Abstract
UHRF1 (Ubiquitin-like, containing PHD and RING Finger domains, 1) is one of the essential components of mammalian DNA methylation machinery. Chromatin association of UHRF1 is controlled via an interplay between its intramolecular interaction and dual recognition of histone H3 trimethylated at lysine 9 (H3K9me3) and hemimethylated DNA. Here, we report the crystal structure of the N-terminal tandem Tudor domain (TTD) of UHRF1 in complex with the C-terminal polybasic region (PBR). Structural analysis reveals that PBR binding leads to displacement of the TTD – Plant homeodomain (PHD) linker, as well as blockage of the H3K9me3-engaging cage, both of which contribute to a chromatin-occluded UHRF1 conformation. Disruption of the TTD – PBR interaction, which is facilitated by the binding of UHRF1 to hemimethylated DNA or regulatory protein USP7, shifts the UHRF1 conformation toward an open state, allowing for efficient H3K9me3 binding. Together, this study provides structural basis for the allosteric regulation of UHRF1.
eTOC Blurb
Gao et al report the crystal structure of the UHRF1 TTD domain bound to the C-terminal polybasic region, revealing how the intramolecular interaction of UHRF1 gives rise to its chromatin-occluded conformation. Combined with in vitro biochemical analysis, this structural study provides insights into the allosteric regulation of UHRF1.
Introduction
DNA methylation represents one of the major epigenetic mechanisms that control silencing of retrotransposons, genomic imprinting, X-chromosome inactivation and genome integrity (Bird, 2002). In mammals, DNA methylation mainly occurs at the C-5 position of cytosine within CpG dinucleotide context, creating distinct epigenetic patterns across various tissue cells (Ehrlich et al., 1982). Mammalian DNA methylation is installed by de novo DNA methyltransferases DNMT3A and DNMT3B during gametogenesis and early embryonic development, and subject to stable propagation by maintenance DNA methyltransferase DNMT1 in a replication-dependent manner (Goll and Bestor, 2005; Law and Jacobsen, 2010). DNMT1-mediated maintenance DNA methylation is further regulated by UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) (Bostick et al., 2007; Sharif et al., 2007), which controls the recruitment of DNMT1 to target sites in a cell cycle-dependent manner (Du et al., 2010; Nishiyama et al., 2013; Qin et al., 2015).
UHRF1 is a multi-domain protein that contains an N-terminal ubiquitin-like (UBL) domain, a tandem Tudor domain (TTD), a plant homeodomain (PHD), a SET- and RING-associated (SRA) domain, and a C-terminal RING-finger domain. It has been established that the TTD and PHD domains recognize histone H3 trimethylated at lysine 9 (H3K9me3) (Arita et al., 2012; Cheng et al., 2013; Karagianni et al., 2008; Rothbart et al., 2013; Rothbart et al., 2012; Xie et al., 2012) and unmodified at arginine 2 (Hu et al., 2011; Lallous et al., 2011; Rajakumara et al., 2011), respectively, generating a coordinated reader cassette for the multivalent silencing marks at the H3 tail. The SRA domain binds to hemimethylated CpG DNA (Arita et al., 2008; Avvakumov et al., 2008; Hashimoto et al., 2008), which, together with the H3K9me3 recognition, mediates the association of UHRF1 with the replicating heterochromatin region during the S phase of cell cycle (Liu et al., 2013). The C-terminal RING finger domain serves as an E3 ubiquitin ligase that mediates the ubiquitylation of histone H3 lysine 18 and/or 23, which in turn recruits DNMT1 to the replication foci (Nishiyama et al., 2013; Qin et al., 2015). In addition to a regulatory role in maintenance DNA methylation, UHRF1 has been associated with heterochromatin replication and assembly (Miura et al., 2001; Papait et al., 2007; Taylor et al., 2013; Uemura et al., 2000), DNA damage response (DDR) (Liang et al., 2015; Mistry et al., 2010; Muto et al., 2002; Tian et al., 2015), and gene transcription (Jin et al., 2010; Kim et al., 2009; Rajakumara et al., 2011).
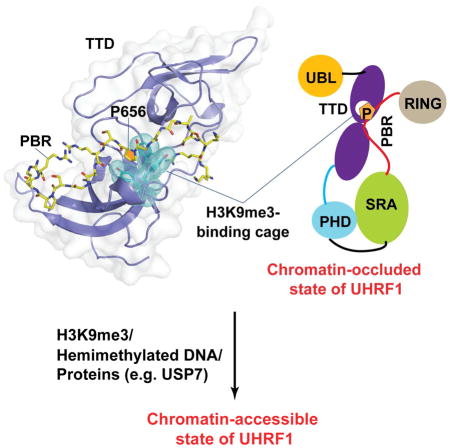
Recent studies have revealed that the chromatin association and modifying activities of UHRF1 are controlled by a conformational switch, in which alternative conformations of UHRF1 regulate its bindings to H3K9me3 and/or hemimethylated DNA (Fang et al., 2016; Gelato et al., 2014; Harrison et al., 2016; Zhang et al., 2015). An intramolecular interaction between the TTD domain and a C-terminal polybasic region (PBR) drives UHRF1 into a “closed” conformation, which occludes the binding of TTD to H3K9me3. Intermolecular association of the PBR with PI5P (Gelato et al., 2014), USP7 (Ma et al., 2012; Zhang et al., 2015) or hemimethylated DNA (Fang et al., 2016; Harrison et al., 2016) helps transit UHRF1 into an “open” conformation, thereby promoting the TTD – H3K9me3 binding and consequent association of UHRF1 with heterochromatin (Fang et al., 2016; Gelato et al., 2014; Zhang et al., 2015), as well as UHRF1-mediated H3 ubiquitylation (Harrison et al., 2016). Consistently, a recent nuclear magnetic resonance (NMR) study provided structural evidence on the interaction between the TTD domain and PBR (Fang et al., 2016). However, due to lack of high-resolution structural characterization, the mechanism by which the H3K9me3 epigenetic mark “unlocks” the UHRF1 TTD – PBR interaction was not explained. Therefore, the functional regulation of UHRF1 – chromatin association remains unclear.
