proportion of subjects alloimmunized from LAPS and TRAP cohorts in relation to rs62163199 genotype
| LAPS | Class I | Class II | |||
|---|---|---|---|---|---|
| Genotype | Number | Immunized | % immunized | Immunized | % immunized |
| AA | 47 | 26 | 55% | 18 | 38% |
| AG | 416 | 145 | 35% | 156 | 38% |
| GG | 1042 | 47 | 26% | 351 | 34% |
| TRAP | Class I | Class II | |||||
|---|---|---|---|---|---|---|---|
| Genotype | Number | Prior Ab | New Ab | % immunized | Prior Ab | New Ab | % immunized |
| AA | 9 | 2 | 1 | 14% | 0 | 2 | 22% |
| AG | 99 | 17 | 27 | 33% | 17 | 20 | 24% |
| GG | 265 | 47 | 78 | 36% | 56 | 65 | 31% |
HLA associations with alloimmunization
Given the centrality of HLA variation itself in the production of antibodies, and the prominent association signals in the HLA region for autoimmune diseases that are characterized by the presence of particular autoantibodies (e.g., rheumatoid arthritis and systemic lupus erythematosus), we might have expected to see association at the HLA region on chr6. This hypothesis, and the failure to observe a simple association for SNPs spanning the HLA region prompted us to look in more detail at the HLA region using fully imputed HLA alleles for HLA-A, -B, -C, -DP, -DQ, and -DR. We first compared imputed alleles at HLA-DRB1 with those determined directly by conventional DNA sequencing methods. Concordance at the 2-digit level, which corresponds roughly with the serological reactivities of the HLA alleles, reached 98%, and dropped to only 87% at the 4-digit level. This result provided sufficient confidence to examine more detailed associations with particular HLA alleles among cases and controls (Tables 2 and 3, which examine homozygous alleles only). Looked at in this way, particular allelic pairings do appear to be associated, despite the much smaller counts for specific alleles. None surpass genome-wide significance, but do surpass thresholds of significance for a 6 locus test of just the HLA region itself. The most significant association is for HLA-B7 homozygosity, which is greatly over-represented in cases versus controls.
Table 2A.
Allelic associations at HLA class I loci for HLA class I positive cases vs. controls.
| Allele | Case alleles (n) | Control alleles (n) | P-value | Direction | HLA class of allele |
|---|---|---|---|---|---|
| HLA A3 | 155 | 197 | 0.0033 | Causative | 1 |
| HLA B7 | 150 | 176 | 0.0003 | Causative | 1 |
| HLA B15 | 83 | 98 | 0.0104 | Causative | 1 |
| HLA B38 | 8 | 42 | 0.0016 | Protective | 1 |
| HLA C7 | 327 | 451 | 0.0003 | Causative | 1 |
Only significant results (P< 0.05) are shown.
Table 3A.
Allelic associations at HLA class I or II loci for HLA class II positive cases vs. controls. Only significant results (P< 0.05) are shown.
