Abstract
Technical advances in Aspergillus nidulans enable relatively easy deletion of genomic sequences, insertion of sequences into the genome and alteration of genomic sequences. To extend the power of this system we wished to create strains with several selectable markers in a common genetic background to facilitate multiple, sequential transformations. We have developed an approach, using the recycling of the pyrG selectable marker, that has allowed us to create new deletions of the biA, pabaA, choA, and lysB genes. We have deleted these genes in a strain that carries the commonly used pyrG89, riboB2, and pyroA4 mutations as well as a deletion of the sterigmatocystin gene cluster and a deletion of the nkuA gene, which greatly reduces heterologous integration of transforming sequences. The new deletions are fully, easily and cheaply supplementable. We have created a strain that carries seven selectable markers as well as strains that carry subsets of these markers. We have identified the homologous genes from Aspergillus terreus, cloned them and used them as selectable markers to transform our new strains. The newly created strains transform well and the new deletion alleles appear to be complemented fully by the A. terreus genes. In addition, we have used deep sequencing data to determine the sequence alterations of the venerable and frequently used pyrG89, riboB2 and pyroA4 alleles and we have reannotated the choA gene.
Keywords: Aspergillus, gene targeting, transformation, deletions, selectable markers
1. Introduction
Progress in transformation methods and in production of transforming DNA fragments has greatly facilitated molecular genetic manipulations of Aspergillus nidulans (Nayak et al., 2006; Szewczyk et al., 2006; Oakley et al., 2012; Nødvig et al., 2015). These advances allow rapid, sequential molecular genetic manipulations that would have been impossible a few years ago. To extend the power of this system and facilitate complex molecular genetic manipulations [e.g. multiple promoter replacements in a single strain (Yeh et al., 2016)], it would be advantageous to have strains carrying several selectable markers as well as cloned genes that complement the markers.
Many selectable mutations have been isolated in A. nidulans by conventional mutagenesis and multiply marked strains have been created by crosses. However, many of these mutations were created by heavy mutagenesis (often more than 90% of the mutagenized conidia were killed). This almost certainly creates a heavy background of single nucleotide polymorphisms and may create mutations that are silent under most conditions but manifest themselves under particular conditions or interact synthetically with other mutations [e.g. (Oakley and Morris, 1981)]. In addition, UV mutagenesis, which was used extensively in the isolation of A. nidulans mutations, can result in chromosomal abnormalities. Strain G191, the ancestor of many strains that carry pyrG89, a mutation that is used extensively as a selectable marker for transformation in A. nidulans, carries duplications of portions of chromosomes I and VIII and a likely translocation of a portion of chromosome VIII to chromosome VI (our unpublished data). It is also advantageous to have complementing markers from species other than A nidulans because they allow selection but have sufficiently low nucleotide homology to the corresponding A nidulans selectable marker that Anidulans sequences can be used to target the selectable marker to the desired location in the genome (Nayak et al., 2006). Finally, it is useful for selectable markers to be deletions to minimize or eliminate reversion.
We have used a molecular genetic approach to create new selectable markers by deleting target genes. Using this approach, we have created strains with as many as seven selectable markers in a common genetic background. We have also amplified the corresponding homologs from Aspergillus terreus, shown that they complement the deletions and cloned them into a plasmid vector.
2. Materials and Methods
2.1 Media and supplements
Our minimal medium consisted of 6 g/L NaNO3, 0.52 g/L KCl, 0.52 g/L MgSO4 7H2O, 1.52 g/L KH2PO4, 10 g/L d-glucose, and 400 μl/L of a trace element solution (Vishniac and Santer, 1957), 15 g/L agar and appropriate nutrients to supplement nutritional markers carried by the strains. YG (5 g/L yeast extract, 20 g/L d-glucose, supplemented with 400 μl/L of the same trace element solution) was used as a liquid complete medium and YAG (YG plus 15 g/L agar) was used as a complete solid medium. To support growth of our newly created mutant strains we supplemented minimal media with the following, as required, (final concentrations are listed) p-aminobenzoic acid (PABA), 1.0 μg/mL; biotin 20 ng/mL; L-lysine, 200 μg/mL and choline, 20 μg/mL. Supplements were not required for YAG or YG media.
