Abstract
Scope
Body weight responds variably to the intake of dairy foods. Genetic variation may contribute to inter-individual variability in associations between body weight and dairy consumption.
Methods and results
A genome-wide interaction study to discover genetic variants that account for variation in BMI in the context of low-fat, high-fat and total dairy intake in cross-sectional analysis was conducted. Data from nine discovery studies (up to 25 513 European descent individuals) were meta-analyzed. Twenty-six genetic variants reached the selected significance threshold (p-interaction<10−7), and six independent variants (LINC01512-rs7751666, PALM2/AKAP2-rs914359, ACTA2-rs1388, PPP1R12A-rs7961195, LINC00333-rs9635058, AC098847.1-rs1791355) were evaluated meta-analytically for replication of interaction in up to 17 675 individuals. Variant rs9635058 (128 kb 3′ of LINC00333) was replicated (p-interaction = 0.004). In the discovery cohorts, rs9635058 interacted with dairy (p-interaction = 7.36 × 10−8) such that each serving of low-fat dairy was associated with 0.225 kg m−2 lower BMI per each additional copy of the effect allele (A). A second genetic variant (ACTA2-rs1388) approached interaction replication significance for low-fat dairy exposure.
Conclusion
Body weight responses to dairy intake may be modified by genotype, in that greater dairy intake may protect a genetic subgroup from higher body weight.
Keywords: body mass index, CHARGE consortium, dairy intake, genome-wide interaction study, meta-analysis
1. Introduction
Dairy foods represent an important food group across many cultures and geographic regions, and are recommended as part of several healthy diets. The extent to which dairy foods influence anthropometric traits such as BMI and waist circumference has been a longstanding and still unresolved focus of investigation. Observational and intervention studies, meta-analyses and systematic reviews have reported protective, detrimental and neutral associations of different types of dairy foods (e.g., low-fat, high-fat) with anthropometric outcomes.[1–8]
Genetic variability may contribute to reported inter-individual variation in associations between dairy foods and anthropometric traits. Strong positive selection near the lactase locus favored genetic variants that enabled lifelong dairy consumption,[9,10] but whether other variants modulate responses to dairy intake is largely unexplored. The few studies that simultaneously considered both genetic variation and dairy intake for anthropometric traits have been limited to a small set of loci, including lactase (LCT), insulin-like growth factor II (IGF) and apolipoprotein AII (APOA2).[11–14]
As genome-wide association studies provide an agnostic tool for discovery of new variants, genome-wide interaction studies (GWIS) have the potential to uncover additional loci that may be modulated by specific foods. Findings from GWIS focused on dairy foods and BMI may broaden biological understanding of the genetic risk of obesity, and may eventually be relevant to dietary recommendations. Therefore, the objective of the current study was to conduct a GWIS to investigate low-fat, high-fat and total dairy intake for the outcome of BMI.
2. Experimental Section
2.1. Discovery Cohorts
Participating cohorts included up to 25 513 individuals of European ancestry from nine studies in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Nutrition Working Group (Table 1; Supporting Information Table 1): Atherosclerosis Risk in Communities Study (ARIC), Cardiovascular Health Study (CHS), Family Heart Study (FamHS), Genetics of Lipid Lowering Drugs and Diet Network (GOLDN), Health, Aging and Body Composition (Health ABC), Invecchiare in Chianti, aging in the Chianti area (InCHIANTI), Multi-Ethnic Study of Atherosclerosis (MESA), Rotterdam Study, and the Young Finns Study (YFS). All individuals participating in the studies provided written informed consent, and approval for all study protocols was granted by local institutional review boards.
Table 1.
Discovery and replication cohorts characteristics.
| n | Age (years) | Sex (% women) | Dairy consumption (servings/day)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| BMI (kg m−2) | Total energy (kcal day−1) | Total dairy | Low-fat dairy | High-fat dairy | Milk | Cheese | Yogurt | ||||
| Discovery cohorts | |||||||||||
| Atherosclerosis Risk in Communities Study (ARIC) | 9105 | 54 ± 6 | 53 | 27.0 ± 4.8 | 1644 ± 607 | 1.83 ± 1.42 | 0.88 ± 1.04 | 0.86 ± 0.95 | 0.95 ± 1.05 | 0.54 ± 0.59 | 0.09 ± 0.24 |
| Cardiovascular Heart Study (CHS) | 3094 | 72 ± 5 | 61 | 26.3 ± 4.4 | 2026 ± 652 | 1.67 ± 0.87 | 0.57 ± 0.54 | 1.10 ± 0.79 | 0.75 ± 0.45 | 0.38 ± 0.36 | 0.10 ± 0.20 |
| Family Heart Study (FamHS) | 2983 | 54 ± 14 | 53 | 27.7 ± 5.5 | 1478 ± 619 | 2.38 ± 1.73 | 1.27 ± 1.23 | 1.11 ± 1.22 | 1.05 ± 1.14 | 0.42 ± 0.52 | 0.14 ± 0.33 |
| Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) | 784 | 49 ± 16 | 51 | 28.5 ± 6 | 2010 ± 785 | 2.3 ± 1.6 | 0.95 ± 1.2 | 1.3 ± 1.3 | 1.3 ± 1.3 | 0.53 ± 0.45 | 0.1 ± 0.16 |
| Health Aging and Body Composition (Health ABC) | 1497 | 74 ± 3 | 48 | 26.6 ± 4.1 | 1818 ± 606 | 1.36 ± 1.29 | 0.65 ± 1.11 | 0.71 ± 0.95 | 0.96 ± 1.22 | 0.2 ± 0.27 | 0.06 ± 0.14 |
| Invecchiare in Chianti, aging in the Chianti area (InCHIANTI) | 1090 | 67 ± 15 | 55 | 27.2 ± 4.2 | 2011 ± 588 | 1.31 ± 0.9 | 0.06 ± 0.16 | 1.24 ± 0.86 | 0.55 ± 0.63 | 0.60 ± 0.47 | 0.06 ± 0.16 |
| Multi-Ethnic Study of Atherosclerosis (MESA) | 1713 | 70 ±9 | 51 | 28 ± 5.4 | 1709 ± 735 | 1.51 ± 1.39 | 1.07 ± 1.27 | 0.45 ± 0.42 | 0.73 ± 1.12 | 0.30 ± 0.34 | 0.24 ± 0.34 |
| Rotterdam Study | 3563 | 66 ± 7 | 59 | 26.8 ± 4 | 1978 ± 501 | 3.95 ± 2.37 | 2.47 ± 2.19 | 1.49 ± 1.37 | 1.13 ± 0.97 | 0.85 ± 0.54 | 0.29 ± 0.33 |
| Young Finns Study (YFS) | 1684 | 38 ± 5 | 56 | 25.7 ± 4.6 | 2386 ± 765 | 4.24 ± 2.53 | 2.06 ± 1.49 | 2.20 ± 1.89 | 1.57 ± 1.34 | 0.50 ± 0.44 | 0.67 ± 0.7 |
| Replication cohorts | |||||||||||
| Diet, Cancer and Health (Danish EPIC) | 6583 | 57 ± 4 | 57 | 26.8 ± 4.3 | 2369 ± 667 | 2.59 ± 1.48 | 1.05 ± 1.19 | 1.54 ± 0.93 | 1.12 ± 1.2 | 0.86 ± 0.6 | 0.35 ± 0.44 |
| Malmö Diet and Cancer Study (MDCS) | 4446 | 57 ± 6 | 59 | 25.7 ± 4 | 2328 ± 675 | 5.8 ± 4.8 | 0.98 ±1 | 4.83± 4.83 | 1.18 ± 0.99 | 0.98 ± 0.7 | 0.37 ± 0.46 |
| Netherlands Epidemiology of Obesity (NEO) | 5688 | 56 ± 6 | 52 | 30.0 ± 4.8 | 2275 ± 708 | 2.78 ± 2.09 | 0.52 ± 0.50 | 2.26 ± 2.06 | 0.77 ± 0.72 | 0.79 ± 0.66 | 0.31 ± 0.29 |
| Western Australian Pregnancy Cohort Study (Raine) | 958 | 20 ± 0.4 | 49 | 24.5 ± 5.1 | 2059 ± 936 | 2.44 ± 1.83 | 0.36 ± 0.59 | 2.04 ± 1.88 | 1.32 ± 0.86 | 0.28 ± 0.29 | 0.13 ± 0.04 |
2.2. Dietary Assessment
Dietary intakes for most of the cohorts were estimated using food-frequency questionnaires (FFQs) (Table 2, Supporting Information) that were adapted to each cohort. In Malmö Diet and Cancer study (MDCS), a modified diet history method combining a 7-d food record, an FFQ and a 45 minute interview were used to estimate intakes. Dairy intake was evaluated as low-fat dairy, high-fat dairy and total dairy servings/day, according to published definitions.[15,16] Low-fat dairy foods included 1%milk, skim milk, cottage cheese, and yogurt. High-fat dairy foods included cream or half and half, whole milk, 2% milk, cheese, butter, and ice cream.
