Abstract
Introduction
Shiga toxin-producing Escherichia coli (STEC) is an emerging infectious pathogen which could lead to haemolytic uremic syndrome. Even though previous studies have compared the performance of CHROMagarTMSTEC to real-time polymerase chain reaction (PCR) in Europe, no study has been done to assess its performance on African isolates.
Objectives
This project aimed to validate and test an in-house-developed duplex real-time PCR and use it as a reference standard to determine the performance of CHROMagarTMSTEC on African isolates from diarrhoeic stool samples.
Methods
This study evaluated STEC diagnostic technology on African isolates. An in-house-developed duplex real-time PCR assay for detection of stx1 and stx2 was validated and tested on diarrhoeic stool samples and then used as a reference standard to assess the performance of CHROMagarTMSTEC. Real-time PCR was used to screen for stx in tryptic soy broth and the suspected STEC isolates, while conventional PCR was used to detect the other virulence genes possessed by the isolates.
Results
The real-time PCR limit of detection was 5.3 target copies/μL of broth. The mean melting temperature on melt-curve analysis for detection of stx1 was 58.2 °C and for stx2 was 65.3 °C. Of 226 specimens screened, real-time PCR detected stx in 14 specimens (6.2%, 95% confidence interval = 3.43% – 10.18%). The sensitivity, specificity, negative predictive value and positive predictive value of the CHROMagarTMSTEC were 33.3%, 77.4%, 95.3% and 11.3%.
Conclusions
The in-house developed real-time PCR assay is a sensitive and specific option for laboratory detection of STEC as compared to CHROMagarTMSTEC in this setting.
Introduction
Globally, food- and water-borne outbreaks of both O157 and non-O157 Shiga toxin-producing Escherichia coli (STEC) have been successfully detected due to the availability of good baseline data and effective active laboratory-based surveillance systems.1,2,3,4 Early detection of outbreaks is important to minimise morbidity, mortality and associated economic losses.5 There is a lack of good baseline data on STEC in Africa, which can be attributed to a lack of laboratory resources and the surveillance strategy employed. STEC has been implicated in outbreaks of bloody diarrhoea in sub-Saharan countries;6,7,8 however, these have been difficult to track and manage due to laboratory weakness.9,10 Furthermore, typical haemolytic uremic syndrome, which is overwhelmingly caused by STEC, was reported as the leading contributor to acute renal failure in paediatric patients at a South African academic hospital.11 Even though several studies have evaluated the performance of CHROMagarTMSTEC by comparison to molecular and antigen detection methods in developed countries,12,13 no study has so far evaluated its performance in Africa. This is necessary, especially given that there are geographical differences in characteristics of STEC that are dependent on the index of suspicion for the different STEC serotypes and on the availability of suitable laboratory methods to detect them.14
In many South African (and African) laboratories, stool specimens are not routinely tested for STEC, although physicians may request tests specific for E. coli serotype O157:H7, if it is clinically suspected. Testing is based on the non-sorbitol fermenting property using sorbitol MacConkey and only on request by a physician. This practice is of concern, is misleading and underestimates the real magnitude of STEC, since not all serotype O157 strains are non-sorbitol fermenting (O157: NM), and over 470 non-O157 serotypes have been attributed to clinical disease.15
Laboratory capacity for molecular detection is increasingly available in African countries and may, in some cases, be simpler than culture-based detection. This project, therefore, aimed to validate and test diarrhoeic stool samples by using an in-house developed duplex real-time polymerase chain reaction (PCR) and use it as a reference standard to determine the performance of CHROMagarTMSTEC on African isolates. The duplex assay was used to screen tryptic soy broth (TSB) for stx following overnight stool enrichment, and conventional PCR was used to screen for the other diarrhoeagenic E. coli virulence genes. Diarrhoeagenic E. coli were serotyped, and stx-positive isolates were tested for Shiga toxin production using immunochromatography.
Methods
Ethical approval
This study was approved by the ethics and research committee of the Faculty of Health Sciences at the University of Cape Town (HREC REF: 014/2015).