2.2 Molecular genetic manipulations and transformation
Construction of transforming linear DNA molecules by fusion PCR was as previously described (Szewczyk et al., 2006; Oakley et al., 2012). Phusion [New England Biolabs (M0536)], Q5 Hot Start High-Fidelity [New England Biolabs (M0494)] were used for gene amplification and fusion PCR. OneTaq [New England Biolabs (M0488)] was used for diagnostic PCR. DNA for diagnostic PCR was prepared from conidia using a miniprep procedure (Edgerton-Morgan and Oakley, 2012). Transformation was as previously described (Szewczyk et al., 2006; Oakley et al., 2012) except that 1.0 M sucrose was often used to osmotically balance selection plates instead of 0.6 M KCl. The protoplasting enzyme was VinoTaste Pro (Novo Nordisk) used at 100 mg/mL. Selection of transformants in which the AtpyrG gene had been evicted was on osmotically balanced YAG plates containing 1.0 mg/mL 5-fluoroorotic acid (5FOA) (Gold Biotechnology) as well as 1 g/L uracil and 10 mM uridine. Genomic DNA from A. terreus strain NIH2624 was supplied by Dr. Kenneth Bruno (Pacific Northwest National Laboratory). A. terreus pyrG, biA, lysB, pabaA, choA and riboB genes were amplified by PCR from genomic DNA and cloned into the pCR-BluntII-TOPO plasmid vector (Invitrogen) following the manufacturer's instructions. The resulting plasmids (Table 1) have been deposited at the Fungal Genetics Stock Center (www.fgsc.net) and Addgene (www.addgene.org). Multi-marker A. nidulans strains (listed in section 3.4) have been deposited at the Fungal Genetics Stock Center.
Table 1.
A. nidulans genes corresponding to previously identified nutritional markers, A. terreus homologs of these genes and plasmids carrying the cloned A. terreus genes.
| Aspergillus nidulans gene | Aspergillus terreus homolog | E value | Plasmid carrying A. terreus gene |
|---|---|---|---|
| pyrG (AN6157) | ATET_09675 | 2.0E-121 | pLO103 |
| biA (AN6644) | ATET_07102 | 0.0E+00 | pLO101 |
| lysB (AN5206) | ATET_03691 | 0.0E+00 | pLO99 |
| pabaA (AN6550) | ATET_06966 | 0.0E+00 | pLO100 |
| choA (AN2154) | ATET_02347 | 0.0E+00 | pLO102 |
| riboB (AN0670) | ATET_00531 | 3.0E-167 | pLO104 |
3. Results and Discussion
3.1 Identification of pyrG89, riboB2 and pyroA4 mutations
Our starting strain was LO4389 (Ahuja et al., 2012) which carries pyrG89, riboB2, pyroA4, a replacement of nkuA by argB, and a deletion of the sterigmatocystin gene cluster. We used a strain carrying at deletion of the sterigmatocystin gene cluster because sterigmatocystin is a potent toxin and carcinogen. Since we and others have used pyrG89, riboB2 and pyroA4 extensively as selective markers for transformation, we were interested in determining the nature of the mutations in these alleles. For other projects, we have deep sequenced strains carrying these mutations (Oakley et al., 2017) so we compared the sequences of the mutant alleles with the sequences of the same genes in the reference strain. With respect to pyrG89, we confirmed the finding of Dr. Miguel Peñalva (personal communication) that the mutation is a G to A transition at nucleotide 511 (with the A of the start codon being nucleotide 1) resulting in a substitution of glycine for serine. With respect to riboB2, we found that nucleotide 856 is deleted resulting in a frame shift and premature termination. With respect to pyroA4, we found that nucleotides 319-321 (GGA) are deleted and nucleotide 322 is changed from C to T. These changes result in the loss of a glycine and the replacement of a histidine with a tyrosine.