In MDCS, 3% yogurt was included as a high-fat dairy food. A serving of milk was 240 mL (1 cup or 8 fluid oz). Milk serving equivalents[17] were used to calculate servings of other dairy products including 43 g (1½ oz) hard cheese (cheddar, mozzarella, Swiss, parmesan), 480 mL (2 cups) cottage cheese, 57 g (2 oz) processed cheese, 360 mL (1½ cups) ice cream. Butter was included as a high-fat dairy food with one serving consisting of 1 pat (5 grams or 1 inch sq × 1/3 inch high) or 1 teaspoon). Total dairy included low-fat and high-fat forms. Non-dairy milks (soy, nut, rice, coconut) were not included as dairy foods.
2.3. Genotyping and Imputation
Genome-wide genotyping was conducted using Illumina and Affymetrix platforms as described in Supporting Information Table 3. All cohorts contributing to the current study have participated previously in genome-wide association studies for cardiometabolic traits. Phased haplotypes from HapMap II in the CEU populations were used to impute approximately 2.5 million autosomal SNPs using algorithms that were implemented in Bim-Bam, IMPUTE2 and MACH.
2.4. BMI Measurements
BMI was calculated as weight in kg, divided by height in meters squared. Methods for measuring body weight and height are described in Supporting Information Table 4.
2.5. Cohort-Specific Genome-Wide x Dairy Analyses: Discovery
Analysts from each participating cohort conducted statistical analyses using the same centrally designed analysis plan that was developed in the CHARGE Nutrition Working Group. Individuals with implausible dietary data such as very high or very low energy intake (as defined by the cohort), non-European ancestry or missing phenotype or genotype data were excluded from analysis. The interaction between genome-wide single nucleotide polymorphisms (SNPs) and dairy foods as described in Dietary assessment (low-fat, high-fat, and total) for the outcome of BMI was examined in each cohort using linear or linear mixed effects regression with an additive genetic model. Genotyped SNPs were assigned the values 0,1, or 2 representing the number of copies of the effect allele. Imputed SNPs reflected the expected dosage as output from imputation software, ranging between 0 and 2. Additional details of genotyping and imputation are included in the Genotyping and imputation section above. Software for the genome-wide interaction analysis was cohort–specific based on the pipeline established by each cohort, and included software such as QUICKTEST, ProbABEL and R (gee.test function and others). Two models were evaluated in the nine discovery cohorts: Model 1 included adjustment by age, sex, field center and familial relationships (as needed) and principal components (as needed to adjust for population substructure). Model 2 included all covariates applied in Model 1, with additional adjustment for total energy (kcal day−1), physical activity (sedentary/non-sedentary),[ 18] and the CHARGE diet score (a food groups-based indicator of dietary quality that was developed in CHARGE populations, where it was reported to be highly predictive for BMI).[19] Each cohort generated an output file that was meta-analyzed. Data elements of the output file included: SNP id, call rate, sample size for the SNP, coded and non-coded alleles, allele frequencies, strand, regression coefficient and standard errors, Hardy-Weinberg equilibrium p value, imputation method, and imputation quality.
2.6. Meta-Analyses of Discovery Cohorts: Fixed and Random Effects
Prior to meta-analysis of the linear regression output (regression coefficients and standard errors) generated by each discovery cohort, EasyQC software was used to perform quality control. We eliminated SNPs that contained any of the following: invalid allele value, low minor allele frequency (<1%), low call rate (<0.95), or low imputation quality (MACH: R2 <0.3; or IMPUTE: proper info <0.4).[20] We conducted inverse-variance weighted, fixed-effect meta-analyses using METAL software (http://umich.edu/csg/abecasis/Metal/) for genome-wide interactions with dairy intake for the outcome of BMI. Additional quality control filtering was applied by METAL software to exclude SNPs with high heterogeneity (p-value <1 × 10−6), low number of contributing cohorts (<4) or low sample size (<5000 individuals). Genomic control was applied to account for population stratification. A threshold significance of p < 10−7 was selected for evaluating SNPs for replication. We selected p < 10−7 as a threshold based on its use in several earlier GWAS,[21–23] and empirical evidence from the Genome-Wide Significance Project that a large proportion of associations at p>5 × 10−8 and < 10−7 were replicable.[24] We assessed heterogeneity across studies using Cochran’s Q statistic, which was quantified using the I2 statistic. Guidelines for quantifying heterogeneity rely on I2 with low, moderate and high corresponding to 25, 50, and 75%, respectively.[25] For SNPs that reached the selected significance threshold (Table 2) we also conducted random effects meta-analysis.
Table 2.
Meta-analysis of genome-wide (<10−7) interactions with dairy intake for BMI.