Study design
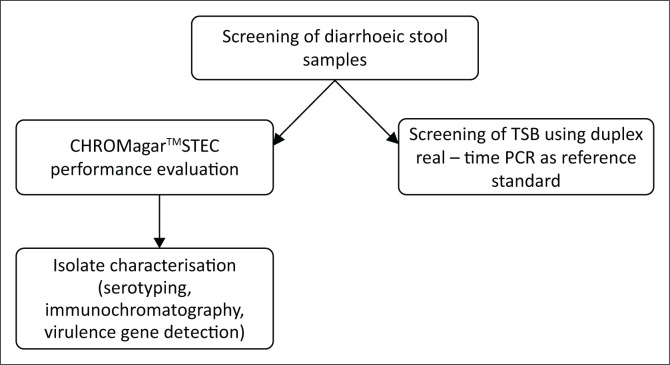
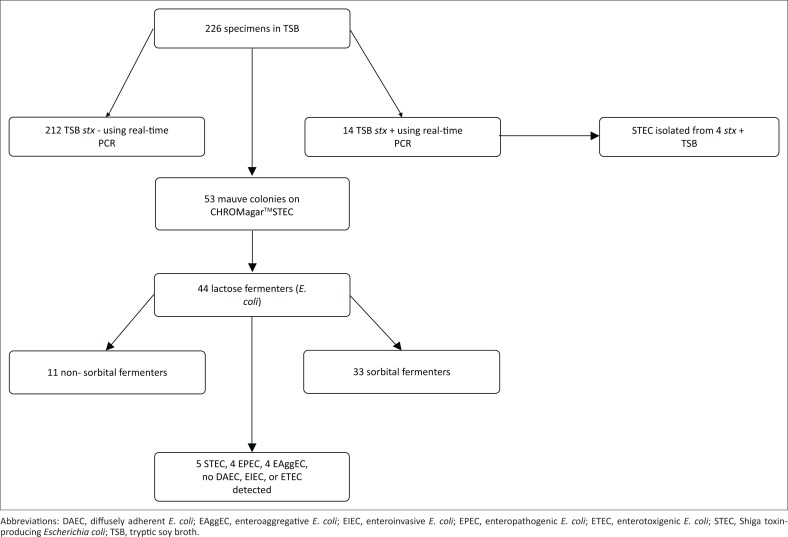
This study validated an in-house-developed duplex real-time PCR assay for detection of stx1 and stx2. The assay was then tested on diarrhoeic stool samples at a tertiary referral hospital and was used as a reference standard to assess the performance of a commercial chromogenic medium (CHROMagarTMSTEC) for STEC screening (Figure 1).
FIGURE 1.
Summary of methods used in this study.
Target plasmid preparation
The real-time PCR previously described by Grys et al.16 was used to amplify stx1 and stx2 gene targets from a STEC O157:H7 NCTC control strain (C4193-1) with both stx1 (subtype 1a) and stx2 (subtype 2a). PCR amplicon size was confirmed visually by agarose gel detection (~208 bp for stx1 and ~204 bp for stx2) before confirmation by sequencing using the Big Dye® Terminator v3.1 Cycle Sequencing Kit (Life Technologies Corporation, Carlsbad, California, United States). We used primers 1a and 2a (Table 1) for unidirectional Sanger sequencing of the amplicons. Resultant sequences were then trimmed and submitted for BLAST analysis against the NCBI database and confirming stx1 or stx2 target sequences in comparison to O157:H7 EDL933 (NCBI Reference: NC_002655.2).17 Purified amplicons (Mini Elute Gel extraction kit, Qiagen, Madrid, Spain) were cloned using CloneJet PCR cloning kit (Thermofisher Scientific, Austin, Texas, United States) into a pJet 1.2/blunt vector using the sticky end cloning protocol and transfected into the JM109 competent cells by calcium chloride transformation. Plasmids containing stx1 and stx2 were separately extracted using a Genopure plasmid Maxi kit (Roche Life Sciences, Rotkreuz, Switzerland) and quantified by spectrophotometry. To verify successful preparation purified plasmids were subjected to PCR amplification using primers 1a and 1b for stx1 and 2a and 2b for stx2 with amplicon size visually confirmed by agarose gel detection and subsequent sequence analysis. Plasmid quantification was determined spectrophotometrically employing the BioDrop-μLite (Isogen Life Science, B.V, Veldzigt, Netherlands). The A260 was used to calculate the plasmid concentration expressed as the number of molecules/μL.
TABLE 1.