3.2 Creation of strains carrying deletions of selectable genes
In choosing genes to target for deletion, we looked for genes in which strongly selectable mutations had been isolated through classical genetics. In A. nidulans most such mutations confer nutritional requirements, and we specifically looked for genes in which classically isolated alleles largely or completely prevent growth on unsupplemented minimal medium and which are supplemented with relatively inexpensive nutrients. To find suitable candidates we relied on the list of A. nidulans gene list compiled by Dr. A. J. Clutterbuck (www.fgsc.net/Aspergillus/gene_list/loci.html, accessed Dec. 12, 2017) and the Aspergillus genome database (www.aspgd.org, accessed Dec. 12, 2017). We chose biA (AN6644, biotin requirement) (Roper, 1950; Clutterbuck, 1994; Magliano et al., 2011), lysB (AN5206, lysine requirement) (Käfer, 1958; Clutterbuck, 1994; Mogensen et al., 2006) (David et al., 2008), and pabaA (AN6550, p-aminobenzoic acid requirement) (Pontecorvo et al., 1953; Clutterbuck, 1994; Tüncher et al., 2005). In addition, Dr. A. J. Clutterbuck had tentatively identified AN2154 as choA (choline requirement) (Käfer, 1958; Arst, 1968) although it is annotated in the Aspergillus genome database as a possible pseudogene. We compared the AspGD AN2154 sequence to DNA sequences and RNAseq data we have obtained (Oakley et al., 2017) and found that there is a sequence error in the AspGD sequence, an extra G at nucleotide 2277 (with the A of the start codon being nucleotide 1). This error resulted in an erroneous early termination and a consequent mis-annotation of the gene in the current AspGD annotation. Curiously, the stop codon was correctly annotated in an earlier AspGD annotation. We have reannotated AN2154 and the revised sequence is given in Fig. S1. Our results (below) reveal, moreover, that AN2154 is a real gene and its deletion causes a choline requirement. Our results confirm, therefore, the identification of AN2154 as choA.
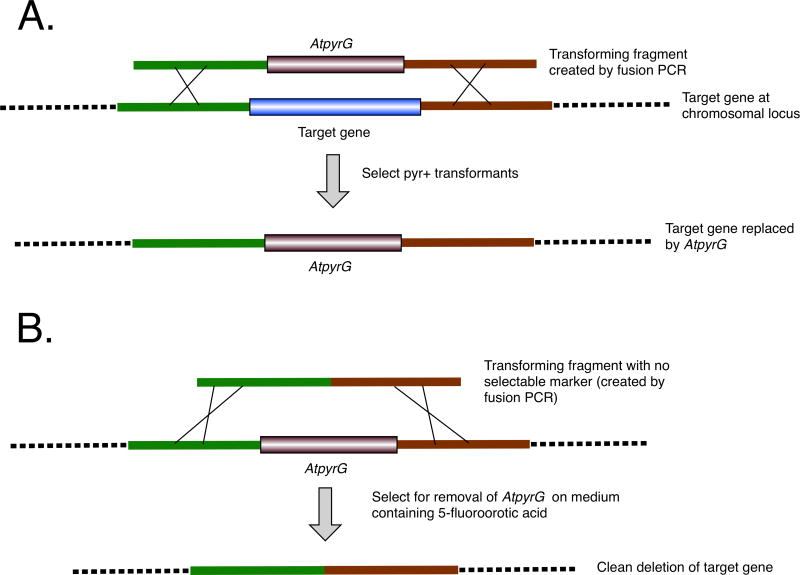
Our strategy for deleting target genes is shown in Fig. 1A. In this strategy, the target gene is initially deleted by replacement with the pyrG gene from A. terreus strain NIH 2624 (ATEG_09675 using the AspGD designation, herein designated AtpyrG for brevity). The transforming fragments were created by fusion PCR and consisted of a 1491 bp fragment from A. terreus containing the AtpyrG gene flanked by approximately 1 kb of A. nidulans genomic sequences that flank the target gene on each side. Transformants carrying a correct replacement of the target gene were verified by diagnostic PCR.