| Allele | Case alleles (n) | Control alleles (n) | P-value | Direction | HLA class of allele |
|---|---|---|---|---|---|
| DPB1 11 | 29 | 21 | 0.007 | Causative | 2 |
| DPB1 11 | 85 | 154 | 0.039 | Protective | 2 |
| DQA 2 | 206 | 221 | 0.0004 | Causative | 2 |
| DQB 6 | 283 | 351 | 0.026 | Causative | 2 |
| DRB 1 | 89 | 167 | 0.019 | Protective | 2 |
| DRB 7 | 187 | 211 | 0.005 | Causative | 2 |
| HLA C 17 | 2 | 15 | 0.007 | Protective | 1 |
Effect of mother/child HLA disparity on alloimmunization rate
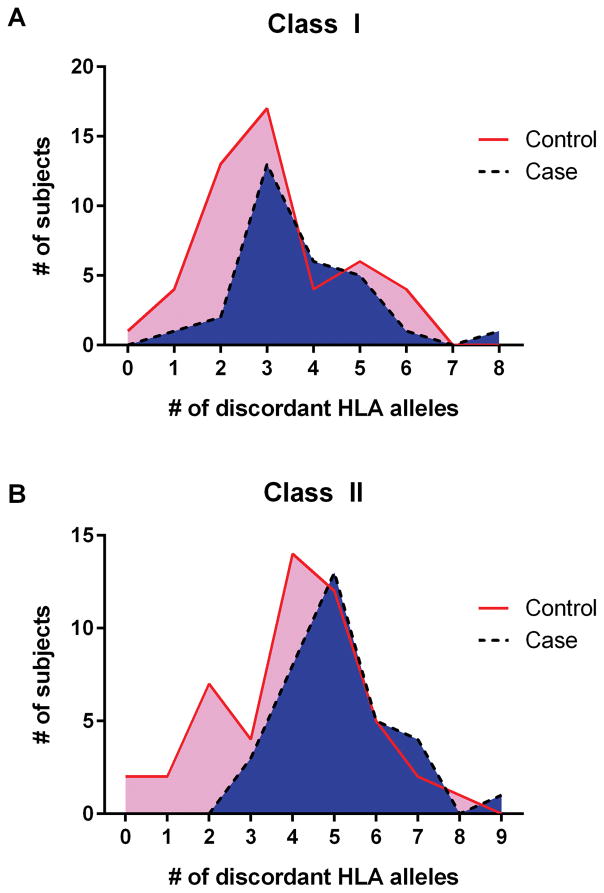
Finally, for a subset of mothers in the current study, we had imputed (or, for DRB1, directly typed) HLA allele data for one or more of her children and her husband. This enabled us to look for the frequency with which mothers may have been exposed to mismatching alloantigens (from the father) via the fetus during pregnancy. One of the most obvious reasons for a failure to produce alloantibodies during pregnancy is if both parents are matched at most or all HLA loci. Given the sheer diversity of HLA variation in this population, it would be rare statistically for spouses to share many HLA alleles, and there are compelling data to indicate that some degree of disassortative mating occurs in humans for HLA variants, such that spouses are mismatched even beyond what is expected on the basis of allele frequencies in the population (37). The degree of HLA discordance between mother and spouse did not differ between women who were or were not alloimmunized (Table 4A). However, women who developed HLA class I antibodies showed a trend to a higher degree of HLA mismatch with their children than non-alloimmunized women, and for development of HLA class II antibodies, those who were alloimmunized were significantly more mismatched with their progeny (Figure 3, Table 4B). Based on Table 4A, even in pregnancies that do not result in alloantibody formation in the mother, the fetus presents non-self HLA antigens that have the potential to result in alloantibody production in the mother.
Table 4A.
Degree of HLA incompatibility across HLA loci between mother and spouse.
| Allele discordance | Number of discordant | |
|---|---|---|
| Class 1 alleles | Class 2 alleles | |
| Case - HLA class 1 positive | 4.56 (NS) | 5.38 (NS) |
| Control | 4.53 | 5.82 |
| Case - HLA class 2 positive | 4.28 (NS) | 5.95 (NS) |
Figure 3.
Distribution of HLA mismatches between mother and children for controls (mothers without detectable alloantibodies) and cases (those mothers with alloantibodies) for Class I [A] and Class II [B].
Table 4B.