| SNP | Chr | Position | Nearest gene | Effect allele/ other allele | Minor allele frequency | Dairy exposure/model | β ± SE | P-interact | Cohort-specific beta directions | I2 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs7751666 | 6 | 43919724 | LINC01512 | G/T | G(0.03) | Highfat/Model 1 | −0.8356 ± 0.154 | 5.30E-08 | -?+------ | 45.1 |
| rs914359 | 9 | 110160839 | PALM2/AKAP2 | T/G | G(0.13) | Highfat/Model 1 | 0.307 ± 0.0564 | 5.09E-08 | ++--+++++ | 41.3 |
| rs1388 | 10 | 88942942 | ACTA2 | C/A | C (0.01) | Total/Model 2 | −0.4982 ± 0.0916 | 5.44E-08 | -?-+----? | 11.9 |
| rs7970890 | 12 | 79906599 | PPP1R12A | A/C | C(0.02) | Highfat/Model 2 | −0.8688 ± 0.134 | 9.06E-11 | -?-+++--? | 74.7 |
| rs17006080 | 12 | 79870485 | PPP1R12A | T/C | C(0.02) | Highfat/Model 2 | −0.8673 ± 0.134 | 9.76E-11 | -?-+++--? | 74.7 |
| rs12831031 | 12 | 79904615 | PPP1R12A | T/C | C(0.02) | Highfat/Model 2 | −0.8649 ± 0.134 | 1.06E-10 | -?-+++--? | 74.7 |
| rs3898515 | 12 | 79887660 | PPP1R12A | A/G | A(0.02) | Highfat/Model 2 | 0.8651 ± 0.134 | 1.09E-10 | +?+---++? | 75 |
| rs7310740 | 12 | 79881403 | PPP1R12A | C/G | C(0.02) | Highfat/Model 2 | 0.865 ± 0.134 | 1.09E-10 | +?+—++? | 75 |
| rs7305173 | 12 | 79912942 | PPP1R12A | T/C | C(0.02) | Highfat/Model 2 | −0.8632 ± 0.134 | 1.12E-10 | -?-+++--? | 75.6 |
| rs12817698 | 12 | 79855923 | PPP1R12A | A/G | G(0.02) | Highfat/Model 2 | −0.8661 ± 0.135 | 1.26E-10 | -?-+++--? | 75.9 |
| rs12811009 | 12 | 79928288 | PPP1R12A | A/G | G(0.02) | Highfat/Model 2 | −0.8644 ± 0.1358 | 1.93E-10 | -?-+++--? | 75.4 |
| rs7298596 | 12 | 79936051 | PPP1R12A | T/C | T(0.02) | Highfat/Model 2 | 0.864 ± 0.1359 | 2.03E-10 | +?+---++? | 75.1 |
| rs17006192 | 12 | 79949733 | RP11-84G21.1 | T/C | T(0.02) | Highfat/Model 2 | 0.844 ± 0.1351 | 4.12E-10 | +?+---++? | 76.2 |
| rs7309266 | 12 | 79951225 | RP11-84G21.1 | A/G | A(0.02) | Highfat/Model 2 | 0.8425 ± 0.1355 | 4.98E-10 | +?+---?+? | 80.5 |
| rs7978089 | 12 | 79847493 | PPP1R12A | A/T | A(0.02) | Highfat/Model 2 | 0.8556 ± 0.1388 | 7.09E-10 | +?+---++? | 76.5 |
| rs17005986 | 12 | 79815144 | PPP1R12A | T/C | C(0.02) | Highfat/Model 2 | −0.8421 ± 0.1399 | 1.76E-09 | -?-+++--? | 76 |
| rs7961195 | 12 | 79792285 | PPP1R12A | T/C | C(0.02) | Highfat/Model 2 | −0.8219 ± 0.139 | 3.37E-09 | ---+++--? | 71.6 |
| rs7295269 | 12 | 79981139 | RP11-84G21.1 | A/G | G(0.02) | Highfat/Model 2 | −0.7589 ± 0.1301 | 5.41E-09 | -?--+++-? | 79.2 |
| rs12833727 | 12 | 79773581 | PPP1R12A | C/G | G(0.02) | Highfat/Model 2 | −0.8054 ± 0.1392 | 7.23E-09 | ---++++-? | 72.4 |
| rs4403864 | 12 | 79981332 | RP11-84G21.1 | C/G | C(0.02) | Highfat/Model 2 | 0.7493 ± 0.1301 | 8.40E-09 | +?++---+? | 80 |
| rs9565965 | 13 | 84689543 | LINC00333 | A/T | T (0.15) | Lowfat/Model 1 | 0.2248 ± 0.0417 | 7.17E-08 | +++++-+++ | 0 |
| rs9635058 | 13 | 84691029 | LINC00333 | A/G | A (0.15) | Lowfat/Model 1 | -0.2246 ± 0.0417 | 7.36E-08 | -----+--- | 0 |
| rs9565960 | 13 | 84671780 | LINC00333 | A/C | A (0.15) | Lowfat/Model 1 | −0.2235 ± 0.0416 | 7.93E-08 | -----+--- | 0 |
| rs9565961 | 13 | 84671796 | LINC00333 | T/C | T (0.15) | Lowfat/Model 1 | −0.2238 ± 0.0417 | 8.19E-08 | -----+--- | 0 |
| rs9575642 | 13 | 84684207 | LINC00333 | A/G | A (0.15) | Lowfat/Model 1 | −0.2238 ± 0.0417 | 8.19E-08 | -----+--- | 0 |
| rs1791355 | 18 | 59780080 | AC098847.1 | C/T | C (0.02) | Highfat/Model 2 | 0.8159 ± 0.153 | 9.70E-08 | +++-++++? | 41.8 |
Additive allele model. Model 1 = adjusted for by age, sex, and study-specific covariates (field center, familial relationships and principal components for population stratification, when applicable). Model 2 = adjusted for Model 1 plus total energy, physical activity (sedentary versus non-sedentary) and diet quality (CHARGE diet score). Interaction coefficients are shown as β ± SE. β represents the difference in BMI (kg m−2) with each additional serving of dairy, per each additional copy of the effect allele. I2 represents the heterogeneity statistic, presented as %. Order of cohorts for beta directions: ARIC, CHS, FamHS, GOLDN, Health ABC, InCHIANTI, MESA, Rotterdam, YFS.
chr, chromosome; SNP, single nucleotide polymorphism.
2.7. Selection of SNPs from the Discovery Meta-Analyses for Evaluation of Replication
Twenty-six SNPs encompassing six loci reached the selected significance threshold in the discovery cohorts (Table 2). From among these 26 SNPs, six independent (r2<0.8) SNPs captured the six regions (rs7751666, rs914359, rs1388, rs7961195, rs9635058, rs1791355). These SNPs were chosen for meta-analytic evaluation of replication to reduce the multiple testing burden, and a threshold p value of 0.008 (0.05/6) was used to correct for multiple comparisons. Based on the hypothesis that the direction of the meta-analyzed replication regression coefficient would be the same as the meta-analyzed discovery regression coefficient, one-sided replication p values were compared to the replication significance threshold. For four loci at which variants reached the threshold (LINC01512, PALM2/AKAP2, ACTA2 and AC098847.1) only one SNP was available at each locus for evaluation of replication. For one locus where multiple, linked SNPs were available (PPP1R12A), the SNP with lowest heterogeneity (I2) and the highest availability in discovery cohorts (8/9) was selected. For the LINC00333 locus at which I2 heterogeneity was 0 for all linked SNPs, a SNP with in silico support for functionality was selected.[26]
2.8. Cohort-Specific Analyses of Selected SNPs: Replication
Replication cohorts consisted of up to 17 675 individuals of European ancestry from four studies participating in the CHARGE Consortium Nutrition Working Group (Table 3: Diet, Cancer and Health (Danish EPIC), MDCS, Netherlands Epidemiology of Obesity (NEO) and Western Australian Pregnancy Cohort (Raine) Study. All individuals participating in the replication studies provided written informed consent, and approval for all study protocols was granted by local institutional review boards.
Table 3.
Meta-analysis of replication interactions of six independent SNPs with dairy intake for BMI.
| SNP | Effect allele/ Other allele | Minor allele (frequency) | Dairy exposure/model | β ± SE | P-interact (one-sided) | Cohort-specific beta directions | I2 |
|---|---|---|---|---|---|---|---|
| rs7751666 | T/G | G (0.03) | Lowfat/model 3 | −0.186 ± 0.166 | 0.132 | ---- | 65.9 |
| rs914359 | T/G | G (0.13) | Highfat/model 3 | 0.033 ± 0.022 | 0.069 | -+++ | 0 |
| rs1388 | A/C | C (0.01) | Lowfat model 3 | −0.592 ± 0.320 | 0.032 | --?+ | 0 |
| rs7961195 | T/C | C (0.02) | Lowfat/model 3 | −0.265 ± 0.187 | 0.078 | ---- | 0 |
| rs9635058 | A/G | A (0.15) | Highfat/model 2 | −0.053 ± 0.022 | 0.004 | +--+ | 7.8 |
| rs1791355 | T/C | C (0.02) | Total/model 3 | 0.088 ± 0.062 | 0.077 | -+++ | 0 |
Additive allele model. Model 2 = adjusted for age, sex, cohort-specific covariates (field center, family and principle components for population stratification, as applicable), physical activity (sedentary versus non-sedentary) and diet quality (CHARGE diet score) Model 3 = Model 2 with diet quality covariate removed. Interaction coefficients are shown as β ± SE. β represents the difference in BMI (kgm−2) with each additional serving of dairy, per each additional copy of the effect allele. I2 represents the heterogeneity statistic, presented as %. Order of cohorts for beta directions: Danish EPIC, MDC, NEO, Raine.