Fig. 1.
Gene deletion strategy. A. Transformation with a linear molecule containing the Aspergillus terreus pyrG gene (AtpyrG) flanked by left and right flanking sequences from the target gene to be deleted results in replacement of the target gene by AtpyrG. B. Transformation with a linear molecule containing only the left and right flanking sequences from the target gene results in the removal of the AtpyrG gene. Transformants in which AtpyrG has been removed can be selected on 5-fluoroorotic acid which selects against strains carrying a functional pyrG gene. The result is a deletion of the target gene with no selectable marker sequences left behind. The AtpyrG gene can be reused in subsequent transformations.
To remove AtpyrG and, thus, allow pyrG to be used as a selectable marker for subsequent transformations, a fragment was created by fusion PCR that contained only the flanking sequences fused together. This fragment was used to transform a strain in which the target gene had been replaced by AtpyrG (Fig. 1B). 5-fluoroorotic acid was used to select transformants in which the AtpyrG gene had been replaced (Dunne and Oakley, 1988). The net result was a deletion of the entire coding sequences of the target genes along with a small amount of flanking DNA on each side of the coding sequences. (The deleted regions are given in Table S1). This approach removed all of the AtpyrG sequences leaving clean deletions and allowed AtpyrG to be used again as a selectable marker.
Using this strategy, we created strain LO9478 by consecutive deletion of the biA and pabaA genes. In parallel, we created strain LO9487 by consecutive deletion of the lysB and choA genes. We next deleted the wA gene in LO9478 by replacing it with the A. fumigatus riboB gene (Nayak et al., 2006), thereby creating strain LO9548, with white conidia caused by the wA replacement being a useful color marker for crosses. We then crossed LO9548 with strain LO9487 and tested segregants for nutritional requirements. Among the segregants, we were able to obtain strains carrying all combinations of the newly created selectable markers as shown in Table 2. Notably, strain LO9771 carries all seven selectable markers as well as the nkuA deletion and a deletion of the sterigmatocystin gene cluster.
Table 2.
Strains carrying multiple selectable markers. Check marks indicate that the strain carries the mutation or deletion.