To characterize the TTD – PBR interaction in further detail, we determined the crystal structure of the TTD domain of UHRF1 in complex with the PBR peptide at 1.68 Å resolution. The structure reveals that the TTD – PBR interaction not only competes against the positioning of the TTD-PHD linker, an element modulating the relative orientation of the TTD-PHD tandem domains, but also blocks the H3K9me3-binding cage with a proline ring from the PBR peptide. Both interactions help stabilize UHRF1 in a “closed” conformational state that occludes chromatin binding. Consistently, disruption of the TTD – PBR interaction by site-directed mutagenesis or intermolecular interactions significantly shifts the UHRF1 conformation to an “open” state with enhanced interaction with H3K9me3. Together, these studies provide structural basis for the H3K9me3-speific chromatin association of UHRF1.
Results
Crystal structure of the UHRF1 TTD – PBR complex
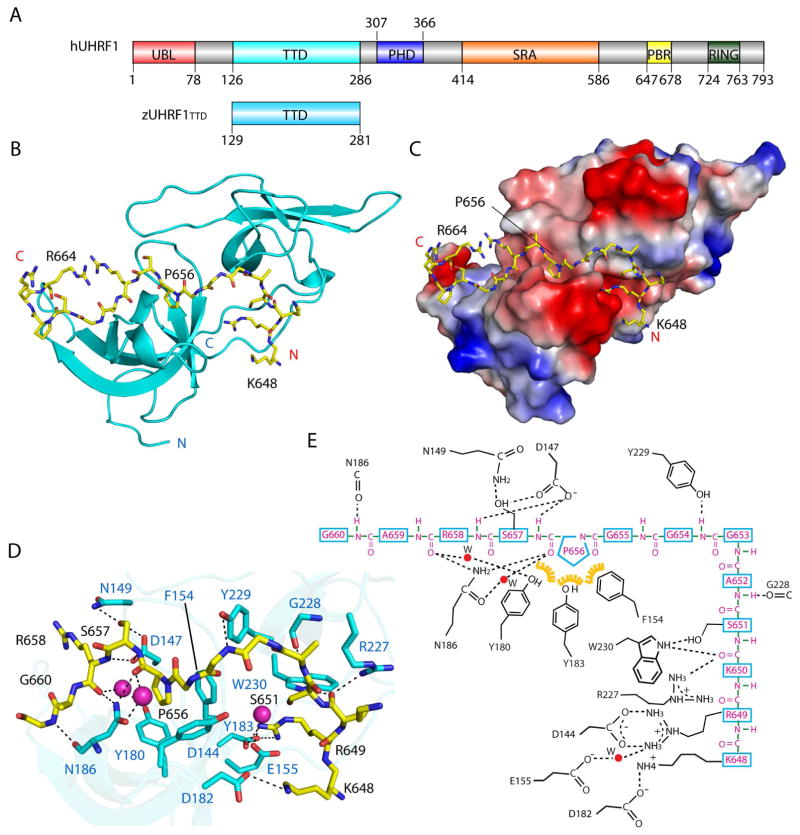
The C-terminal PBR sequence of UHRF1 (Fig. 1A), comprised of residues E634-T665, has been previously shown to mediate an intramolecular interaction with the N-terminal TTD domain (Fang et al., 2016; Gelato et al., 2014; Zhang et al., 2015). To provide structural basis for this interaction, we determined the crystal structure of the TTD domain of zebrafish UHRF1 (zUHRF1TTD) (Fig. 1A) in complex with the PBR peptide from human UHRF1 at 1.68 Å resolution (Fig. 1B,C and Table 1). There are three TTD molecules and one PBR peptide in each asymmetric unit (Fig. S1A). We were able to trace residues K648-R664 of the PBR peptide, which is predominantly associated with one TTD molecule, with its C-terminal tail (residues S661-R664) further extending to make crystallographic contacts with the second TTD molecule (Fig. S1A). Meanwhile, the PBR peptide is occluded from binding to the third TTD molecule by the crystal packing effects (Fig. S1A).
Figure 1. Crystal structure of the UHRF1 TTD – PBR complex.
(A) Domain architecture of human UHRF1, with individual domains labeled with residue numbers. The TTD domain of Zebrafish UHRF1 (zUHRF1TTD) used for structure determination is also shown. (B) Ribbon representation of zUHRF1TTD (cyan) bound to the PBR peptide (yellow sticks). (C) Surface representation of the zUHRF1TTD – PBR complex. (D) Close-up view of the intermolecular interaction between zUHRF1TTD and PBR. The water molecules are shown in sphere representation. The hydrogen bonds are shown as dashed lines. (E) Schematic representation of the zUHRF1TTD – PBR interaction. The residues from zUHRF1TTD and PBR are colored in magenta and black, respectively. Yellow: hydrophobic contact. See also Figure S1.
Table 1.
Crystallographic data collection and refinement statistics of the zUHRF1TTD – PBR complex.
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 81.2, 66.1, 120.5 |
| α,β,γ(°) | 90, 105, 90 |
| Wavelength | 0.9774 |
| Resolution (Å) | 28.76-1.68 (1.74-1.68)a |
| Rmerge | 0.038 (0.47) |
| I/σI | 27 (2.0) |
| Completeness (%) | 98.4 (96.4) |
| Redundancy | 3.0 (2.9) |
| Total reflections | 208438 |
| Unique reflections | 69241 |
| Refinement | |
| Resolution (Å) | 28.76-1.68 (1.74-1.68)a |
| No. reflections | 69239 (6738) |
| Rwork/Rfree | 18.3/20.7 (27.0/29.4) |
| No. atoms | |
| Protein | 3777 |
| Water | 535 |
| B factors (Å2) | |
| Protein | 41.54 |
| Water | 47.40 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.14 |
| Ramachandran | |
| Favored (%) | 97.4 |
| Allowed (%) | 2.6 |
| Outliers (%) | 0 |
Values in parentheses are for highest-resolution shell.