SNP, single nucleotide polymorphism.
Each cohort evaluated interactions between dairy intake (low-fat, high-fat and total) for the 26 SNPs that reached the selected significance threshold. As in the discovery phase, analysis to evaluate replication was conducted by cohort-specific analysts using Model 1 (age, sex, population-specific covariates) and Model 2 (Model 1 + total energy, physical activity and diet quality). For replication, we also applied a third model (Model 3) that consisted of Model 2 with the removal of the diet quality score covariate. Regression coefficients and standard errors for 6 independent SNPs were meta-analyzed using METAL software.
3. Results
3.1. Study Characteristics
The general characteristics and dairy intakes of the nine discovery cohorts and four replication cohorts are shown (Table 1; Supporting Information Table 1). The mean age in discovery cohorts ranged from 38 to 74 years, and 20 to 57 years in replication cohorts. Mean BMI ranged from 25.7 to 28.5 kgm−2 in discovery and 24.5 to 30 kg m−2 in replication cohorts. Distributions of low-fat and high-fat dairy intakes varied across groups. Low-fat dairy intake ranged from a low of 0.06 servings per day in InCHIANTI to a high of 2.47 servings per day in the Rotterdam Study. High-fat dairy ranged from 0.45 servings/day in MESA to 2.2 servings/day in the Young Finns study, and 4.8 in the MDCS (a replication cohort). Total dairy intake ranged from 1.3 to 4.2 servings/day in discovery cohorts, and 2.4 to 5.8 servings/day in replication cohorts.
3.2. Genome-Wide Interactions with Dairy Intake for the Outcome of BMI
Meta-analyzed genome-wide SNP × dairy interactions that reached the selected significance threshold are shown (<10−7; N = 26 SNPs; Table 2). The interaction regression coefficients indicate the difference in the association of dairy intake and BMI in the context of the effect allele. For each SNP × dairy interaction in Table 2, the direction of the regression coefficient is indicated for each of the nine cohorts. Taking the SNP rs9635058 on chromosome 13 as an example, for each additional serving of low-fat dairy, BMI was 0.2246 kgm−2 lower (p interaction=7.36×10−8) per each additional copy of the effect allele (A). Meta-analysis heterogeneity was quantified as I2, and was 0 for rs9635058, indicating no heterogeneity.
We also conducted random effects meta-analysis for the 26 SNPs (Supporting Information Table 6). Of note, the regression coefficient and standard error for rs96350658 (described above, with 0 heterogeneity), were virtually the same for both random effects as for fixed effects. In general, for SNPs that showed moderate to high heterogeneity, random effects meta-analyses yielded somewhat less significant p for interaction. Most SNPs remained at least nominally significant. For the single SNP rs7961195 (and for all linked SNPs in or near PPP1R12A), the p for interaction became completely non-significant with random effects meta-analysis (p interaction = 0.402 for rs7961195).
Manhattan plots are provided for low-fat, high-fat and total dairy interactions for Models 1 and 2 (Supporting Information Figures 1–6). The 26 SNPs are located in or closest to the following genes: LINC01512, AKAP2/PALM2-AKAP2, ACTA2, PPP1R12A, LINC00333, and AC098847.1.
3.3. Evaluation of Replication of Six Independent SNPs
For meta-analytic evaluation of replication, we selected 6 independent (r2<0.8) SNPs (rs7751666, rs914359, rs1388, rs7961195, rs9635058, rs1791355) from the 26 SNPs that reached the selected P-interaction threshold (<10−7) in discovery (Table 2). Criteria for choosing the 6 SNPs from the 26 SNPs are described in Methods. The most significant SNP × dairy intake interaction is shown (Table 3) with one SNP (rs9635058) reaching the replication significance threshold of 0.008. We hypothesized that the direction of the meta-analyzed replication beta would be the same as the meta-analyzed discovery beta, so one-sided replication P values were compared to the replication significance threshold. For SNP rs9635058 (closest gene LINC00333), for each reported serving of high-fat dairy, BMI was 0.053 kg m−2 lower (one-sided interaction p = 0.004) per one additional copy of the effect allele (A), with low heterogeneity (I2 = 7.8) and MAF~0.15. A second SNP, rs1388 (closest gene ACTA2; MAF 0.01), approached significance such that for each reported serving of low-fat dairy, BMI was 0.592 kg m−2 lower (one-sided interaction p = 0.032; SE = 0.32) per one additional copy of the effect allele (A), with heterogeneity I2 of 0 in replication. Interaction p values for the four remaining SNPs were not significant (Table 3).
3.4. Consistency of Low-Fat Dairy x rs9635058 Interaction for BMI in Discovery Cohorts
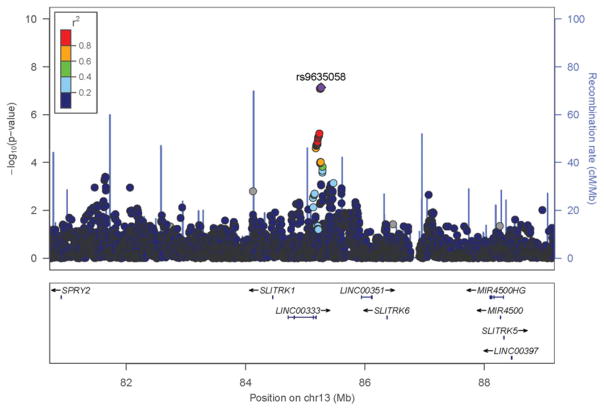
Following validation of the discovery SNP rs963058 in replication, we generated a forest plot to illustrate the direction, magnitude and consistency of interaction among discovery cohorts (Figure 1). With the exception of the InCHIANTI study (the group with lowest low-fat dairy (0.06 servings/day) and total dairy (1.3 servings/day), the direction of the low-fat dairy interaction regression coefficient was negative in all groups and the P-interaction was <0.2 for 7 out of 9 cohorts and < 0.1 for 6 of those. Of note, low-fat intake in CHS was also small (0.57 servings/day), and the P-interaction was similarly insignificant in CHS (p-interaction >0.4). A regional plot with rs963058 indicated as an index SNP, illustrates its position on chromosome 13, relative to LINC00333 (Figure 2).[27]
Figure 1.
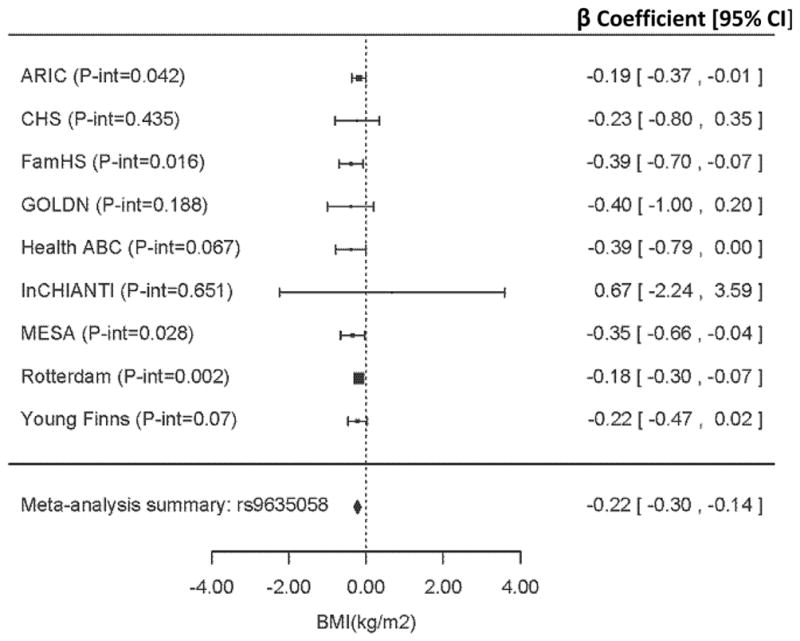
Meta-analysis of the interaction between rs9635058 and low-fat dairy intake in the nine discovery cohorts. Forest plot shows adjusted regression coefficients (β [95% CI] representing expected change in body mass index (kg m−2) per one-daily-serving–greater low-fat dairy intake per one additional copy of the effect allele (A). Data are adjusted for model 1 covariates: age, sex, energy intake, field center, family relationships and population structure (as needed). Abbreviations: ARIC, Atherosclerosis Risk in Communities; CHS; Cardiovascular Health Study; FAMHS, Family Heart Study; GOLDN, Genetics of Lipid Lowering Drugs and Diet Network; Health ABC, Health, Aging, and Body Composition; InCHIANTI, Invecchiare in Chianti, aging in the Chianti area; MESA, Multi-Ethnic Study of Atherosclerosis.