| Strains | Mutations carried | Deletions carried | |||||||
|---|---|---|---|---|---|---|---|---|---|
| pyrG89 | riboB2 | pyroA4 | Sterigmatocystin cluster (AN7804-7825) | biA (AN6644) | pabaA (AN6550) | lysB (AN5206) | choA (AN2154) | nkuA∷argB | |
| LO4389 | ✓ | ✓ | ✓ | ✓ | ✓ | ||||
| LO9368-LO9370 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| LO9451 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| LO9452 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| LO9478 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| LO9487-LO9489 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| LO9734-LO9736 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| LO9743 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| LO9749-LO9751 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |||
| LO9768 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| LO9770 | ✓ | ✓ (AfriboB∷w A) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| LO9771 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| LO9772-LO9774 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| LO10831 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
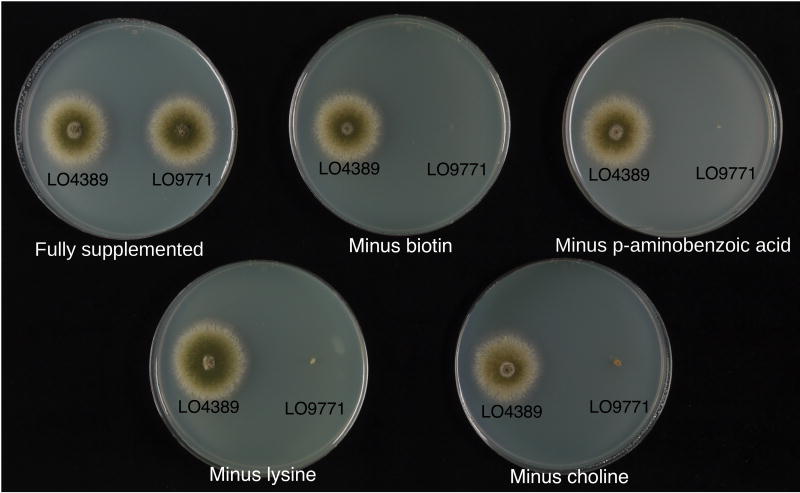
We next tested the “tightness” of the newly created selectable markers and the health of the strains on supplemented media. Results for LO9771, the seven-marker strain, are shown in Fig. 2. LO9771 grew as well as the parental strain LO4389 on fully supplemented minimal medium. It also grew as well as the parental and wild-type strains on complete medium (Fig. S1). Growth was strongly inhibited on medium lacking the appropriate supplement (Fig. 2). Interestingly, although the biA deletion completely inhibited growth, we note that the biA marker (our deletion or the classically isolated mutation, biA1) was particularly susceptible to cross feeding. If a biA+ colony was nearby, enough biotin was released by the biA+ strain to partially restore growth to the biotin requiring strain. This result is consistent with the fact that biA1 and the biA deletion are each supplemented fully with very small amounts of biotin (see Materials and Methods).
Fig. 2.