Primers and probes used for real-time polymerase chain reaction.
| Primers/probes | 5! | Sequence | 3! | Reference |
|---|---|---|---|---|
| stx1a-primer | CAAGAGCGATGTTACGGT | (16) | ||
| stx1b-primer | AATTCTTCCTACACGAACAGA | (16) | ||
| stx1f-probe | CTGGGGAAGGTTGAGTAGCG | Fluorescein | (16) | |
| stx1r-probe | CAL Fluor 610 | CCTGCCTGACTATCATGGACA | 3′ phosphor | (16) |
| stx2a-primer | GGGACCACATCGGTGT | (16) | ||
| stx2b-primer | CGGGCACTGATATATGTGTAA | (16) | ||
| stx2f-probe | CTGTGGATATACGAGGGCTTGATGTC | Fluorescein | (16) | |
| stx2r-probe | CAL Fluor 610 | ATCAGGCGCGTTTTGACCATCT | 3′ phosphor | (16) |
Polymerase chain reaction assay validation
To assess the potential for PCR cross-reactivity and assess the analytical specificity of the hybridisation probe-based real-time PCR described by Grys et al.,16 the primer and probe sequences were subjected to BLAST analysis on the NCBI database. The PCR reaction was optimised for use on the LightCycler®480 Instrument II (Roche Life Sciences, Rotkreuz, Switzerland) employing the LightCycler® 480 Probes Master mastermix (Roche Life Sciences, Rotkreuz, Switzerland) with modification to the thermal cycling conditions for amplification consisting of denaturation at 95 °C for 10 min followed by 45 cycles of 95 °C for 5 s, 56 °C for 5 s and 72 °C for 15 s. A positive amplification signal was defined as an increase in fluorescence signal that crossed the threshold before 30 cycles. Amplicon identity was determined using the melt-curve analysis program of 95 °C for 30 s, 40 °C for 60 s and 85 °C for 5 s with continuous fluorescence acquisition. The Multi-color HybProbe detection format was used for analysis, combining the Red 610, Red 640 and FAM filter pairs (LightCycler®480 Instrument II Manual, Roche Life Sciences, Rotkreuz, Switzerland). The resulting amplicon size was visualised using agarose gel electrophoresis and subjected to DNA sequencing and BLAST alignment to reference stx1a and stx2a sequences (NC_002655.2). To mimic the sample matrix for sensitivity determination, TSB was inoculated with a pea-size amount of stool (from a single donor) shown to be stx-negative by PCR. To this inoculated broth 1 mL of plasmid stock (5.3*106 copies/μL) containing both stx1 and stx2 was added and serially diluted eight times in 9 mL of TSB, to a lowest dilution of 1:108 (53 plasmid copies/ml). Nucleic acid extraction was performed on 200 μL broth employing the MagNApure LC instrument (Roche Diagnostics, Rotkreuz, Switzerland) to yield 100 μL of extract. Initially, real-time PCR was performed in triplicate using a template from each of the eight dilutions to estimate a limit of detection (LOD). Subsequently, real-time PCR was performed in eight replicates on the dilution with the estimated LOD, as well as one dilution above and one dilution below the estimate. The LOD was defined as the lowest plasmid concentration spiked into TSB, before nucleic acid extraction, yielding a positive signal, as described above in all eight replicates. Nucleic acid extractions from STEC subtypes 1d (Reference strain MH1813, GenBank accession No. AY170851), 2b (Reference strain EH250, GenBank accession No. AF043627), 2c (Reference strain 031, GenBank accession No. L11079), 2d (Reference strain C165-02, GenBank accession No. DQ059012), 2e (Reference strain S1191, GenBank accession No. M21534), 2f (Reference strain T4/97, GenBank accession No. AJ010730) and 2g (Reference strain 7V, GenBank accession No. AY286000) were also subjected to PCR amplification to assess impact of strain variation on detection. The reproducibility of melting temperature assessment for stx1 and stx2 differentiation was determined by testing 24 replicates of TSB spiked with cloned stx1 and stx2 plasmids. To further assess the reproducibility of melting temperature across the subtypes, three stx1 subtypes and seven stx2 subtypes were tested similarly.
Clinical specimen testing
Between September 2014 and May 2015, we collected same day residual stool after routine testing from 226 consecutive stool specimens (the stool samples were transported in a temperature regulated box and processed within 12 h of collection) from the National Health Laboratory Services located at the Groote Schuur Hospital in Cape Town, South Africa, a tertiary care academic teaching laboratory affiliated with the University of Cape Town. This tertiary academic hospital serves the greater Cape Town area.