Figure 2.
Regional association plots for SNPs near LINC00333 on chromosome 13. The figure shows −log10 P values for SNPs from the discovery meta-analysis of low-fat dairy interactions. The SNPs shown are within 4000 kb of the index SNP: rs9635058. LD is indicated in color in relation to the highlighted marker. The scheme is red for strong LD, orange for moderately strong LD, green for moderate LD and blue for weak LD. Abbreviations: chr, chromosome; cM, centimorgan; LD, linkage disequilibrium; Mb, mega base pairs; SNP, single-nucleotide polymorphism.
3.5. Cohort-Specific Forest Plots of Dairy × SNP Interactions for Independent SNPs in Discovery and Replication Cohorts
We also generated forest plots to illustrate the direction and magnitude of interactions for the five independent SNPs that reached the specified significance threshold among 9 discovery cohorts but were not replicated (Supporting Information Figures 8–12). For several SNPs (rs7751666, rs1791355, rs1388, rs91439) the regression coefficients for either the Family Heart Study or the GOLDN Study, or both, differed in direction from the other cohorts. For the SNP rs7961195, the Rotterdam Study appears to be the primary driver of the significant interaction. Similarly, we generated forest plots for the six independent SNPs based on data from the four replication cohorts (Supporting Information Figures 13–18). Again, overall patterns are difficult to establish, but differences in the direction and magnitude were evident, reflecting cohort-specific variability in responses to dairy.
3.6. Sensitivity Analyses for Five Independent SNPs that Did Not Achieve Significance for Replication
We conducted sensitivity analyses to investigate potential sources of variability among the discovery cohorts (Supporting Information Table 5). Stratified analysis with countries divided by geographic region (Europe, n = 3 cohorts; USA, n = 6 cohorts) showed that for 4/5 SNPs (all except rs7961195), regression coefficient directions were consistent (all positive or all negative) in all European (but not USA) cohorts. Stratified analysis with countries divided into older/younger according to mean cohort age (younger, age<65, n = 4 cohorts; older, age ≥65 years, n = 5 cohorts) similarly showed that for the same 4/5 SNPs (all except rs7961195), the sign of the regression coefficient was either all positive or all negative in the older (but not the younger) cohorts.
3.7. Comparative Minor Allele Frequencies (MAF) for Six SNPs Selected for Evaluation of Replication, Implications for Positive Selection
One premise underlying our analyses is that while the LCT locus is the strongest recent positive selection signal in the human genome, other loci, including those related to body weight, may have undergone selection. Among the six SNPs identified in the discovery cohorts, based on integrated haplotype score (iHS) analysis in the 1000 Genomes Selection Browser (http://hsb.upf.edu/),[28,29] two SNPs: rs7961195 and rs9635058 appear to be subject to positive selection in European ancestry, with iHS of 2.47 and 3.02, respectively. In addition, the wide spectrum of MAF observed across populations further supports the likelihood of positive selection. Variation is shown in a graph of MAF for seven 1000 Genomes populations for each of the six replication SNPs, as compared to two other common SNPs: (i) the best-studied CEU lactase persistence SNP (MCM6 rs4988235) and (ii) a well-established obesity SNP (FTO rs9939609) (Supporting Information Figure 7). The figure shows wide-ranging MAF for several of the investigated SNPs and the common SNPs: from close to 0 to approximately 0.4 for 3 of the 6 investigated SNPs, from 0 to 0.8 for the MCM6 SNP, and from approximately 0.1 to>0.5 for the FTO SNP.
4. Discussion
On the basis of the strong positive selection signal at the LCT locus, and the considerable inter-individual variability in response to dairy foods, we conducted a GWIS to identify genetic loci that may modulate the relationship between dairy intake and BMI. Of six loci identified as potential interactors in the discovery phase, one locus closest to the gene LINC00333 (long intergenic non-protein coding RNA 333) was confirmed through evaluation of replication. The MAF of 0.15 indicates a common SNP, with approximately 28% of the population carrying at least 1 copy of the minor allele). The overall consistency in direction and considerable magnitude of the rs9635058 interaction effect (Figure 1, forest plot, regression coefficient = 0.22) and low meta-analytic heterogeneity across discovery cohorts increases confidence in its plausibility as a responder to dairy intake. These findings provide evidence that dairy intake may protect certain genetic subgroups from greater body weight.
The intergenic location of the interacting variants challenges mechanistic understanding of how the variants could be affecting body weight. The replicated SNP (rs9635058) falls within a large linkage disequilibrium (LD) block and is located 84 kb downstream of LINC00333 and 481 kb upstream of SNORA107, two novel and uncharacterized non-coding RNAs. As reported in the results section, the iHS score for rs9635058 suggests that this SNP may have recently undergone positive selection in Europeans. Several SNPs within this LD block have extreme iHS values. The highest is 3.19 for the SNP rs9575649 (r2 = 0.96 with rs9635058). While available epigenomic data do not support a functional role for the sequence surrounding rs9575649, it does map within a mammalian-wide interspersed repeat (MIR) sequence, an ancient form of transposable element, based on the GENCODE project data.[30] Interestingly, MIRs have been implicated in both enhancer and suppressor activity in the human genome (https://mobilednajournal.biomedcentral.com/articles/10.1186/1759-8753-5-14).
In the same LD block, the iHS of 2.56 for rs9575642 (a proxy for rs9635058 with r2 = 0.95) also reflected an outlier. Further investigation of the proxy rs9575642 using the Epigenome Road Map mapped it to several sites of chromatin modification (for enhancers) and DNase hypersensitivity sites in several of the same tissues. Specifically, DNAse hypersensitivity sites to which rs9575642 maps include stomach, pancreas, and small intestine tissues (Roadmap Epigenomics Consortium, 2015). Chromatin modification sites to which rs9575642 maps include not only gastrointestinal (liver, stomach, duodenum, pancreas, colon) but also skeletal muscle and adipose, which are constituents of lean body mass and fat mass.[31] However, given the relatively large number of SNPs within this region, particularly in this LD block, it is impossible to determine which SNPs may contribute in one or more tissues to the observed interaction modulating body weight. The in silico evidence for this locus supports a potential regulatory role in processes related to digestion and absorption of nutrients, and body size and body composition, but does not reveal which constituents (e.g., lactose, fat, protein, oligosaccharides, minerals or other bioactive components) or properties (such as fermented versus non-fermented) of dairy foods could be physiologically relevant.
Our evaluation of low-fat and high-fat as well as total dairy was based on previous studies that report differential health impacts for dairy foods with different proportions of fat.[6,32,33] In the current study, the form of dairy that modulated the LINC00333 variant’s association with BMI was low-fat in the discovery cohorts and high-fat in the replication cohorts. These differences in exposures may be related to actual differences in patterns of intake across groups (e.g., the proportion of dairy consumed in high-fat versus low-fat forms in the discovery versus replication cohorts), or may be related to imprecision in the measurement and categorization of dairy intake as estimated by FFQs. In any case, we are unable to conclude whether the fat content of the dairy foods, or the form (e.g., milk, cheese, yogurt) is relevant to the observed interaction near the LINC00333 locus.