Newly created deletions are fully supplementable and strongly selectable. LO9771 carries seven selectable markers. When minimal medium is supplemented for all seven markers, growth is essentially the same as the parental strain LO4389. Selections against the four newly created deletions are shown. Omission of biotin selects against the biA deletion, omission of p-aminobenzoic acid selects against the pabaA deletion, omission of lysine selects against the lysB mutation and omission of choline selects against the choA deletion. In the absence of each of these supplements, the growth of the strain is strongly inhibited. For simplicity only the seven-marker strain is shown but similar results were obtained with strains carrying fewer markers.
3.3 Cloning and testing of A. terreus homologs of deleted A. nidulans genes
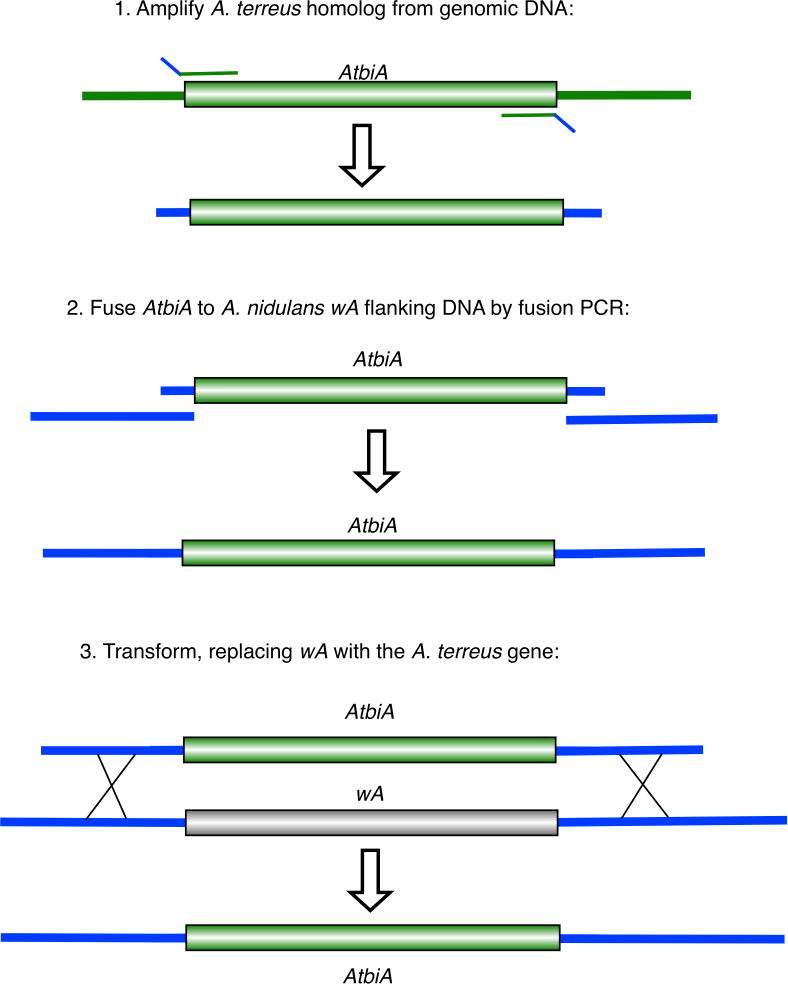
We identified and cloned genes from A. terreus strain NIH 2624 that complement the newly created deletions. We performed BLAST searches of the A. terreus genome using the sequences of the deleted A. nidulans genes as queries. A. terreus homologs of the deleted genes were easily identified (Table 1). We tested them for complementation of the A. nidulans deletions by amplifying them from genomic DNA, fusing them to flanking sequences from the wA gene and transforming appropriate strains (Fig. 3). Each of the A. terreus genes complemented the corresponding A. nidulans deletion fully (Fig. 4). Although we have not yet attempted to transform all of the strains we have created, we have encountered no difficulties with the strains we have attempted to transform (LO4389, LO9368, LO9451, LO9478, LO9487, LO9743, LO9770 and LO9771). We discovered quirks, however, in complementing the choA deletion by transformation. In attempting to select transformants on plates containing 1.0 M sucrose as an osmotic balancer, we found that the choA deletion strain grew too much to allow selection of transformants. We were able to select choA+ transformants on plates using 0.6 M KCl as an osmotic balancer, but the transformant colonies grew very slowly. These slow-growing colonies did form conidia, however, and when these conidia were streaked onto plates containing no osmotic balancer normal colonies were formed. The choA, marker, was thus quirky but perfectly usable.
Fig. 3.
Strategy for introducing A. terreus homologs of the deleted A. nidulans genes at the wA locus. The A. terreus biA gene (AtbiA) is shown as example. This strategy results in replacement of the wA gene with the A. terreus gene of interest. Correct transformant colonies will be white as shown in Figure 4.
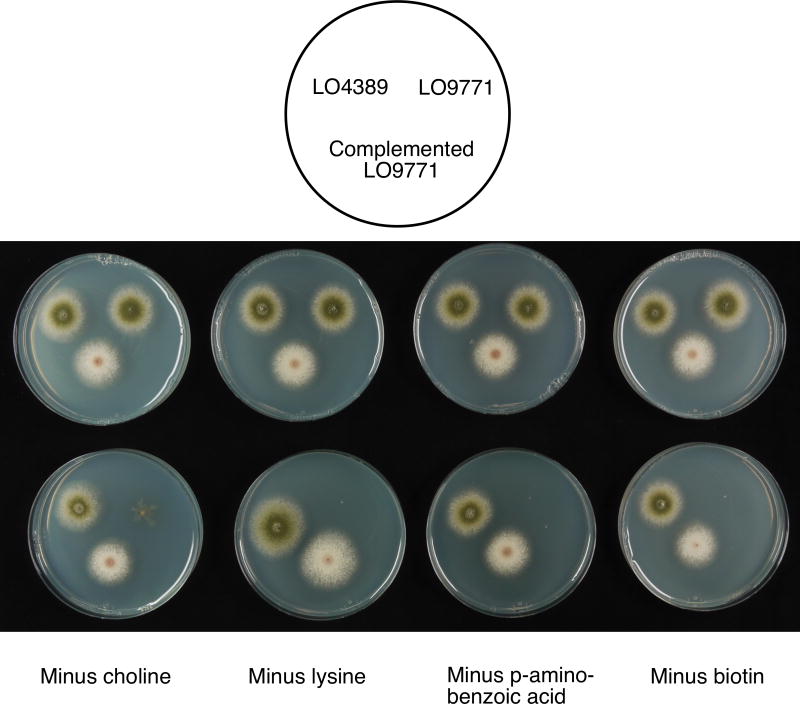
Fig. 4.