A pea-sized stool sample was inoculated in 90 mL of TSB and vortexed before incubation at 37 °C for 18 h. Two hundred microlitres of broth were subsequently extracted employing the MagNA Pure LC Total Nucleic Acid isolation kit (Roche Diagnostics, Rotkreuz, Switzerland) using the total variable elution volume protocol and following the manufacturer’s manual (version 14) to yield 100 μL of nucleic acid extract.
In addition, CHROMagarTMSTEC (CHROMagar Microbiology, Paris, France) was inoculated with a loop full of overnight inoculated broth and incubated at 37 °C for 18 h. Bright mauve colonies (up to five mauve colonies were picked per sample, depending on the number of mauve colonies formed) were sub-cultured onto MacConkey agar with crystal violet, sorbitol MacConkey agar, and 2% blood agar (Green point Media, National Health Laboratory Service, Albertynshof, South Africa). E. coli was presumptively identified as lactose-positive, oxidase-negative, spot indole-positive and pyrrolidonyl arylamidase (PYR)-negative with confirmatory identification using VITEK 2 (bioMerieux, Inc., Durham, North Carolina, United States).
Isolate characterisation
Isolates yielding mauve colonies on CHROMagarTMSTEC and presumptively identified as E. coli were subjected to stx characterisation employing the real-time PCR assay characterised herein. Other diarrhoeic E. coli virulence genes, including the fimbrial adhesion gene for diffusely adherent E. coli, the anti-aggregation protein transporter gene for enteroaggregative E. coli, heat-stable and heat-labile enterotoxin genes of enterotoxigenic E. coli, the intimin coding gene eae for enteropathogenic E. coli (EPEC) and the bundle-forming pili gene for the typical EPEC, were determined using standard gel-based PCR as previously described using primers as shown in Table 2.18
TABLE 2.
Primers used for detection of virulence genes by conventional polymerase chain reaction.
| Genes | Primers | Primer sequence | Product size | |
|---|---|---|---|---|
| A | eae | eae-F | TCAATGCAGTTCCGTTATCAGTT | 482 bp |
| eae-R | GTAAAGTCCGTTACCCCAACCTG | |||
| bfp | bfp-F | GGAAGTCAAATTCATGGGGGTAT | 298 bp | |
| bfp-R | GGAATCAGACGCAGACTGGTAGT | |||
| stx1 | stx1-F | CAGTTAATGTGGTGGCGAAGG | 348 bp | |
| stx1-R | CACCAGACAATGTAACCGCTG | |||
| stx2 | stx2-F | ATCCTATTCCCGGGAGTTTACG | 584 bp | |
| stx2-R | GCGTCATCGTATACACAGGAGC | |||
| B | Est | ST-F | ATTTTTCTTTCTGTATTGTCTT | 190 bp |
| ST-R | CACCCGGTACAAGCAGGATT | |||
| Elt | LT-F | GGCGACAGATTATACCGTGC | 440 bp | |
| LT-R | CGGTCTCTATATTCCCTGTT | |||
| C | ipa | ipaH-F | CTCGGCACGTTTTAATAGTCTGG | 933 bp |
| ipaH-R | GTGGAGAGCTGAAGTTTCTCTGC | |||
| aat | pCVD432-F | CTGGCGAAAGACTGTATCAT | 630 bp | |
| pCVD432-R | CAATGTATAGAAATCCGCTGTT | |||
| DaaC | daaC-F | CAGGTCATCCGGTCAGTCGG | 212 bp | |
| daaC-R | CAATGCCACGTACAACCGGC |
ST, heat-stable; LT, heat-labile.
To confirm Shiga toxin production among stx-positive isolates, the Immunocard STAT!® EHEC (Meridian Biosciences, Inc., Cincinnati, Ohio, United States) was used to detect Shiga toxin 1 and 2 (by employing immunochromatography with toxin-directed monoclonal antibodies labelled with red-coloured gold particles). All mauve isolates found to carry virulence genes were serotyped at the Centre for Enteric Diseases, National Institute of Communicable Disease, Johannesburg, by employing antisera (Statens Serum Institut, Copenhagen, Denmark) and the detection of somatic O-antigens as previously described.19,20 H-antigen serotyping was not undertaken.