Degree of HLA incompatibility across HLA loci between mother and child. For mothers for whom data on multiple children were available, each child’s discordance was counted separately
| Allele discordance | Number of discordant | |
|---|---|---|
| Class 1 alleles | Class 2 alleles | |
| Case - HLA class 1 positive | 3.69 (P=0.072) | 4.52 (NS) |
| Control | 3.08 | 4 |
| Case - HLA class 2 positive | 3.29 (NS) | 5.08 (P=0.0029) |
Discussion
Our primary finding shows an association between SNPs in and near the NXPH2 gene and the presence of HLA class I alloantibodies in a population of previously pregnant women. Members of this gene family were first described in the context of neural signaling, but are expressed widely and probably have functions well beyond the neural system (38). Neurexophilins are secreted and have been described as detectable in plasma; and a member of the same family, NXPH1, has been shown to influence growth of hematopoietic progenitor cells (39). In a preliminary GWAS using sliding windows, a SNP within NXPH2 has been associated with rheumatoid arthritis (40). SNPs in NXPH2 have also be associated with male infertility and alcohol dependency (41,42). Given the large effect size of variation in the NXPH2 gene on the risk for alloimmunization and its known role in cell signaling in the neural system, it is plausible that this protein would have an effect on the immune system and the propensity to develop alloantibodies. A parallel example is found in neuropilin-1, which is important for axon guidance but also has been identified as an important marker of regulatory T cells (43). Neurexophilins are known to bind to neurexins and other ligands, and NXPH1 has been shown to bind to and influence the growth of hematopoietic progenitor cells (39). Whether or not NXPH2 can interact with cells of the immune system has not yet been studied, but our genetic association data and the prior literature suggests that such an interaction is possible. The implicated SNPs within NXPH2 are contained in the midst of a single, long intron. How the SNPs are related to the effect on alloimmunization is potentially via modulation of expression of NXPH2 or other mechanisms dependent on intragenic sequence such as long, noncoding RNA (lncRNA) modulation of neighboring genes (44).
While the minor allele variant of NXPH2 appeared to confer a modest to strong increased risk for the development of HLA class I antibodies in pregnancy [Odds Ratio (OR) = 1.96], there was no association between NXPH2 SNPs and the risk of becoming sensitized to HLA class II antibodies. If this finding were to be confirmed, it would help design further studies about the pathway of allosensitization during pregnancy and for the identification of candidate genes that are necessary for the responder status. From epidemiologic studies of pregnant women it is known that of those who become alloimmunized, nearly equal proportions will develop antibodies against HLA class I antigens only, class II antigens only, or both class I and II antigens, with the last category slightly less common than the first two (18). Why women develop class I versus II allosensitization is not clear, though it is known that there is differential regulation of expression of these antigens on fetal cells. The fetal cell with the most direct exposure to the maternal immune system is the trophoblast. In these cells HLA class II expression is suppressed due to lack of class II transactivator (CIITA) expression (45,46) that is mediated via methylation of the CIITA interferon-γ-responsive promoter (PIV) (47). Trophoblasts also lack expression of most HLA class I molecules, with exception of the non-classical molecules HLA-C (48), HLA-E (49), and HLA-G (50). It is possible that NXPH2 could interact differentially with the epigenetic pathways that silence HLA class I versus class II molecules in trophoblasts. In addition to direct exposure to trophoblasts, fetal DNA and cells can be detected in maternal circulation during gestation (51). Finally, a major potential source of fetal interaction with maternal blood is during parturition. For these last two routes of exposure, most of the cells that would be seen by the maternal immune system would express HLA class I but not class II, with the exception of professional antigen presenting cells (APCs). There are several differences between HLA class I and II antigen presentation in pregnancy and in non-pregnant subjects, and it is not clear which mechanism would underlie a differential ability of NXPH2 to protect from HLA class I but not class II alloimmunization.
A second cluster of SNPs that approached but did not achieve genome-wide significance was 5′ of the E2F7 gene. E2F transcription factors comprise a family that is important in regulation of cell cycle, apoptosis, and cellular differentiation (52). Expression of E2F7 is cell cycle regulated, the protein binds DNA and localizes to the nucleus, and it blocks E2F-dependent activation (53,54). The net result of E2F7 suppression of cellular proliferation is cells accumulating in G1 phase of cellular division (54). E2F7 has not been previously reported to be associated with alloimmunization risk or immune function, and presumably if it did affect alloimmunity it would be through its influence on cellular proliferation.