Complementation of newly created deletions with homologous A. terreus genes. The plates in the top row contain minimal medium fully supplemented for all seven nutritional requirements of LO9771. LO9771 grows as well as the parental strain LO4389. The bottom strain on each plate is LO9771 transformed with a fragment carrying an A. terreus gene homologous to one of the genes deleted in LO9771. The bottom row of plates contain minimal medium lacking a supplement required due to one of the newly created deletions. Thus, in the minus choline plate, LO9771 does not grow because of the choline requirement created by the choA deletion. However, the white strain at the bottom of the plate is LO9771 that has been transformed with the AtchoA gene (inserted at the wA locus), and it grows as well as the parental strains. Similarly, the AtlysB, AtpabaA, and AtbiA genes fully complement the corresponding deletions.
Finally, to create stable and easily accessible clones of the complementing genes, we ligated PCR fragments carrying each of the genes into the pCR-BluntII-TOPO vector (Table 1, sequences in Fig. S4-S7). To verify that each of the resultant plasmids carries a fully functional gene, we amplified the genes from each plasmid, created transforming molecules by fusion PCR, transformed appropriate deletion strains with them and verified that the deletions were fully complemented. We also cloned AtpyrG and the A. terreus riboB gene (AtriboB) into pCR-BluntII-TOPO (Table 1, sequences in Fig. S3 and S8).
3.4 Construction of a multi-marker pyrG+ strain
The pyrG selectable marker is extremely useful in that it affords both positive and negative selections. It has, however, two large liabilities. One is that it requires large amounts of uridine and uracil to complement it, resulting in considerable expense. A second is that it is not fully complemented by uridine and uracil at low temperatures, resulting in a weakly cold sensitive phenotype. We consequently created a multi-marker pyrG+ strain by simply amplifying a 2902 bp sequence containing the pyrG allele from A. nidulans wild-type DNA and transforming LO9771 with it, selecting for pyrimidine prototrophs. We designated this strain LO10831. Finally, to make these strains generally accessible to the A. nidulans research community, we have deposited the following strains at the Fungal Genetics Stock Center: LO4389, LO9368, LO9451, LO9452, LO9749, LO9478, LO9487, LO9734, LO9743, LO9755, LO9768, LO9770, LO9771, LO9772 and LO10831.
Supplementary Material
Highlights.
We have created four new selectable markers through targeted deletions.
We have determined the sequence changes in three frequently used mutants.
We have created strains of Aspergillus nidulans with up to seven selectable markers.
We have cloned Aspergillus terreus genes that complement each new selectable marker.
These advances will facilitate multiple, sequential transformations.