The structure of the zUHRF1TTD domain reveals two five-stranded β-barrels abutting each other through an interface formed by two protruding loops from the N- and C-terminal subdomains, respectively, as previously observed for its human counterpart (Fig. 1B). Notably, the linker sequence between the two subdomains joins in antiparallel with the C-terminal tail, together supporting the TTD as one integrated structural unit (Fig. 1B). The PBR peptide fills in an acidic groove extending from the subdomain interface, followed by a 90° turn, toward one end of the N-terminal β-barrel (Fig. 1B and Fig. S1B). Accordingly, association of the PBR peptide with the TTD domain spans two discrete binding surfaces: one involves PBR K648-G654, while the other involves PBR P656-G660 (Fig. 1D,E). At one interface, the side chains of PBR K648 and R649 interact with residues D144, E155 and D182 from the TTD N-terminal subdomain through direct and/or water-mediated hydrogen bonding interactions (Fig. 1D,E). Meanwhile, PBR K650, S651, A652 and G654 interact with residues R227, G228, Y229 and W230 from the C-terminal subdomain through main-chain or side-chain hydrogen bonding interactions (Fig. 1D,E). At the second interface, the proline ring of PBR P656 slides into the aromatic cage formed by F154, Y180 and Y183 from the TTD N-terminal subdomain, with its backbone carbonyl group interacting with the side chain of TTD N186 through direct and water-mediated hydrogen bonds (Fig. 1C–E). Additional intermolecular interactions involve hydrogen bonds formed between PBR S657 and R658 and a number of TTD residues (D147, N149, Y180 and N186), which further contribute to the TTD – PBR association (Fig. 1D,E). Notably, the PBR-interacting sites on the TTD domain are highly conserved across species (Fig. S1C), supporting that zUHRF1TTD is a valid model for its human orthologue.
Mutational analysis of the TTD – PBR interaction
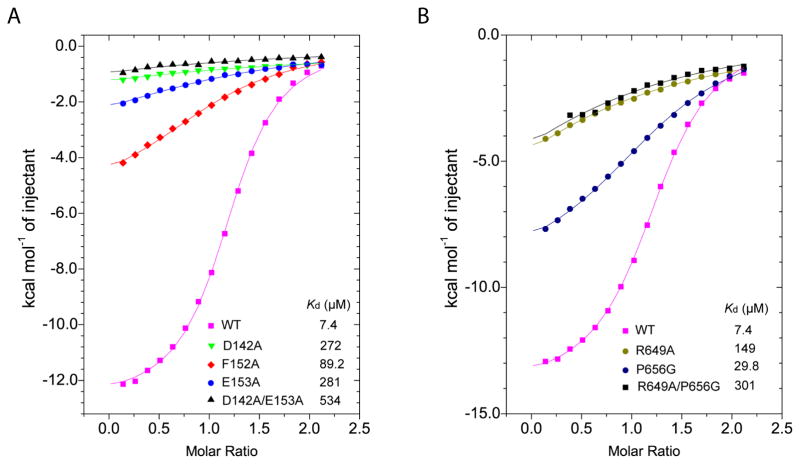
To evaluate the structural observations, we selected a number of residues from human UHRF1 for mutagenesis, followed by Isothermal Titration Calorimetry (ITC) analysis (Table S1). Notably, titration of wild-type PBR peptide over the TTD domain yielded a dissociation constant (Kd) of 7.4 μM (Fig. 2A, B), consistent with the previous observations that TTD binds strongly to PBR (Fang et al., 2016; Gelato et al., 2014; Zhang et al., 2015). By contrast, mutations of D142 (equivalent to D144 in zUHRF1), D145 (equivalent to D147 in zUHRF1), F152 (equivalent to F154 in zUHRF1) and E153 (equivalent to E155 in zUHRF1) in the TTD domain into alanine all reduced the binding affinity by > 3-fold, with Kds of 272 μM, 23.5 μM, 89.2 μM and 281 μM, respectively (Fig. 2A and Fig. S2A). In line with these observations, introduction of D142A/E153A double mutation nearly abolished the binding, with a Kd of 534 μM (Fig. 2A). On the PBR side, introduction of R649A and S657A mutation reduces the TTD – PBR binding by ~20-fold (Kd = 149 μM) and ~2-fold (Kd = 13.9 μM), respectively, while replacement of P656 by glycine, which eliminates the effect of amino side chain, reduced the TTD – PBR binding affinity to a Kd of 29.8 μM (Fig. 2B and Fig. S2B). Consistently, introduction of R649A/P656G double mutation further decreases the binding affinity, with a Kd of 301 μM (Fig. 2B). Together, these data lend a strong support to the bipartite interaction observed for the TTD – PBR complex. On the other hand, mutation of either N147 (equivalent to N149 in zUHRF1) or N194 (equivalent to N186 in zUHRF1) into alanine did not lead to appreciable change in the TTD – PBR binding (Fig. S2C, D), suggesting that the intermolecular interactions involving these sites likely arise from the crystal packing effects.
Figure 2. ITC binding assays for human UHRF1 TTD and PBR.
(A) Mutational effects of the UHRF1 TTD domain on the TTD – PBR interaction. (B) Mutational effects of the UHRF1 PBR peptide on the TTD – PBR interaction. See also Table S1.