Interestingly, in the initial GWAS analysis we did not find genome wide association for alloresponse to HLA region genes. But on more detailed analysis of imputed alleles for HLA-A, -B, -C, -DR, -DQ located in the HLA region, we found that HLA-B7 homozygosity was over-represented in responders as compared to the non-responders. Similar to our findings, over-representation of HLA-B35 in sickle cell anemia patients with red blood cell alloimmunization has been shown by others (55). We were able to detect antigen disparity between the mother and the child by performing HLA typing of mother-child-father combinations in a subset of the study subjects. As has been demonstrated previously, we found a greater degree of antigen disparity between the mother and the child than would be expected by the known antigen frequencies in the population (56). This indicates that most pregnancies result in alloexposures, supporting a role for genetic variation and other factors in the susceptibility to becoming alloimmunized. It has been described in the transplant literature that certain HLA mismatch pairs are more likely to result in generation of alloantibodies (57). It was beyond the scope of the current study to perform detailed analysis of maternal-fetal HLA mismatches, but such an analysis could complement the extensive data available in the transplant field (58).
There were several limitations of the study that bear mentioning. The current study was limited to women who identified as white or European ancestry, so further study would be needed to demonstrate whether or not the identified genetic variations also influenced alloimmunization across other ethnic groups. The assay cutoff used to determine allosensitization was three standard deviations above the mean found in non-transfused males, so associations between novel genetic polymorphisms and generation of low-level antibodies in previously pregnant females could have been missed. Imputation of HLA alleles was performed, which has a high degree of accuracy, but a small percentage of imputed alleles may have been incorrect.
The findings reported here should motivate larger genetic studies in search of variants associated with HLA alloimmunization, as well as red blood cell and platelet derived alloimmunization in other transfusion medicine contexts, although our modestly sized study of 373 individuals from the TRAP study did not reveal an association between NXPH2 and alloimmunization due to platelet transfusion. This will enable a better understanding of the functional basis for alloimmunization, as well as identifying the degree to which genetic associations are shared or different according to the type of alloexposure. In addition, the current study was conducted using only subjects reporting European ancestry, so replication studies would ideally be performed using an ethnically diverse population. Finally, functional studies to elucidate the role of NXPH2 in the alloimmunization process should be a priority, and could suggest novel therapeutic targets for the modulation of alloimmunization risk during pregnancy and other alloexposures such as blood transfusion.
Supplementary Material
Table 2B.
Genotypic associations at HLA class I loci for HLA class I positive cases vs. controls.
| Genotype | Case Homozygotes | Control Homozygotes | P-value | Direction |
|---|---|---|---|---|
| HLA A3/A3 | 21 | 14 | 0.001 | Causative |
| HLA B7/B7 | 21 | 7 | 2.18E-06 | Causative |
| HLA C7/C7 | 68 | 69 | 0.0002 | Causative |
Only significant results (P< 0.05) are shown.
Table 3B.
Genotypic associations at HLA class II loci for HLA class II positive cases vs. controls. Only significant results (P< 0.05) are shown.
| Genotype | Case Homozygotes | Control Homozygotes | P-value | Direction |
|---|---|---|---|---|
| DQA 2/2 | 26 | 13 | 0.0003 | Causative |
| DRB 7/7 | 23 | 9 | 0.0001 | Causative |
Acknowledgments
Funding. This work was supported by NHLBI contracts HHSN268201100001I, HHSN268201100002I, HHSN268201100003I, HHSN268201100004I, HHSN268201100005I, and HHSN268201100006I and NIH grant HL124260.
We thank Anne Guiltinan for assistance with the coordination and recruitment of study subjects; and Leilani Montalvo for extracting and preparing DNA samples for genotyping. Genotyping was performed at Centrillion Biosciences, Inc., Palo Alto, CA; and we acknowledge the assistance of Rui Meng, Guochon Liao, Robert Feldman, and Wei Zhou of Centrillion.