Statistical analysis
Data on the possession of virulence genes, cultural characteristics on different media, serotypes and Shiga toxin production was entered in Microsoft Office Excel 2010 (Microsoft Corp.,Redmond, Washington, United States) and then exported to EPIINFOTM 7.1.5.2 (Centers for Disease Control and Prevention, Atlanta, Georgia, United States) for analysis. Using the LightCycler 480 II software, efficiency of the in-house real-time PCR assay was determined. The amplification curves and the melting peaks were used to differentiate between stx1 and stx2.
Results
Real-time polymerase chain reaction validation
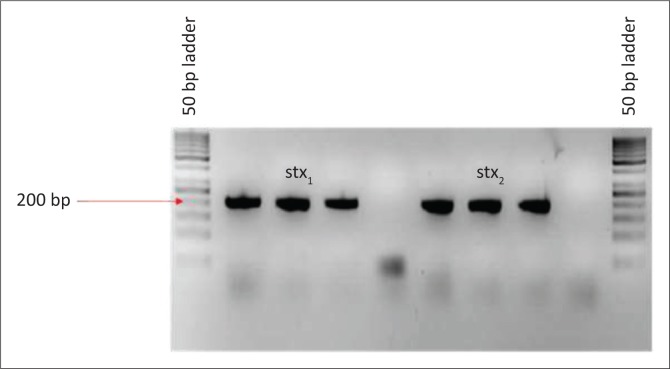
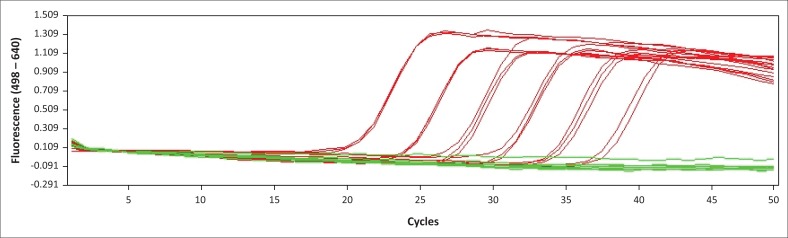
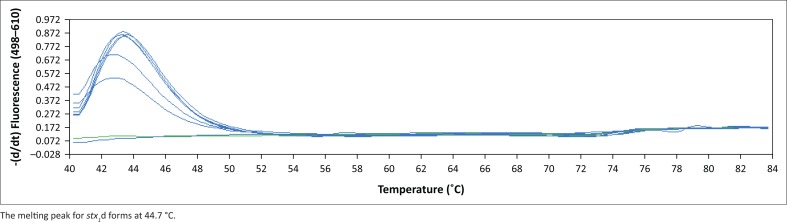
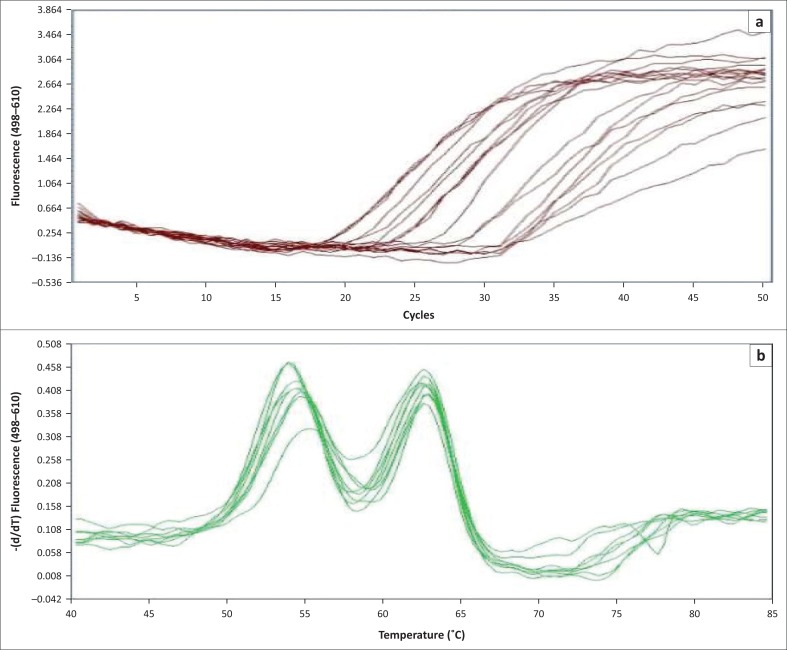
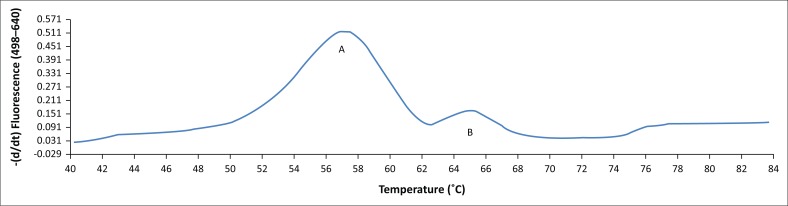
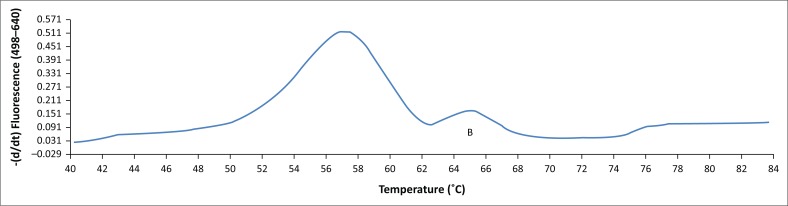
The BLAST analysis of the primers and probe sequence specificity yielded no significant homology to non-stx targets (data not shown). Real-time PCR amplicons generated were confirmed as 208 bp for stx1 and 204 bp for stx2 (Figure 2). Sequencing and BLAST analysis confirmed the identity of both stx1 and stx2 amplicons. The serially diluted plasmid-stool-TSB was successfully amplified in 8/8 replicates in the sixth dilution, whereas the seventh dilution yielded an amplification signal in 3/8 replicates, yielding a LOD of 5.3 target copies/μL of broth. All other stx subtypes investigated (stx1a, stx1b, stx2a, stx2b, stx2c, stx2d, stx2e, stx2f, and stx2g) were successfully amplified by this assay (data not shown). stx1 and stx2 were successfully distinguished by a melting temperature of 58.2 °C (SD = 0.033) and 65.3 °C (SD = 0.037) (Figures 3–6). The Tm for stx2 subtypes 2a, 2b, 2c, 2d, 2e, 2f and 2g were the same at 65.3 °C (SD = 