Acknowledgments
This work was supported by the National Institutes of Health (grant number P01GM084077), by US Department of Energy funds awarded through Pacific Northwest National Laboratory and by the Irving S. Johnson Fund of the Kansas University Endowment Association. Alexander Grubbs was supported by a Research Experiences for Undergraduate award from the National Science Foundation. We thank Dr. Kenneth Bruno (Pacific Northwest National Laboratory) for the A. terreus DNA. We thank Dr. Tomohiro Akashi (Nagoya University School of Medicine) for CAGE RNAseq data and Dr. Adrian Tsang (Concordia University) for assistance in reannotating choA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, Lo HC, Yeh HH, Oakley BR, Wang CCC. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–8221. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst HN. Genetic analysis of the first steps of sulphate metabolism in Aspergillus nidulans. Nature. 1968;219:268–270. doi: 10.1038/219268a0. [DOI] [PubMed] [Google Scholar]
- Clutterbuck AJ. Linkage map and locus list. Prog Ind Microbiol. 1994;29:791–824. [PubMed] [Google Scholar]
- David H, Ozcelik IS, Hofmann G, Nielsen J. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics. 2008;9:163. doi: 10.1186/1471-2164-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne PW, Oakley BR. Mitotic gene conversion, reciprocal recombination and gene replacement at the benA, beta-tubulin locus of Aspergillus nidulans. Mol Gen Genet. 1988;213:339–345. doi: 10.1007/BF00339600. [DOI] [PubMed] [Google Scholar]
- Edgerton-Morgan H, Oakley BR. γ-Tubulin plays a key role in inactivating APC/CCdh1 at the G1-S boundary. J Cell Biol. 2012;198:785–791. doi: 10.1083/jcb.201203115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käfer E. An 8-chromosome map of Aspergillus nidulans. Adv Genet. 1958;9:105–145. [PubMed] [Google Scholar]
- Magliano P, Flipphi M, Sanglard D, Poirier Y. Characterization of the Aspergillus nidulans biotin biosynthetic gene cluster and use of the bioDA gene as a new transformation marker. Fungal Genet Biol. 2011;48:208–215. doi: 10.1016/j.fgb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Mogensen J, Nielsen HB, Hofmann G, Nielsen J. Transcription analysis using high-density micro-arrays of Aspergillus nidulans wild-type and creA mutant during growth on glucose or ethanol. Fungal Genet Biol. 2006;43:593–603. doi: 10.1016/j.fgb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nødvig CS, Nielsen JB, Kogle ME, Mortensen UH. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS One. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A β-tubulin mutation in Aspergillus nidulans that blocks microtubule function without blocking assembly. Cell. 1981;24:837–845. doi: 10.1016/0092-8674(81)90109-4. [DOI] [PubMed] [Google Scholar]
- Oakley CE, Ahuja M, Sun WW, Entwistle R, Akashi T, Yaegashi J, Guo CJ, Cerqueira GC, Russo Wortman J, Wang CC, Chiang YM, Oakley BR. Discovery of McrA, a master regulator of Aspergillus secondary metabolism. Mol Microbiol. 2017;103:347–365. doi: 10.1111/mmi.13562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley CE, Edgerton-Morgan H, Oakley BR. Tools for manipulation of secondary metabolism pathways: rapid promoter replacements and gene deletions in Aspergillus nidulans. Methods Mol Biol. 2012;944:143–161. doi: 10.1007/978-1-62703-122-6_10. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons DW, Macdonald DK, Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Roper JA. Search for linkage between genes determining a vitamin requirement. Nature. 1950;166:956–957. doi: 10.1038/166956b0. [DOI] [PubMed] [Google Scholar]
- Szewczyk E, Nayak T, Oakley CE, Edgerton H, Xiong Y, Taheri-Talesh N, Osmani SA, Oakley BR. Fusion PCR and gene targeting in Aspergillus nidulans. Nat Protoc. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Tüncher A, Spröte P, Gehrke A, Brakhage AA. The CCAAT-binding complex of eukaryotes: evolution of a second NLS in the HapB subunit of the filamentous fungus Aspergillus nidulans despite functional conservation at the molecular level between yeast, A.nidulans and human. J Mol Biol. 2005;352:517–533. doi: 10.1016/j.jmb.2005.06.068. [DOI] [PubMed] [Google Scholar]
- Vishniac W, Santer M. The thiobacilli. Bacteriol Rev. 1957;21:195–213. doi: 10.1128/br.21.3.195-213.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh HH, Ahuja M, Chiang YM, Oakley CE, Moore S, Yoon O, Hajovsky H, Bok JW, Keller NP, Wang CC, Oakley BR. Resistance Gene-Guided Genome Mining: Serial Promoter Exchanges in Aspergillus nidulans Reveal the Biosynthetic Pathway for Fellutamide B, a Proteasome Inhibitor. ACS Chem Biol. 2016;11:2275–2284. doi: 10.1021/acschembio.6b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.