The TTD – PBR interaction perturbs the conformation and H3K9me3 binding of the TTD-PHD dual domains
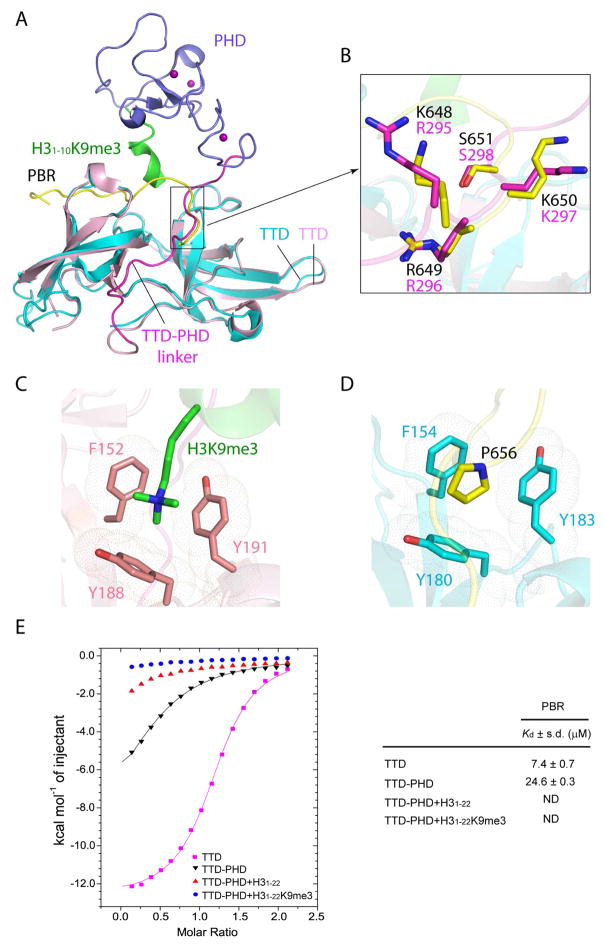
Given that the TTD and PHD domains serve as an integrated histone-binding cassette (Arita et al., 2012; Cheng et al., 2013), we asked whether the TTD – PBR interaction affects the conformation and H3K9me3 binding of the TTD-PHD dual domains. Structural superposition of the TTD – PBR complex with the previously determined structure of UHRF1 TTD-PHD tandem domains in complex with histone H3K9me3 peptide shows that the TTD domains are well aligned (Fig. 3A), with a root-mean-square deviation (RMSD) of 0.73 Å over 122 Cα atoms, suggestive of structural rigidity of the UHRF1 TTD domain. On the other hand, it is apparent that the PBR-binding sites overlap significantly with the binding surface for the TTD-PHD linker or the H3K9me3 peptide (Fig. 3A). In particular, consistent with a recent NMR observation (Fang et al., 2016), the N-terminal segment of PBR (residues K648-S651) superimposes well with a segment of the TTD-PHD linker (residues R295-S298), a region that facilitates the UHRF1 – H3K9me3 association through regulation of the TTD-PHD domain orientation (Arita et al., 2012; Cheng et al., 2013). These two segments, with similar backbone and side-chain conformations (Fig. 3B), compete on the same binding site on TTD, suggesting that the TTD – PBR association would lead to displacement of the TTD-PHD domain linker. Furthermore, structural superimposition of the TTD – PBR and TTD-PHD – H3K9me3 complexes reveals that the PBR peptide-binding site also partially overlaps with the H3K9me3-binding site, with residue P656 occupying the H3K9me3-engaging aromatic cage of the TTD domain (Fig. 3C,D). Together, these structural observations suggest that the TTD – PBR intramolecular interaction may affect the UHRF1 – H3K9me3 binding through a two-layered mechanism: First, binding of PBR to the TTD domain dislodges the TTD-PHD linker from the TTD surface, triggering a conformational rearrangement of the TTD-PHD dual domains that would in turn impede the UHRF1 – H3K9me3 binding. Second, the PBR sequence directly inhibits the binding of H3K9me3 with TTD through embedding a proline ring inside the H3K9me3-binding pocket.
Figure 3. UHRF1 PBR and histone H3K9me3 compete on TTD binding.
(A) Structural comparison of the UHRF1 TTD – PBR complex with the UHRF1 TTD-PHD – H3K9me3 complex (PDB 4GY5), with individual domains and peptides labeled. The zinc ions are shown as purple spheres. (B) Structural overlay the TTD-bound PBR peptide (yellow) and the TTD-PHD linker (magenta), with the aligned residues shown in stick representation. (C) The side chain of H3K9me3 inserted into the aromatic cage formed by residues F152, Y188 and Y191 of human UHRF1 TTD (PDB 4GY5). (D) The side chain of P656 embedded in the equivalent aromatic cage formed by residues F154, Y180 and Y183 of zUHRF1TTD. (E) ITC binding curves of the PBR peptide over the TTD domain or the TTD-PHD dual domains of human UHRF1, in the absence or presence of unmodified H31-22 or H31-22K9me3 peptides. The average and standard deviation of the dissociation constants were derived from two independent measurements. ND, not determinable due to undetectable or non-stoichiometric binding. See also Figure S1, S2 and Table S1.
We then performed ITC binding assays to evaluate how the presence of the TTD-PHD linker, histone H3 or H3K9me3 each affects the TTD – PBR binding (Table S1). Consistent with a previous observation (Fang et al., 2016), binding of the PBR peptide with TTD-PHD is weaker than with TTD by ~3 fold (Fig. 3E), confirming that the TTD-PHD linker and PBR compete on TTD binding. Likewise, the presence of unmodified H3 (residues 1-22, H31-22) and H3K9me3 (residues 1-22, H31-22K9me3) peptides both significantly impairs the TTD – PBR binding, with a relatively more severe effect associated with the H31-22K9me3 peptide (Fig. 3E). Conversely, the presence of excess of the PBR peptide (molar ratio of PBR over TTD-PHD = 3:1) led to a reduction of the TTD-PHD – H31-22K9me3 binding, with the apparent Kd increasing from 0.7 μM to 2.4 μM. By contrast, the presence of the same excess of P656G-mutated PBR reduces the TTD-PHD – H31-22K9me3 binding only to an apparent Kd of 1.3 μM (Fig. S2E). Together, these observations support that the TTD – PBR binding also competes against the interaction of UHRF1 TTD-PHD with histone H3, in particular H3K9me3.