In addition, while only the LINC00333 locus was replicated, several other discovery loci are of potential interest based on functions of the proteins they encode (e.g., ACTA2 (Actin, Alpha 2, Smooth Muscle, Aorta), PPP1R12A (Protein Phosphatase 1 Regulatory Subunit (a subunit of myosin phosphatase), and PALM2/AKAP2 (where AKAP2 (A kinase anchoring protein 2) is associated with actin). These loci all share functional links to actin and myosin, the proteins that comprise the molecular motor unit that accomplishes muscle contraction in vertebrates. Interestingly, muscle is not only a constituent of the lean body mass that would have been crucial for survival at the time of selection for lactase variants, but muscle protein synthesis has also been reported to be particularly responsive to whey proteins, and whey proteins are uniquely supplied by dairy foods.[34,35]
The study exhibits strengths and limitations. Important strengths include its novelty, large meta-analyzed sample, multinational cohorts and the implementation of a single analytic plan with harmonized dietary data. Moreover, the magnitude of the gene-diet interaction suggests that these findings may be relevant to understanding of body weight regulation. Limitations must also be considered. The most important limitation is the lack of functional knowledge for the replicated SNP and the closest gene (LINC00333). Another intergenic SNP (rs9546711) located closest to LINC00333 was implicated in coronary artery disease in a large scale GWAS of African Americans, but functional knowledge is lacking in that case as well.[36] Laboratory studies are needed to address these gaps in understanding, and provide understanding of how a particular food group modifies the genetic risk of obesity.
Other important challenges to gene x diet interaction studies arise from the complications of quantifying and categorizing dietary exposures. Imprecise measurement of dietary intakes can obscure interactions.[37] Moreover, true differences in dairy consumption and the food supplies across geographic regions and life stages may further complicate the detection of differences. Forest plots illustrated variability in genetic modulation of response to dairy, even within a single country (USA). Regression cohorts for two USA cohorts (Family Heart Study and GOLDN) differed in direction from other cohorts for several SNPs. Both of these studies are family-based, and include centers across a wide range of US regions (Minnesota, Utah, Massachusetts and North Carolina). Either the family structures, or the variability in regional field centers, could have contributed to differential responses in these studies. To improve understanding of the sources of variability in response to dairy, we conducted sensitivity analyses with cohorts divided by geographic region and mean age. These analyses were revealing, in that greater consistency was observed in European (but not American) cohorts and in older individuals (but not in younger). Dietary patterns, including dairy intakes, might be more similar among European groups and among older individuals. These observations could be used to inform the choice of cohorts for future investigation of gene x diet interactions related to dairy intake.
In summary, we performed a GWIS for dairy intake for BMI and identified gene × diet interactions in nine meta-analyzed cohorts, one of which was replicated in meta-analysis of four independent cohorts. Although biological understanding of this intergenic locus near the LINC00333 gene is lacking, in silico epigenetic evidence is consistent with a possible role of these variants in digestion, absorption or transport of nutrients, and body composition. Future studies of additional phenotypes, including adiposity and glucose metabolism, may extend and refine understanding of metabolic variability in dairy responses, and improve strategies for identifying genetic subsets of individuals who may benefit most from greater dairy intake.
Supplementary Material
Acknowledgments
Complete funding information is located in the Supporting Information, Table 8. B. P. serves on the DSMB of a clinical trial funded by the manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. M. N.’ participation is supported by a consulting contract between Kelly Services and the National Institute on Aging, NIH, Bethesda, MD, USA. Young Finns Study: The expert technical assistance in the statistical analyses by Irina Lisinen and Mika Helminen is gratefully acknowledged.
Abbreviations
- chr
chromosome
- cM
centimorgan
- LD
linkage disequilibrium
- Mb
mega base pair
- SNP
single-nucleotide polymorphism
Footnotes
Conflict of Interest
The authors have declared no conflict of interest.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Dr. Caren E. Smith, Nutrition and Genomics Laboratory, Jean Mayer-US Department of Agriculture Human Nutrition Research, Center on Aging, Tufts University, Boston, MA
Dr. Jack L. Follis, University of St Thomas, Houston, TX, USA
Hassan S. Dashti, Center for Human Genetic Research, Massachusetts General Hospital, Boston, MA, USA. Program in Medical and Population Genetics, Broad Institute, Cambridge, MA, USA
Dr. Toshiko Tanaka, Translational Gerontology Branch, National Institute on Aging, Baltimore, MD, USA
Dr. Mariaelisa Graff, Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA
Dr. Amanda M. Fretts, Department of Epidemiology, University of Washington, Seattle, WA, USA
Dr. Tuomas O. Kilpeläinen, Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2100, Denmark
Dr. Mary K. Wojczynski, Department of Genetics, Washington University School of Medicine, St. Louis, MO, USA
Dr. Kris Richardson, Nutrition and Genomics Laboratory, Jean Mayer-USDA Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA
Dr. Mike A. Nalls, Laboratory of Neurogenetics, National Institute on Aging, Bethesda, MD, USA. Contractor/consultant with Kelly Services, Rockville, MD, USA
Dr. Christina-Alexandra Schulz, LUDC, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden
Dr. Yongmei Liu, Department of Epidemiology & Prevention, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, NC, USA
Dr. Alexis C. Frazier-Wood, USDA / ARS Children’s Nutrition Research Center, Baylor College of Medicine, Houston, Texas, USA
Dr. Esther van Eekelen, Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands
Carol Wang, School of Women’s and Infants’ Health, University of Western Australia, Perth, Australia.
Dr. Paul S. de Vries, Human Genetics Center, University of Texas Health Science Center at Houston, Houston, TX, USA. Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands
Dr. Vera Mikkilä, Division of Nutrition, Department of Food and Environmental Sciences, University of Helsinki, Helsinki. Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland
Dr. Rebecca Rohde, Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA
Dr. Bruce M. Psaty, Departments of Medicine, Epidemiology and Health Services, University of Washington, Seattle, WA, USA. Group Health Research Institute, Group Health Cooperative, Seattle, WA, USA
Dr. Torben Hansen, Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2100, Denmark
Dr. Mary F. Feitosa, Department of Genetics, Washington University School of Medicine, St. Louis, MO, USA
Dr. Chao-Qiang Lai, USDA ARS, Nutrition and Genomics Laboratory, Jean Mayer-USDA Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA
Dr. Denise K. Houston, Department of Internal Medicine, Wake Forest School of Medicine, Wake Forest University, Winston-Salem, NC, USA
Dr. Luigi Ferruci, Translational Gerontology Branch, National Institute on Aging, Baltimore, MD, USA
Dr. Ulrika Ericson, LUDC, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden
Dr. Zhe Wang, Human Genetics Center, School of Public Health, University of Texas Health Science Center at Houston, Houston, Texas, USA
Dr. Renée de Mutsert, Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands
Dr. Wendy H. Oddy, Menzies Institute for Medical Research, University of Tasmania, Australia
Dr. Ester A. L. de Jonge, Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands. Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands
Dr. Ilkka Seppälä, Department of Clinical Chemistry, Fimlab Laboratories, Tampere University School of Medicine, Tampere, Finland
Dr. Anne E. Justice, Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA
Dr. Rozenn N. Lemaitre, Department of Medicine, University of Washington, Seattle, WA, USA
Dr. Thorkild I. A. Sørensen, Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2100, Denmark. Department of Clinical Epidemiology (formerly Institute of Preventive Medicine), Bispebjerg and Frederiksberg Hospitals, The Capital Region, Copenhagen 2000, Denmark. MRC Integrative Epidemiology Unit & School of Social and community Medicine, University of Bristol, Bristol, BS82BN, UK
Dr. Michael A. Province, Department of Genetics, Washington University School of Medicine, St. Louis, MO, USA
Laurence D. Parnell, USDA ARS, Nutrition and Genomics Laboratory, Jean Mayer-USDA Human Nutrition Research Center on Aging, Tufts University, Boston, MA, USA
Dr. Melissa E. Garcia, National Institute of Aging, Bethesda, MD, USA
Dr. Stefania Bandinelli, Geriatric Unit, Azienda Sanitaria Firenze (ASF), Florence, Italy
Dr. Marju Orho-Melander, LUDC, Department of Clinical Sciences Malmö, Lund University, Malmö, Sweden
Dr. Stephen S. Rich, Center for Public Health Genomics, University of Virginia, Charlottesville, VA, USA
Dr. Frits R. Rosendaal, Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands
Dr. Craig E. Pennell, School of Women’s and Infants’ Health, University of Western Australia, Perth, Australia
Dr. Jessica C. Kiefte-de Jong, Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands
Dr. Mika Kähönen, Department of Clinical Physiology, University of Tampere and Tampere University Hospital, Tampere, Finland
Dr. Kristin L. Young, Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA
Oluf Pedersen, Novo Nordisk Foundation Center for Basic Metabolic Research, Section of Metabolic Genetics, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen 2100, Denmark.