0.037, 0.041, 0.035, 0.039, 0.034, 0.033 and 0.032, respectively), whereas that of 1d was 44.7 °C (SD = 0.042). The efficiency of the assay was 1.99 as calculated from the amplification curves generated using the Light Cycler® 480 software. The duplex assay detected both targets in the same run, and these could be differentiated by the melt curve with two distinct peaks at 58.2 °C for stx1 and 65.3 °C for stx2 (Figure 7).
FIGURE 2.
Polymerase chain reaction amplicons of stx1 and stx2 after cloning.
FIGURE 3.
Amplification curves for the validated duplex real-time polymerase chain reaction for detection of the stx gene
FIGURE 6.
Melting peaks for the detection of stx1d using the duplex real-time polymerase chain reaction assay.
FIGURE 7.
Amplification curves and melting peaks obtained on screening tryptic soy broth using the in-house optimised real-time polymerase chain reaction assay. (A) Amplification curves. (B) Melting peaks for the duplex real-time polymerase chain reaction assay.
FIGURE 4.
Melting peaks for stx1 – peak A (subtypes other than 1d – 55.7 °C) and stx2 – peak B (65.3 °C) detected in the same run.
FIGURE 5.
Melting peaks for optimised duplex real-time polymerase chain reaction showing stx2 peak B (65.3°C).
Clinical specimens
Of the 226 specimens screened, real-time PCR detected Shiga toxin genes in 14 samples (6.2%), comprising 8 stx1, 5 stx2 and 1 specimen containing both stx1 and stx2. CHROMagarTMSTEC yielded mauve colonies from 23.45% (53/226) of the stool broth cultures (Figure 8).
FIGURE 8.
Summary of isolate characterisation results.
Performance of CHROMagarTMSTEC
Of the 53 mauve isolates, 48 were negative for stx genes using the validated real-time PCR assay. Of the 14 broths that were positive on PCR, nine did not yield any mauve colonies on CHROMagarTMSTEC culture.