Competing intramolecular and intermolecular interactions govern the conformational states of UHRF1
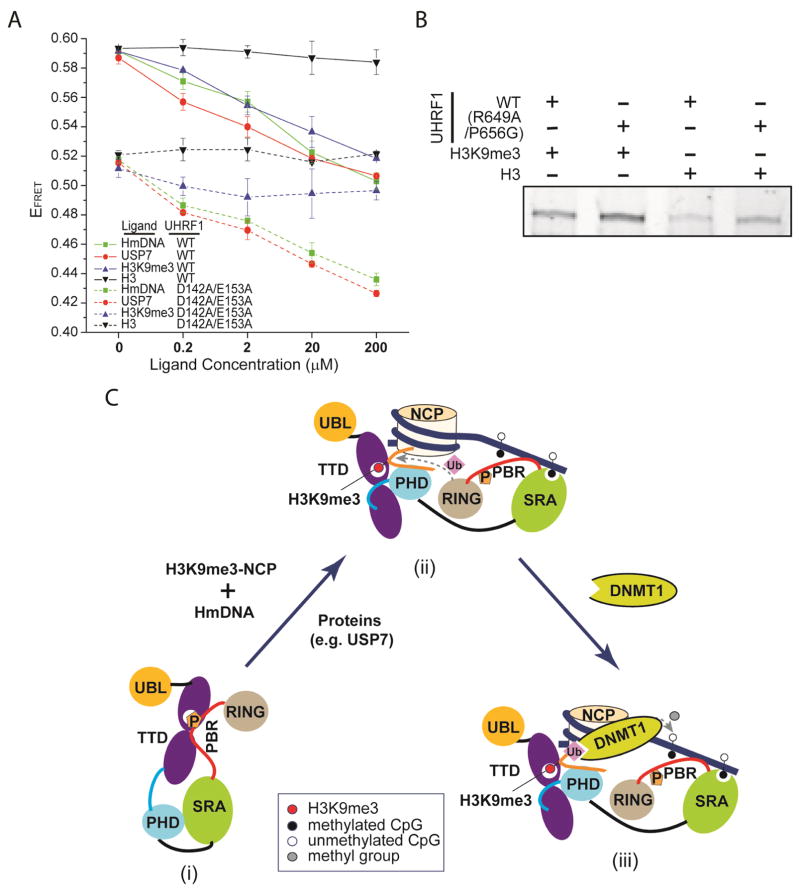
To gain a quantitative view on the effect of the intramolecular and intermolecular interactions of UHRF1 on its conformational states, we generated a CFP-UHRF1-YFP construct, in which full-length UHRF1 was N- and C-terminally fused to CFP and YFP proteins, respectively, for FRET measurements (Fig. S3). Wild-type CFP-UHRF1-YFP fusion protein generated a clear FRET signal (increased acceptor signal associated with decreased donor signal), with IAcceptor/(IDonor+IAcceptor) ratio of 0.6, indicating that the CFP and YFP tags, separated by the ~800 amino acid long UHRF1 protein, are spatially close enough for the FRET signal. In contrast, the CFP-UHRF1 (D142A/E153A)-YFP mutant, in which the TTD – PBR interaction was largely disrupted (Fig. 2B), decreased the FRET ratio to 0.52, suggesting that disruption of the TTD – PBR interaction leads to a more open conformational state for UHRF1 (Fig. 4A). Consistently, addition of increasing amount of the H31-22K9me3 peptide gradually decreased the FRET signal for wild-type, but not D142A/E153A-mutated, CFP-UHRF1-YFP fusion protein. Note that the D142A/E153A mutation, in addition to disrupting the TTD – PBR interaction, reduced the H31-22K9me3-binding affinity of UHRF1 TTD-PHD dual domains by ~4 fold (Fig. S4). In the same vein, this mutation may also affect the association of CFP-UHRF1-YFP with the H31-22K9me3 peptide. Nevertheless, the observation that the FRET signal of D142A/E153A-mutated CFP-UHRF1-YFP remained largely unaffected even in the presence of 1,000-fold molar excess of the H31-22K9me3 peptide (Fig. 4A) suggests that the lack of response to the H31-22K9me3 titration mostly arises from a more “open” conformation of this mutant. These data together support a notion that the H3K9me3 binding “opens” up the conformation of UHRF1 through disruption of the TTD – PBR interaction. On the other hand, titration of the unmodified H31-22 peptide failed to perturb the FRET signals for either wild-type or D142A/E153A-mutated CFP-UHRF1-YFP fusion protein, highlighting the important role of the H3K9me3-binding cage in the TTD – PBR interaction. Furthermore, titration with hemimethylated DNA resulted in even lower FRET signals for both wild-type and D142A/E153A-mutated CFP-UHRF1-YFP (Fig. 4A), suggesting that the interaction of hemimethylated DNA with SRA and PBR synergistically contributes to the conformational “opening” of UHRF1. Finally, we measured the effect of the intermolecular interaction between UHRF1 PBR and USP7 UBL domains, which were previously shown to allosterically regulate the chromatin association of UHRF1 through interaction with PBR (Fig. S5) (Zhang et al., 2015), on the FRET effect of CFP-UHRF1-YPF fusion proteins (Fig. S3). Similar to the effect of hemimethylated DNA, titration of the USP7 UBL domains not only decreased the FRET signal for wild-type UHRF1 fusion protein, but also led to even lower FRET signal for the D142A/E153A-mutated UHRF1 fusion protein (Fig. 4A), further supporting that the PBR-engaged intermolecular interactions synergize with the UHRF1 TTD – H3K9me3 binding to “open up” the conformation of UHRF1.
Figure 4. Coupling between the conformational transition of UHRF1 and its chromatin or USP7 binding.
(A) FRET ratio, EFRET = Iaccetpor/(Iaccetpor+Idonor), of CFP-UHRF1-YFP as function of the concentrations of hemimethylated DNA (HmDNA), USP7 UBL domains (USP7), H31-22K9me3 (H3K9me3) and H31-22 (H3) peptides. (B) Pull-down assays of wild-type (WT) or R649A/P656G-mutated UHRF1 (residues 126-793) with H3K9me3 or unmodified H3 peptides. The gel image cropped from a full gel is boxed. (C) A model for the conformational transition of UHRF1. (i) UHRF1 in free state is dominated by a “closed” conformation. (ii) Association of UHRF1 with hemimethylated DNA (HmDNA) and H3K9me3-modified nucleosome (H3K9me3-NCP) transits UHRF1 into an “open” conformation, permitting strong chromatin association and enhanced H3 ubiquitylation activity. (iii) Ubiquitylated histone H3 subsequently recruits DNMT1 for maintenance DNA methylation. See also Figure S3, S4 and S5.
To further understand the functional implication of the “closed” and “open” UHRF1 conformations, we compared the H3-binding affinities between wild-type and R649A/P656G-mutated UHRF1 fragment, spanning from the TTD domain toward the C-terminal RING-finger domain (residues 126-793), through peptide pull-down assays (Fig. 4B). In comparison with wild-type UHRF1, R649A/P656G-mutated UHRF1 significantly increased its binding to histone H3 peptides, in particular H3K9me3 (Fig. 4B), supporting that disruption of the TTD – PBR interaction promotes the chromatin binding of UHRF1.