Appendix
The NHLBI Recipient Epidemiology Donor Evaluation Study - III (REDS-III), domestic component, is composed of the following persons:
Hubs:
A.E. Mast and J.L. Gottschall, BloodCenter of Wisconsin (BCW), Milwaukee, WI
D.J. Triulzi and J.E. Kiss, The Institute For Transfusion Medicine (ITXM), Pittsburgh, PA
E.L. Murphy and E.W. Fiebig, University of California, San Francisco (UCSF), San Francisco, CA
E.L. Snyder Yale University School of Medicine, New Haven, CT and R.G Cable, American Red Cross Blood Services, Farmington CT
Data coordinating center:
D. J. Brambilla and M. T. Sullivan, RTI International, Rockville, MD
Central laboratory:
M.P. Busch and P.J. Norris, Blood Systems Research Institute, San Francisco, CA
Publication Committee Chairman:
R. Y. Dodd, American Red Cross, Holland Laboratory, Rockville, MD
Steering Committee Chairman:
S. H. Kleinman, University of British Columbia, Victoria, BC, Canada
National Heart, Lung, and Blood Institute, National Institutes of Health:
S. A. Glynn and A.M. Cristman
Footnotes
A complete list of the members of the NHLBI REDSIII Study Investigators appears in the “Appendix.”
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The authors declare no conflict of interest.
Contribution: This study was designed and planned by M.S., M.L., P.J.N. and M.P.B., in consultation with the NHLBI-REDSIII Steering Committee. G.P.P. and N.G. provided statistical analysis in consultation with M.S. T.H.L. oversaw DNA extractions, sample handling and quality control. R.K. provided expertise on HLA analysis and participated in all stages of the projects execution. L.A. For the sub-study on HLA typing and imputation in the spouse and child (or children) of LAPS study subjects, L.F.B., L.A.C., and D.T. conducted and funded subject enrollments, and most of the SNP genotyping used for the imputations. L.F.B. conducted the HLA-DRB typing in her laboratory. The manuscript was written primarily by M.S., P.J.N. and M.P.B. All authors participated in the analysis and interpretation of the data, the editing of the manuscript, and all approve of the final version.
References
- 1.Payne R. The development and persistence of leukoagglutinins in parous women. Blood. 1962;19:411. [PubMed] [Google Scholar]
- 2.Norris PJ, Lee JH, Carrick DM, et al. Long-term in vitro reactivity for human leukocyte antigen antibodies and comparison of detection using serum versus plasma. Transfusion. 2009;49:243. doi: 10.1111/j.1537-2995.2008.01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Triulzi DJ, Kleinman S, Kakaiya RM, et al. The effect of previous pregnancy and transfusion on HLA alloimmunization in blood donors: implications for a transfusion-related acute lung injury risk reduction strategy. Transfusion. 2009;49:1825. doi: 10.1111/j.1537-2995.2009.02206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer MP, Wiersum-Osselton J, Schipperus M, Vandenbroucke JP, Briët E. Clinical predictors of alloimmunization after red blood cell transfusion. Transfusion. 2007;47:2066. doi: 10.1111/j.1537-2995.2007.01433.x. [DOI] [PubMed] [Google Scholar]
- 5.Blumberg N, Heal JM, Gettings KF. WBC reduction of RBC transfusions is associated with a decreased incidence of RBC alloimmunization. Transfusion. 2003;43:945. doi: 10.1046/j.1537-2995.2003.00443.x. [DOI] [PubMed] [Google Scholar]
- 6.Brantley SG, Ramsey G. Red cell alloimmunization in multitransfused HLA-typed patients. Transfusion. 1988;28:463. doi: 10.1046/j.1537-2995.1988.28588337338.x. [DOI] [PubMed] [Google Scholar]
- 7.Fluit CR, Kunst VA, Drenthe-Schonk AM. Incidence of red cell antibodies after multiple blood transfusion. Transfusion. 1990;30:532. doi: 10.1046/j.1537-2995.1990.30690333485.x. [DOI] [PubMed] [Google Scholar]
