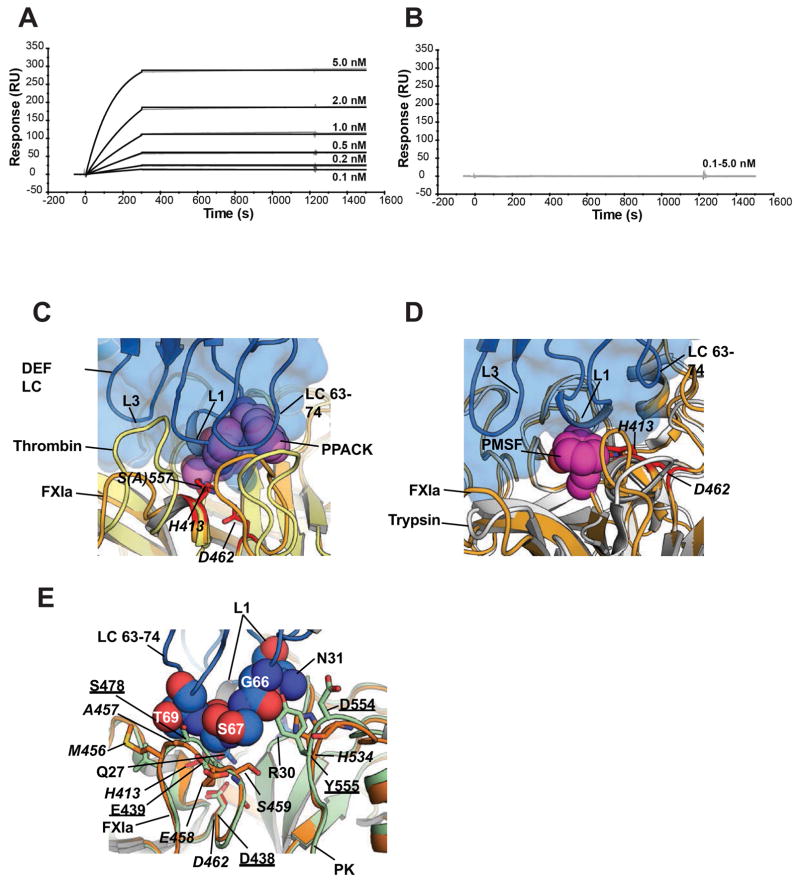
Figure 3. Structure predicts sensitivity of DEF binding to modification of the FXIa active site and specificity for FXIa over plasma kallikrein.
(A, B) Effect of PPACK on C24 Fab binding to FXIa was evaluated by surface plasmon resonance. For each experiment, an equivalent amount of FXIa (A) without and (B) with PPACK prior treatment was captured on two different Biacore chip flow cells. C24 Fab passed over for 3 minutes to allow binding and allowed to dissociate for a further 20 minutes.
(C, D) Structure comparison of the DEF-FXIaiPD complex with the (C) Thrombin-PPACK (1Z8I) and (D) Trypsin-PMSF (1PQA) complexes. DEF light chain is marine and shown as cartoon and surface. FXIa is light orange with the catalytic residues show as red sticks and labeled. Thrombin and Trypsin are yellow and white, respectively. PPACK and PMSF are shown as violet and magenta spheres, respectively. DEF light chain elements are labeled.
(E) DEF-FXIa/kallikrein structure comparison. DEF LC63-74 residues (space filling, marine) likely to clash with plasma kallikrein (PK) (light green, PDB: 4OGY). FXIa is light orange with amino acid labels in italics. Kallikrein amino acids are underlined.