Discussion
Identified as one of the essential components of maintenance DNA methylation machinery, UHRF1 regulates the recruitment of DNMT1 to methylation target sites through its specific association with replicating heterochromatin enriched with H3K9me3 and hemimethylated DNA (Bostick et al., 2007; Liu et al., 2013; Miura et al., 2001; Sharif et al., 2007). The synergistic bindings of UHRF1 to H3K9me3 and hemimethylated DNA presumably provide a redundant mechanism for spatial-temporal control of UHRF1 activity within the nuclei, which in turn contributes to the high-fidelity maintenance of DNA methylation. In this regard, recent evidence has indicated that disruption of the UHRF1 – H3K9me3 interaction leads to a modest reduction of genomic DNA methylation, highlighting a role of the UHRF1 – H3K9me3 recognition in promoting DNA methylation maintenance (Zhao et al., 2016).
Previous studies from others and us have revealed that the intramolecular interaction between TTD and PBR of UHRF1 results in a “closed” conformation with reduced chromatin binding activity (Fang et al., 2016; Gelato et al., 2014; Harrison et al., 2016; Zhang et al., 2015), which can be “opened up” through bindings of UHRF1 to H3K9me3 and hemimethylated DNA (Fang et al., 2016; Harrison et al., 2016), chromatin modifiers such as USP7 (Zhang et al., 2015) or DNMT1 (Fang et al., 2016), or nuclear ligand PI5P (Gelato et al., 2014). Recent studies further demonstrated that conformational opening of UHRF1 elevates both of its chromatin binding and E3 ubiquitin ligase activities (Fang et al., 2016; Harrison et al., 2016), therefore establishing a link between the conformational transition of UHRF1 and its cell cycle-dependent chromatin localization (Papait et al., 2007) and ubiquitylation activity (Harrison et al., 2016; Nishiyama et al., 2013; Qin et al., 2015).
Through high-resolution structure determination, mutagenesis and biochemical characterizations, this study confirmed the previous NMR study (Fang et al., 2016) that the intramolecular TTD – PBR interaction of UHRF1 leads to repositioning of the TTD-PHD linker, which consequently interferes with the UHRF1 – H3K9me3 interaction. Extending from the NMR study, the crystal structure presented herein further demonstrated that the interaction of PBR with TTD also leads to the blockage of the H3K9m3-binding cage, thereby providing a more comprehensive view on the conformational regulation of UHRF1 (Fig. 4C). Disruption of the UHRF1 TTD – PBR interaction would presumably lead to simultaneous exposure of the H3K9m3-binding cage and re-alignment of the TTD-PHD linker, resulting in enhanced UHRF1 – H3K9me3 association (Fig. 4C). This H3K9me3 occlusion-regulated conformational transition may provide a mechanism for the H3K9me3-specific localization of UHRF1, which is important for DNMT1-mediated maintenance DNA methylation (Fig. 4C).
The PBR segment of UHRF1 appears to serve as a central platform for functional regulation of UHRF1. Our previous study indicated that introduction of K644E/K646E mutations in the PBR sequence led to reduced chromatin binding of UHRF1, suggesting an important role of this region in nuclear localization of UHRF1 (Zhang et al., 2015). In fact, the intramolecular interaction between PBR and the TTD domain has been shown to be modulated by its intermolecular interaction with DNA (Fang et al., 2016; Harrison et al., 2016), USP7 (Fig. S5) (Zhang et al., 2015), DNMT1 (Fang et al., 2016) and PI5P (Gelato et al., 2014), resulting in a conformational transition of UHRF1 for differential chromatin accessibility. It is conceivable that interactions of the PBR sequence with different nuclear factors are subject to a dynamic regulation throughout the cell cycle, which might be important for its function in maintenance DNA methylation and other cellular activities. For instance, the PBR-mediated interaction between UHRF1 and DNMT1 might facilitate the recruitment of DNMT1 to its target loci, while opening up the conformation of UHRF1 for efficient histone H3 ubiquitylation (Harrison et al., 2016). Subsequently, association of DNMT1 with ubiquitylated H3 (Nishiyama et al., 2013; Qin et al., 2015) likely leads to release of PBR to engage DNA binding in adjacent regions. The mechanism underlying the dynamic transition between different PBR engagements awaits further investigation.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jikui Song (jikui.song@ucr.edu).
METHOD DETAILS
Protein preparation
DNA sequences encoding UHRF1 TTD domain from zebrafish (residues 129-280) or human (residues 126-284), human UHRF1 TTD-PHD dual domains (residues 126-366), human UHRF1 TTD-RING (residues 126-793), human UHRF1 PBR (residues 634-665) and the UBL domains of USP7 (residues 560-1084) were each inserted in a pRSFDuet-1 vector (Novagen), preceded by a hexahistidine and SUMO tag. All the proteins were overexpressed in E. coli BL21 (DE3) RIL strain. The transformed cells were grown at 37 °C in LB medium and induced by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to 0.1 g/L when the OD600 reached 0.8. After induction, the cells continued to grow at 16 °C overnight. The cells were harvested and lysed in a buffer containing 50 mM Tris-HCl (pH 8.0), 1 M NaCl, 25 mM imidazole, and 0.5 mM DTT. After centrifugation, the fusion proteins were purified from the supernatant by a Ni-NTA column, followed by removal of the hexahistidine-SUMO tag by Ubiquitin-like-specific protease 1 (ULP1) cleavage and subsequent Ni-NTA chromatography. The tag-free proteins were further purified by size exclusion chromatography on a Superdex 200 16/600 column (GE Healthcare) pre-equilibrated with a buffer containing 25 mM Tris-HCl (pH 7.5), 100 mM NaCl and 2 mM DTT. Purified samples were stored in −80°C at a concentr ation of ~ 20 mg/mL for future use.
DNA sequence encoding full-length human UHRF1 was inserted in frame with an N-terminal enhanced cyan fluorescence protein (CFP) sequence and a C-terminal yellow fluorescence protein (YFP) sequence in pBad Vector to generate hexahistidine-CFP-UHRF1-YFP construct. Wild type and D142A/E153A mutant of CFP-UHRF1-YFP proteins were expressed in E. coli BL21 (DE3) RIL strain and purified sequentially through affinity chromatography using a Ni-NTA column, hydrophobicity interaction chromatography using a Phenyl HIC column (GE Healthcare), ion exchange chromatography using a Heparin column (GE Healthcare) and size exclusion chromatography using a Superdex 200 16/600 column (GE Healthcare). The purified protein was dissolved in a buffer containing 25 mM Tris (pH 7.5), 100 mM NaCl and concentrated to 1 mg/mL for Förster resonance energy transfer (FRET) assays.