- 8.Friedman DF, Lukas MB, Jawad A, Larson PJ, Ohene-Frempong K, Manno CS. Alloimmunization to platelets in heavily transfused patients with sickle cell disease. Blood. 1996;88:3216. [PubMed] [Google Scholar]
- 9.Heal JM, Phipps RP, Blumberg N. One big unhappy family: transfusion alloimmunization, thrombosis, and immune modulation/inflammation. Transfusion. 2009;49:1032. doi: 10.1111/j.1537-2995.2009.02182.x. [DOI] [PubMed] [Google Scholar]
- 10.Heddle NM, Soutar RL, O’Hoski PL, et al. A prospective study to determine the frequency and clinical significance of alloimmunization post-transfusion. British Journal of Haematology. 1995;91:1000. doi: 10.1111/j.1365-2141.1995.tb05425.x. [DOI] [PubMed] [Google Scholar]
- 11.McFarland JG. Alloimmunization and platelet transfusion. Seminars in Hematology. 1996;33:315. [PubMed] [Google Scholar]
- 12.Murao M, Viana MB. Risk factors for alloimmunization by patients with sickle cell disease. Brazilian Journal of Medical and Biological Research = Revista Brasileira De Pesquisas Medicas E Biologicas. 2005;38:675. doi: 10.1590/s0100-879x2005000500004. [DOI] [PubMed] [Google Scholar]
- 13.Rosse WF, Gallagher D, Kinney TR, et al. Transfusion and alloimmunization in sickle cell disease. The Cooperative Study of Sickle Cell Disease. Blood. 1990;76:1431. [PubMed] [Google Scholar]
- 14.Schonewille H, Haak HL, van Zijl AM. Alloimmunization after blood transfusion in patients with hematologic and oncologic diseases. Transfusion. 1999;39:763. doi: 10.1046/j.1537-2995.1999.39070763.x. [DOI] [PubMed] [Google Scholar]
- 15.Schonewille H, van de Watering LMG, Loomans DSE, Brand A. Red blood cell alloantibodies after transfusion: factors influencing incidence and specificity. Transfusion. 2006;46:250. doi: 10.1111/j.1537-2995.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 16.Slichter SJ. Platelet refractoriness and alloimmunization. Leukemia. 1998;12(Suppl 1):S51. [PubMed] [Google Scholar]
- 17.Zimring JC, Welniak L, Semple JW, et al. Current problems and future directions of transfusion-induced alloimmunization: summary of an NHLBI working group. Transfusion. 2011;51:435. doi: 10.1111/j.1537-2995.2010.03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middelburg RA, van Stein D, Briët E, van der Bom JG. The role of donor antibodies in the pathogenesis of transfusion-related acute lung injury: a systematic review. Transfusion. 2008;48:2167. doi: 10.1111/j.1537-2995.2008.01810.x. [DOI] [PubMed] [Google Scholar]
- 19.Osby M, Shulman IA. Phenotype matching of donor red blood cell units for nonalloimmunized sickle cell disease patients: a survey of 1182 North American laboratories. Archives of Pathology & Laboratory Medicine. 2005;129:190. doi: 10.5858/2005-129-190-PMODRB. [DOI] [PubMed] [Google Scholar]
- 20.Picascia A, Grimaldi V, Sabia C, Napoli C. Comprehensive assessment of sensitizing events and anti-HLA antibody development in women awaiting kidney transplantation. Transplant Immunology. 2016;36:14. doi: 10.1016/j.trim.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 21.Patel R, Terasaki PI. Significance of the positive crossmatch test in kidney transplantation. The New England Journal of Medicine. 1969;280:735. doi: 10.1056/NEJM196904032801401. [DOI] [PubMed] [Google Scholar]
- 22.Mao Q, Terasaki PI, Cai J, et al. Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five-year longitudinal study. American Journal of Transplantation: Official Journal of the American Society of Transplantation and the American Society of Transplant Surgeons. 2007;7:864. doi: 10.1111/j.1600-6143.2006.01711.x. [DOI] [PubMed] [Google Scholar]
- 23.Vichinsky EP, Earles A, Johnson RA, Hoag MS, Williams A, Lubin B. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. The New England Journal of Medicine. 1990;322:1617. doi: 10.1056/NEJM199006073222301. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickson JE, Desmarets M, Deshpande SS, et al. Recipient inflammation affects the frequency and magnitude of immunization to transfused red blood cells. Transfusion. 2006;46:1526. doi: 10.1111/j.1537-2995.2006.00946.x. [DOI] [PubMed] [Google Scholar]