Dr. Stella Aslibekyan, Department of Epidemiology, University of Alabama at Birmingham, Birmingham, AL, USA
Dr. Jerome I. Rotter, Institute for Translational Genomics and Population Sciences Los Angeles BioMedical Research Institute at Harbor-UCLA Medical Center, Torrance, CA, USA
Dr. Dennis O. Mook-Kanamori, Department of Clinical Epidemiology, Leiden University Medical Center, Leiden, The Netherlands. Department of Public Health and Primary Care, Leiden University Medical Center, Leiden, The Netherlands
Dr. M. Carola Zillikens, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands
Dr. Olli T. Raitakari, Research Centre of Applied and Preventive Cardiovascular Medicine, University of Turku, Turku, Finland. Department of Clinical Physiology and Nuclear Medicine, University of Turku, Turku, Finland
Dr. Kari E. North, Department of Epidemiology, University of North Carolina, Chapel Hill, North Carolina, USA. Carolina Center for Genome Sciences, University of North Carolina, Chapel Hill, NC, 27514, USA
Dr. Kim Overvad, Department of Public Health, Section for Epidemiology, Aarhus University, DK-8000 Aarhus C, Denmark. Aalborg University Hospital, DK-9000 Aalborg, Denmark
Dr. Donna K. Arnett, College of Public Health, University of Kentucky, Lexington, KY, USA
Dr. Albert Hofman, Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands. Department of Nutrition, Harvard School of Public Health, Boston, USA
Dr. Terho Lehtimäki, Department of Clinical Chemistry, Fimlab Laboratories, Tampere University School of Medicine, Tampere, Finland
Dr. Anne Tjønneland, Danish Cancer Society Research Center, Copenhagen 2100, Denmark
André G. Uitterlinden, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands
Dr. Fernando Rivadeneira, Department of Internal Medicine, Erasmus MC, University Medical Center, Rotterdam, the Netherlands. Netherlands Genomics Initiative (NGI)-sponsored Netherlands Consortium for Healthy Aging (NCHA), Leiden, the Netherlands
Oscar H. Franco, Department of Epidemiology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands
Dr. J. Bruce German, Department of Food Science and Technology, University of California, Davis, CA, USA
Dr. David S. Siscovick, New York Academy of Medicine, New York, NY, USA
Dr. L. Adrienne Cupples, Department of Biostatistics, Boston University School of Public Health, Boston, USA
Prof. Dr. José M. Ordovás, Nutrition and Genomics Laboratory, Jean Mayer-US Department of Agriculture Human Nutrition Research, Center on Aging, Tufts University, Boston, MAThe Department of Epidemiology and Population Genetics, Centro Nacional Investigación Cardiovasculares (CNIC) Madrid, Spain, IMDEA Food, Madrid, Spain
References
- 1.Gunther CW, Legowski PA, Lyle RM, McCabe GP, Eagan MS, Peacock M, Teegarden D. Am J Clin Nutr. 2005;81:751. doi: 10.1093/ajcn/81.4.754. [DOI] [PubMed] [Google Scholar]
- 2.Beydoun MA, Gary TL, Caballero BH, Lawrence RS, Cheskin LJ, Wang Y. Am J Clin Nutr. 2008;187:1914. doi: 10.1093/ajcn/87.6.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dove ER, Hodgson JM, Puddey IB, Beilin LJ, Lee YP, Mori TA. Am J Clin Nutr. 2009;90:70. doi: 10.3945/ajcn.2008.27411. [DOI] [PubMed] [Google Scholar]
- 4.Romaguera D, Ängquist L, Du H, Jakobsen MU, Forouhi NG, Halkjaer J, Feskens EJ, van der ADL, Masala G, Steffen A, Palli D, Wareham NJ, Overvad K, Tjonneland A, Boeing H, Riboli E, Sorensen TI. PLoS One. 2011;6:e23384. doi: 10.1371/journal.pone.0023384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abargouei AS, Janghorbani M, Salehi-Marzijarani M, Esmaillzadeh A. Int J Obes. 2012;36:1485. doi: 10.1038/ijo.2011.269. [DOI] [PubMed] [Google Scholar]
- 6.Arnberg K, Molgaard C, Michaelsen KF, Jensen SM, Trolle E, Larnkjaer A. J Nutr. 2012;142:2083. doi: 10.3945/jn.112.161208. [DOI] [PubMed] [Google Scholar]
- 7.Chen M, Pan A, Malik VS, Hu FB. Am J Clin Nutr. 2012;96:735. doi: 10.3945/ajcn.112.037119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwingshackl L, Hoffmann G, Schwedhelm C, Kalle-Uhlmann T, Missbach B, Knuppel S, Boeing H. PLoS One. 2016;11:e0157461. doi: 10.1371/journal.pone.0157461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Am J Hum Genet. 2004;74:1111. doi: 10.1086/421051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, Ibrahim M, Omar SA, Lema G, Nyambo TB, Ghori J, Bumpstead S, Pritchard JK, Wray GA, Deloukas P. Nat Genet. 2007;39:31. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corella D, Arregui M, Coltell O, Portolés O, Guillem-Saiz P, Carrasco P, Sorli JV, Ortega-Azorin C, Gonzalez JI, Ordovas JM. Obesity. 2011;19:1707. doi: 10.1038/oby.2010.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dedoussis GV, Louizou E, Papoutsakis C, Skenderi KP, Yannakoulia M. Eur J Clin Nutr. 2010;64:253. doi: 10.1038/ejcn.2009.124. [DOI] [PubMed] [Google Scholar]
- 13.Smith CE, Tucker KL, Arnett DK, Noel SE, Corella D, Borecki IB, Feitosa MF, Aslibekyan S, Parnell LD, Lai CQ, Lee YC, Ordovas JM. J Nutr. 2013;143:1865. doi: 10.3945/jn.113.179051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamri A, Poli A, Emery N, Bellili N, Velho G, Lantieri O, Balkau B, Marre M, Fumeron F. Metabolism. 2013;62:1323. doi: 10.1016/j.metabol.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Mozaffarian D, Cao H, King IB, Lemaitre RN, Song X, Siscovick DS, Hotamisligil GS. Ann Intern Med. 2010;153:790. doi: 10.1059/0003-4819-153-12-201012210-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu FB, Stampfer MJ, Manson JE, Ascherio A, Colditz GA, Speizer FE, Hennekens CH, Willett WC. Am J Clin Nutr. 1999;70:1001. doi: 10.1093/ajcn/70.6.1001. [DOI] [PubMed] [Google Scholar]
- 17.United States Department of Agriculture (USDA) food groups. [Accessed 26 July 2017];USDA ChooseMyPlate.gov: All about the dairy group. http://www.choosemyplate.gov/food-groups/dairy.htmL#.