Expression and purification of UHRF1 PBR peptide (residues 634-665) were as described previously (Zhang et al., 2015). Mutant UHRF1 TTD and PBR were generated through site-directed mutagenesis, and expressed and purified in the same manner as that for wild-type proteins.
Crystallization, data collection and structure determination
Human UHRF1 TTD – PBR complex failed to generate crystals for high-resolution structure determination. Therefore, we prepared an inter-species TTD – PBR complex, in which zebrafish UHRF1 TTD domain was mixed with human UHRF1 PBR peptide in a molar ratio of 1:1.5 at a total concentration of ~25 mg/mL, for crystallization. Crystals were generated through mixing of the complex with precipitant containing 0.1 M Succinic acid pH7.0, 15% (v/v) Polyethylene glycol 3350 and incubation using hanging-drop diffusion method. Before flash freezing in liquid nitrogen, crystals were soaked in cryo-protectant made of crystallization solution supplemented with 20% (v/v) glycerol. The X-ray diffraction data for the UHRF1 TTD – PBR complex were collected on the BL 5.0.1 beamline at the Advanced Light Source, Lawrence Berkeley National Laboratory. The diffraction data were indexed, integrated and scaled using the HKL2000 program (Otwinowski and Minor, 1997). The structure was solved using the molecular replacement method in PHASER (McCoy et al., 2007) with the structure of UHRF1 TTD (PDB ID: 3DB3) as search model. The resulting electron density revealed that there are three molecules of TTD and one PBR peptide in each asymmetric unit. The structure of TTD – PBR complex was improved by iterative modeling building and refinement with Coot (Emsley et al., 2010) and PHENIX software packages (Afonine et al., 2012). The same R-free test set was used throughout the refinement. The statistics for data collection and structural refinement of the UHRF1 TTD – PBR complex is summarized in Table 1.
ITC measurements
Histone H3 peptides containing residues 1-22 (H31-22), followed by a C-terminal tyrosine, were used for ITC assay. Protein or peptide samples were dialyzed against the ITC buffer (25 mM Tris-HCl, pH 7.5, 100 mM NaCl) at 4°C overnight. A MicroCal iTC200 system (GE Healthcare) was used to conduct the ITC measurements. For the TTD – PBR displacement titration, protein samples for UHRF1 domains were first mixed with unmodified H3 or H3K9me3 peptides in a molar ratio of 1:1, followed by titration with the PBR peptide. For displacement titration of TTD-PHD over H3K9me3, protein samples for the TTD-PHD domains were first mixed with wild-type or mutant PBR in a molar ratio of 1:3, followed by titration with the H31-22K9me3 peptide. A total of 17 injections with a spacing of 180 seconds and a reference power of 5 μcal/s were performed at 25°C. The ITC curves were processed with software ORIGEN (MicroCal) using one-site fitting model. The results are summarized in Table S1.
Peptide pull-down assay
40 μL streptavidin paramagnetic beads (MagneSphere, Promega) were washed three times with 1× PBS (pH 7.4) buffer and resuspended in 100 μL PD buffer (25 mM Tris, pH 7.5, 300 mM KCl, 20% glycerol, 1 mM DTT and 0.2% v/v Triton X-100). To pull down UHRF1, biotin-labeled H31-20K9me3 or unmodified H31-20, recombinant UHRF1 (residues 126-793) wild type or R649A/P656G mutant were mixed with the beads to a final concentration of 2.0 μM each and incubated at 4 °C for 1 hr. The beads were washed five times with PD buffer, followed by releasing the proteins into elution buffer (50 mM Tris, pH7.5, 25 % v/v glycerol, 1 mM EDTA, 2% SDS, 10 mM DTT) by boiling for 10 min. All the samples were finally resolved by SDS-PAGE and stained by Sypro ruby (Bio-rad).
In vitro FRET measurements
For FRET assay, CFP-UHRF1-YFP wild-type and mutant proteins were adjusted to 0.2 μM, titrated with increasing amount of a hemimethylated DNA duplex (upper strand: 5′-GGCCCXGCAGGCCTG-3′; lower strand: 5′-CAGGCCTGCGGGGCC-3′; X = 5-methylcytosine), USP7 UBL domains or histone peptides. Assays were performed in Take3-plate (#259913 BioTek Instruments) in a buffer containing 25 mM Tris (pH 7.5), 100 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP). Emission intensities were scanned using a SynergyMX microplate reader (#255439 BioTek Instruments) from 470 to 600 nm, with 450 nm as the excitation wavelength. The FRET efficiency ratio was calculated as fluorescence intensity at 525 nm divided by the sum of fluorescence intensities at 480 nm and 525 nm. Emission spectra were normalized to the sum of the fluorescence peak intensities at 480 nm and 525 nm. All assays were performed in duplicate, and the data were processed and normalized using Origin version 7.0 (OriginLab).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics of the X-ray crystallographic data processing, refinement and structure validation are summarized in Table 1.
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates and structure factors for the zUHRF1TTD – PBR complex have been deposited in the Protein Data Bank under accession code 6B9M.
Supplementary Material
Highlights.