- 25.Buetens O, Shirey RS, Goble-Lee M, et al. Prevalence of HLA antibodies in transfused patients with and without red cell antibodies. Transfusion. 2006;46:754. doi: 10.1111/j.1537-2995.2006.00793.x. [DOI] [PubMed] [Google Scholar]
- 26.Higgins JM, Sloan SR. Stochastic modeling of human RBC alloimmunization: evidence for a distinct population of immunologic responders. Blood. 2008;112:2546. doi: 10.1182/blood-2008-03-146415. [DOI] [PubMed] [Google Scholar]
- 27.Gatault P, Jollet I, Rabot N, et al. Mothers without HLA antibodies before transplantation have a low risk of alloimmunization post-transplantation. Tissue Antigens. 2011;78:241. doi: 10.1111/j.1399-0039.2011.01757.x. [DOI] [PubMed] [Google Scholar]
- 28.Yazer MH, Triulzi DJ, Shaz B, Kraus T, Zimring JC. Does a febrile reaction to platelets predispose recipients to red blood cell alloimmunization? Transfusion. 2009;49:1070. doi: 10.1111/j.1537-2995.2009.02116.x. [DOI] [PubMed] [Google Scholar]
- 29.Teo YY, Small KS, Fry AE, Wu Y, Kwiatkowski DP, Clark TG. Power consequences of linkage disequilibrium variation between populations. Genetic Epidemiology. 2009;33:128. doi: 10.1002/gepi.20366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Trial to Reduce Alloimmunization to Platelets Study Group. Leukocyte reduction and ultraviolet B irradiation of platelets to prevent alloimmunization and refractoriness to platelet transfusions. N Engl J Med. 1997;337:1861. doi: 10.1056/NEJM199712253372601. [DOI] [PubMed] [Google Scholar]
- 31.Jackman RP, Deng X, Bolgiano D, et al. Low-level HLA antibodies do not predict platelet transfusion failure in TRAP study participants. Blood. 2013;121:3261. doi: 10.1182/blood-2012-12-472779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.1,000 Genomes haplotypes -- Phase I integrated variant set release (v3) in NCBI build 37 (hg19) coordinates [Internet] 2012 Available from: https://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html.
- 33.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dilthey AT, Moutsianas L, Leslie S, McVean G. HLA*IMP--an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics (Oxford, England) 2011;27:968. doi: 10.1093/bioinformatics/btr061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 37.Chaix R, Cao C, Donnelly P. Is mate choice in humans MHC-dependent? PLoS genetics. 2008;4:e1000184. doi: 10.1371/journal.pgen.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Missler M, Hammer RE, Südhof TC. Neurexophilin binding to alpha-neurexins. A single LNS domain functions as an independently folding ligand-binding unit. The Journal of biological chemistry. 1998;273:34716. doi: 10.1074/jbc.273.52.34716. [DOI] [PubMed] [Google Scholar]
- 39.Kinzfogl J, Hangoc G, Broxmeyer HE. Neurexophilin 1 suppresses the proliferation of hematopoietic progenitor cells. Blood. 2011;118:565. doi: 10.1182/blood-2010-12-325381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X, Qin H, Sha Q. Incorporating multiple-marker information to detect risk loci for rheumatoid arthritis. BMC Proceedings. 2009;3:S28. doi: 10.1186/1753-6561-3-s7-s28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aston KI, Carrell DT. Genome-Wide Study of Single-Nucleotide Polymorphisms Associated With Azoospermia and Severe Oligozoospermia. Journal of Andrology. 2009;30:711. doi: 10.2164/jandrol.109.007971. [DOI] [PubMed] [Google Scholar]
- 42.Bierut LJ, Agrawal A, Bucholz KK, et al. A genome-wide association study of alcohol dependence. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:5082. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruder D, Probst-Kepper M, Westendorf AM, et al. Neuropilin-1: a surface marker of regulatory T cells. European journal of immunology. 2004;34:623. doi: 10.1002/eji.200324799. [DOI] [PubMed] [Google Scholar]