- 18.Kilpeläinen TO, Qi L, Brage S, Sharp SJ, Sonestedt E, Demerath E, Ahmad T, Mora S, Kaakinen M, Sandholt CH, Holzapfel C, Autenrieth CS, Hypponen E, Cauchi S, He M, Kutalik Z, Kumari M, Stancakova A, Meidtner K, Balkau B, Tan JT, Mangino M, Timpson NJ, Song Y, Zillikens MC, Jablonski KA, Garcia ME, Johansson S, Bragg-Gresham JL, Wu Y, van Vliet-Ostaptchouk JV, Onland-Moret NC, Zimmermann E, Rivera NV, Tanaka T, Stringham HM, Silbernagel G, Kanoni S, Feitosa MF, Snitker S, Ruiz JR, Metter J, Larrad MT, Atalay M, Hakanen M, Amin N, Cavalcanti-Proenca C, Grontved A, Hallmans G, Jansson JO, Kuusisto J, Kahonen M, Lutsey PL, Nolan JJ, Palla L, Pedersen O, Perusse L, Renstrom F, Scott RA, Shungin D, Sovio U, Tammelin TH, Ronnemaa T, Lakka TA, Uusitupa M, Rios MS, Ferrucci L, Bouchard C, Meirhaeghe A, Fu M, Walker M, Borecki IB, Dedoussis GV, Fritsche A, Ohlsson C, Boehnke M, Bandinelli S, van Duijn CM, Ebrahim S, Lawlor DA, Gudnason V, Harris TB, Sorensen TI, Mohlke KL, Hofman A, Uitterlinden AG, Tuomilehto J, Lehtimaki T, Raitakari O, Isomaa B, Njolstad PR, Florez JC, Liu S, Ness A, Spector TD, Tai ES, Froguel P, Boeing H, Laakso M, Marmot M, Bergmann S, Power C, Khaw KT, Chasman D, Ridker P, Hansen T, Monda KL, Illig T, Jarvelin MR, Wareham NJ, Hu FB, Groop LC, Orho-Melander M, Ekelund U, Franks PW, Loos RJ, Lewis C, editors. PLoS Medicine. 2011;8:e1001116. doi: 10.1371/journal.pmed.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nettleton JA, Follis JL, Ngwa JS, Smith CE, Ahmad S, Tanaka T, Wojczynski MK, Voortman T, Lemaitre RN, Kristiansson K, Nuotio ML, Houston DK, Perala MM, Qi Q, Sonestedt E, Manichaikul A, Kanoni S, Ganna A, Mikkila V, North KE, Siscovick DS, Harald K, Mckeown NM, Johansson I, Rissanen H, Liu Y, Lahti J, Hu FB, Bandinelli S, Rukh G, Rich S, Booij L, Dmitriou M, Ax E, Raitakari O, Mukamal K, Mannisto S, Hallmans G, Jula A, Ericson U, Jacobs DR, Jr, Van Rooij FJ, Deloukas P, Sjogren P, Kahonen M, Djousse L, Perola M, Barroso I, Hofman A, Stirrups K, Viikari J, Uitterlinden AG, Kalafati IP, Franco OH, Mozaffarian D, Salomaa V, Borecki IB, Knekt P, Kritchevsky SB, Eriksson JG, Dedoussis GV, Qi L, Ferrucci L, Orho-Melander M, Zillikens MC, Ingelsson E, Lehtimaki T, Renstrom F, Cupples LA, Loos RJ, Franks PW. Hum Mol Genet. 2015;24:4728. doi: 10.1093/hmg/ddv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE, Luan J, Gustafsson S, Randall JC, Vedantam S, Workalemahu T, Kilpelainen TO, Scherag A, Esko T, Kutalik Z, Heid IM, Loos RJ. Nat Protoc. 2014;9:1192. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. Am J Hum Genet. 2005;77:685. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, Masson G, Jakobsdottir M, Thorlacius S, Helgason A, Aben KK, Strobbe LJ, Albers-Akkers MT, Swinkels DW, Henderson BE, Kolonel LN, Le Marchand L, Millastre E, Andres R, Godino J, Garcia-Prats MD, Polo E, Tres A, Mouy M, Saemundsdottir J, Backman VM, Gudmundsson L, Kristjansson K, Bergthorsson JT, Kostic J, Frigge ML, Geller F, Gudbjartsson D, Sigurdsson H, Jonsdottir T, Hrafnkelsson J, Johannsson J, Sveinsson T, Myrdal G, Grimsson HN, Jonsson T, von Holst S, Werelius B, Margolin S, Lindblom A, Mayordomo JI, Haiman CA, Kiemeney LA, Johannsson OT, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Nat Genet. 2007;39:865. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 23.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Nat Genet. 2007;39:989. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 24.Panagiotou OA, Ioannidis JP. Genome-Wide Significance Project. Int J Epidemiol. 2012;41:273–86. doi: 10.1093/ije/dyr178. [DOI] [PubMed] [Google Scholar]
- 25.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. BMJ. 2003;327:557. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kheradpour P, Kellis M. Nucl Acids Res. 2014;42:2976. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. Bioinformatics. 2010;26:2336. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voight BF, Kudaravalli S, Wen X, Pritchard JK. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pybus M, Dall’olio GM, Luisi P, Uzkudun M, Carreno-Torres A, Pavlidis P, Laayouni H, Bertranpetit J, Engelken J. Nucleic Acids Res. 2014;42:D903. doi: 10.1093/nar/gkt1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. Genome Res Cold Spring Harbor Lab. 2012;22:1760. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Nature. 2015;518:317. [Google Scholar]
- 32.Kratz M, Baars T, Guyenet S. Eur J Nutr. 2013;52:1. doi: 10.1007/s00394-012-0418-1. [DOI] [PubMed] [Google Scholar]
- 33.Poddar KH, Hosig KW, Nickols-Richardson SM, Anderson ES, Herbert WG, Duncan SE. J Am Diet Assoc. 2009;109:1433–8. doi: 10.1016/j.jada.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Hulmi JJ, Lockwood CM, Stout JR. Nutr Metab (Lond) 2010;7:51. doi: 10.1186/1743-7075-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips SM. Nutr Metab. 2016;13:64. doi: 10.1186/s12986-016-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lettre G, Palmer CD, Young T, Ejebe KG, Allayee H, Benjamin EJ, Bennett F, Bowden DW, Chakravarti A, Dreisbach A, Farlow DN, Folsom AR, Fornage M, Forrester T, Fox E, Haiman CA, Hartiala J, Harris TB, Hazen SL, Heckbert SR, Henderson BE, Hirschhorn JN, Keating BJ, Kritchevsky SB, Larkin E, Li M, Rudock ME, McKenzie CA, Meigs JB, Meng YA, Mosley TH, Newman AB, Newton-Cheh CH, Paltoo DN, Papanicolaou GJ, Patterson N, Post WS, Psaty BM, Qasim AN, Qu L, Rader DJ, Redline S, Reilly MP, Reiner AP, Rich SS, Rotter JI, Liu Y, Shrader P, Siscovick DS, Tang WH, Taylor HA, Tracy RP, Vasan RS, Waters KM, Wilks R, Wilson JG, Fabsitz RR, Gabriel SB, Kathiresan S, Boerwinkle E. PLos Genet. 2011;7:e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tucker KL, Smith CE, Lai CQ, Ordovas JM. Annu Rev Nutr. 2013;33:349. doi: 10.1146/annurev-nutr-072610-145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.