The crystal structure of UHRF1 TTD domain bound to the PBR fragment was solved
A proline ring from UHRF1 PBR occupies the H3K9me3-binding cage of the TTD domain
PBR inhibits the histone binding of UHRF1 via a two-layered regulation
The UHRF1 TTD – PBR interaction controls the conformational states of UHRF1
Acknowledgments
This work was supported by Kimmel Scholar Awards from Sidney Kimmel Foundation for Cancer Research (to J.S.), March of Dimes Foundation (1-FY15-345 to J.S.) and NIH (1R35GM119721 to J.S). We would like to thank staff members at the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory for support in access to X-ray beamlines. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
AUTHOR CONTRIBUTIONS
L.G. and X-F.T. contributed equally to this work. L.G. performed protein purification, crystallization, FRET measurements and ITC assays. X.T. performed cloning, peptide pull-down assay and structure determination and analysis. S.Z., T.W. and H-W.A. provided resources and assistance for FRET measurements. Z-M Z assisted protein purification. J.S. conceived the study and experimental design, analyzed experiments. L.G., X-F. T. and J.S. prepared the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita K, Ariyoshi M, Tochio H, Nakamura Y, Shirakawa M. Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism. Nature. 2008;455:818–821. doi: 10.1038/nature07249. [DOI] [PubMed] [Google Scholar]
- Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, et al. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A. 2012;109:12950–12955. doi: 10.1073/pnas.1203701109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature. 2008;455:822–825. doi: 10.1038/nature07273. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, Xu Y. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem. 2013;288:1329–1339. doi: 10.1074/jbc.M112.415398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Song J, Wang Y, Zhao Y, Guda K, Yang S, Kao HY, Xu Y, Willis J, Markowitz SD, et al. DNMT1 stability is regulated by proteins coordinating deubiquitination and acetylation-driven ubiquitination. Science signaling. 2010;3:ra80. doi: 10.1126/scisignal.2001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang LH, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10:2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Cheng J, Wang J, Zhang Q, Liu M, Gong R, Wang P, Zhang X, Feng Y, Lan W, et al. Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition. Nature communications. 2016;7:11197. doi: 10.1038/ncomms11197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelato KA, Tauber M, Ong MS, Winter S, Hiragami-Hamada K, Sindlinger J, Lemak A, Bultsma Y, Houliston S, Schwarzer D, et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol Cell. 2014;54:905–919. doi: 10.1016/j.molcel.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Harrison JS, Cornett EM, Goldfarb D, DaRosa PA, Li ZM, Yan F, Dickson BM, Guo AH, Cantu DV, Kaustov L, et al. Hemi-methylated DNA regulates DNA methylation inheritance through allosteric activation of H3 ubiquitylation by UHRF1. eLife. 2016:5. doi: 10.7554/eLife.17101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature. 2008;455:826–829. doi: 10.1038/nature07280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell research. 2011;21:1374–1378. doi: 10.1038/cr.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W, Liu Y, Xu SG, Yin WJ, Li JJ, Yang JM, Shao ZM. UHRF1 inhibits MDR1 gene transcription and sensitizes breast cancer cells to anticancer drugs. Breast cancer research and treatment. 2010;124:39–48. doi: 10.1007/s10549-009-0683-8. [DOI] [PubMed] [Google Scholar]
- Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol. 2008;28:705–717. doi: 10.1128/MCB.01598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res. 2009;37:493–505. doi: 10.1093/nar/gkn961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallous N, Legrand P, McEwen AG, Ramon-Maiques S, Samama JP, Birck C. The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PloS one. 2011;6:e27599. doi: 10.1371/journal.pone.0027599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang CC, Zhan B, Yoshikawa Y, Haas W, Gygi SP, Cohn MA. UHRF1 Is a Sensor for DNA Interstrand Crosslinks and Recruits FANCD2 to Initiate the Fanconi Anemia Pathway. Cell reports. 2015;10:1947–1956. doi: 10.1016/j.celrep.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nature communications. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, et al. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A. 2012;109:4828–4833. doi: 10.1073/pnas.1116349109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mistry H, Tamblyn L, Butt H, Sisgoreo D, Gracias A, Larin M, Gopalakrishnan K, Hande MP, McPherson JP. UHRF1 is a genome caretaker that facilitates the DNA damage response to gamma-irradiation. Genome integrity. 2010;1:7. doi: 10.1186/2041-9414-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura M, Watanabe H, Sasaki T, Tatsumi K, Muto M. Dynamic changes in subnuclear NP95 location during the cell cycle and its spatial relationship with DNA replication foci. Experimental cell research. 2001;263:202–208. doi: 10.1006/excr.2000.5115. [DOI] [PubMed] [Google Scholar]
- Muto M, Kanari Y, Kubo E, Takabe T, Kurihara T, Fujimori A, Tatsumi K. Targeted disruption of Np95 gene renders murine embryonic stem cells hypersensitive to DNA damaging agents and DNA replication blocks. J Biol Chem. 2002;277:34549–34555. doi: 10.1074/jbc.M205189200. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature. 2013;502:249–253. doi: 10.1038/nature12488. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Molecular biology of the cell. 2007;18:1098–1106. doi: 10.1091/mbc.E06-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Wolf P, Liu N, Link S, Smets M, La Mastra F, Forne I, Pichler G, Horl D, Fellinger K, et al. DNA methylation requires a DNMT1 ubiquitin interacting motif (UIM) and histone ubiquitination. Cell research. 2015 doi: 10.1038/cr.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell. 2011;43:275–284. doi: 10.1016/j.molcel.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev. 2013;27:1288–1298. doi: 10.1101/gad.220467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol. 2012;19:1155–1160. doi: 10.1038/nsmb.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Taylor EM, Bonsu NM, Price RJ, Lindsay HD. Depletion of Uhrf1 inhibits chromosomal DNA replication in Xenopus egg extracts. Nucleic Acids Res. 2013;41:7725–7737. doi: 10.1093/nar/gkt549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Paramasivam M, Ghosal G, Chen D, Shen X, Huang Y, Akhter S, Legerski R, Chen J, Seidman MM, et al. UHRF1 Contributes to DNA Damage Repair as a Lesion Recognition Factor and Nuclease Scaffold. Cell reports. 2015;10:1957–1966. doi: 10.1016/j.celrep.2015.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Kubo E, Kanari Y, Ikemura T, Tatsumi K, Muto M. Temporal and spatial localization of novel nuclear protein NP95 in mitotic and meiotic cells. Cell structure and function. 2000;25:149–159. doi: 10.1247/csf.25.149. [DOI] [PubMed] [Google Scholar]
- Xie S, Jakoncic J, Qian C. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J Mol Biol. 2012;415:318–328. doi: 10.1016/j.jmb.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Zhang ZM, Rothbart SB, Allison DF, Cai Q, Harrison JS, Li L, Wang Y, Strahl BD, Wang GG, Song J. An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell reports. 2015;12:1400–1406. doi: 10.1016/j.celrep.2015.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang J, Chen R, Wang L, Li B, Cheng H, Duan X, Zhu H, Wei W, Li J, et al. Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nature communications. 2016;7:12464. doi: 10.1038/ncomms12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.