- 44.Villegas VE, Zaphiropoulos PG. Neighboring gene regulation by antisense long non-coding RNAs. International Journal of Molecular Sciences. 2015;16:3251. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morris AC, Riley JL, Fleming WH, Boss JM. MHC class II gene silencing in trophoblast cells is caused by inhibition of CIITA expression. American journal of reproductive immunology (New York, NY: 1989) 1998;40:385. doi: 10.1111/j.1600-0897.1998.tb00423.x. [DOI] [PubMed] [Google Scholar]
- 46.Murphy SP, Tomasi TB. Absence of MHC class II antigen expression in trophoblast cells results from a lack of class II transactivator (CIITA) gene expression. Molecular reproduction and development. 1998;51:1. doi: 10.1002/(SICI)1098-2795(199809)51:1<1::AID-MRD1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 47.van den Elsen PJ, van der Stoep N, Viëtor HE, Wilson L, van Zutphen M, Gobin SJ. Lack of CIITA expression is central to the absence of antigen presentation functions of trophoblast cells and is caused by methylation of the IFN-gamma inducible promoter (PIV) of CIITA. Human immunology. 2000;61:850. doi: 10.1016/s0198-8859(00)00159-2. [DOI] [PubMed] [Google Scholar]
- 48.King A, Boocock C, Sharkey AM, et al. Evidence for the expression of HLAA-C class I mRNA and protein by human first trimester trophoblast. Journal of immunology (Baltimore, Md: 1950) 1996;156:2068. [PubMed] [Google Scholar]
- 49.Guillaudeux T, Rodriguez AM, Girr M, et al. Methylation status and transcriptional expression of the MHC class I loci in human trophoblast cells from term placenta. Journal of immunology (Baltimore, Md: 1950) 1995;154:3283. [PubMed] [Google Scholar]
- 50.Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science (New York, NY) 1990;248:220. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- 51.Ariga H, Ohto H, Busch MP, et al. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion. 2001;41:1524. doi: 10.1046/j.1537-2995.2001.41121524.x. [DOI] [PubMed] [Google Scholar]
- 52.Lammens T, Li J, Leone G, De Veylder L. Atypical E2Fs: new players in the E2F transcription factor family. Trends in Cell Biology. 2009;19:111. doi: 10.1016/j.tcb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. The Journal of Biological Chemistry. 2003;278:42041. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- 54.Di Stefano L, Jensen MR, Helin K. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. The EMBO journal. 2003;22:6289. doi: 10.1093/emboj/cdg613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alarif L, Castro O, Ofosu M, Dunston G, Scott RB. HLA-B35 is associated with red cell alloimmunization in sickle cell disease. Clinical Immunology and Immunopathology. 1986;38:178. doi: 10.1016/0090-1229(86)90136-4. [DOI] [PubMed] [Google Scholar]
- 56.Ober C, Elias S, O’Brien E, Kostyu DD, Hauck WW, Bombard A. HLA sharing and fertility in Hutterite couples: evidence for prenatal selection against compatible fetuses. American journal of reproductive immunology and microbiology: AJRIM. 1988;18:111. doi: 10.1111/j.1600-0897.1988.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 57.Claas FHJ, Dankers MK, Oudshoorn M, et al. Differential immunogenicity of HLA mismatches in clinical transplantation. Transplant Immunology. 2005;14:187. doi: 10.1016/j.trim.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 58.Kramer CSM, Roelen DL, Heidt S, Claas FHJ. Defining the immunogenicity and antigenicity of HLA epitopes is crucial for optimal epitope matching in clinical renal transplantation: KRAMER et al. HLA. 2017;90:5. doi: 10.1111/tan.13038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.