Isolate characteristics
Forty-four (83%) of the 53 mauve colonies fermented lactose on MacConkey agar with crystal violet. Eleven (25%) of the 44 lactose fermenters were non-sorbitol-fermenting. Real-time PCR on the 44 E. coli confirmed the presence of stx genes in five (11%), whereas 39 were negative for the stx gene. Real-time PCR was not done on the nine non-lactose fermenting isolates as these were found not to be E. coli on biochemical testing. Four of the five stx positive E. coli colonies were also positive in the real-time PCR broth assay. Of the 39 stx-negative E. coli, only four (12.5%) carried eae genes, whereas four possessed aat genes. Of the four eae positive isolates, two also had the bfp genes and were typical EPEC. The other two eae positive isolates did not possess the bfp genes and were classified as atypical EPEC. The four enteroaggregative stx-negative isolates all belonged to E. coli serotype O104. All the typical EPEC belonged to serotype O55, whereas one of the two atypical EPEC belonged to serotype O101. The atypical EPEC serotype O101 was from stx2 positive broth. The other atypical EPEC isolate was untypeable. No diffusely adherent E. coli, enteroinvasive E. coli or enterotoxigenic E. coli were detected. None of the 53 E. coli isolates that were screened by immunochromatography was positive for Shiga toxins. For the CHROMagarTMSTEC, sensitivity was 33.3%, specificity was 77.4%, negative predictive value was 95% and positive predictive value was 9.4% (Table 3).
TABLE 3.
Performance of CHROMagarTMSTEC compared with real-time polymerase chain reaction assay.
| In-house developed assay |
||||
|---|---|---|---|---|
| CHROMagarTMSTEC | + | _ | Total | |
| + | 5 | 48 | 53 | |
| _ | 9 | 164 | 173 | |
| Total | 14 | 212 | 226 | |
CHROMagarTMSTEC sensitivity = 5/14*100 = 33.3%
CHROMagarTMSTEC specificity = 164/212*100 = 77.4%
CHROMagarTMSTEC positive predictive value = 5/53*100 = 9.4%
CHROMagarTMSTEC negative predictive value = 164/173*100 = 95%
Discussion
We validated the use of a previously described duplex real-time PCR assay with modification able to detect and differentiate stx1 (melting temperature = 58.2 °C) and stx2 (melting temperature = 65.3 °C) from overnight broth enrichment with a LOD of 5.3 target copies/μL broth. This assay was able to detect both stx1 and stx2 in the same run, thus potentially reducing process turn-around time in a busy laboratory setting. Timely reporting of STEC infections is important, because use of certain antibiotics is contraindicated in STEC infections.
Compared to the validated duplex real-time PCR, CHROMagarTMSTEC showed a sensitivity of 33%, specificity of 77.4%, negative predictive value of 9.4% and positive predictive value of 95% for detection of STEC in stool following TSB enrichment. Of the 53 mauve isolates, 48 were negative for stx genes on use of the validated real-time PCR, whereas nine of the 14 PCR-positive broths did not yield any mauve colonies when cultured on CHROMagarTMSTEC. Reasons for the poor performance of this medium in relation to the in-house developed duplex real-time PCR include the following: (1) delays in reporting of diarrhoea cases to a tertiary hospital (where samples were collected) may have led to loss of stx genes; STEC numbers are sharply reduced in stool after one week of illness, and the Shiga toxin genes might be lost by the bacteria.21 (2) CHROMagarTMSTEC selects for tellurite resistant strains but misses the tellurite susceptible STEC whose prevalence in this setting is not known.
For a chromogenic medium to be considered for routine screening purposes, it must have high specificity so as not to waste scarce laboratory resources on false positives. The false positivity rate in this setting (48/53 [90.6%]) is higher than has previously been reported in Europe (16.3% reported by Gouali et al., 2013 and 18.3% by Wylie et al., 2013).12,22
Similar studies to evaluate this medium were done in Canada, Finland, and Germany, all of which reported high sensitivities for STEC serotypes O26, O111, O121, O145, O118, and O157.13,22,23 The specificity of 77.4% noted in this study was low compared to values (between 95.8% and 98.9%) reported in similar studies done in Europe. The difference in sensitivity and specificity could be explained by the differences in the patient characteristics (whether they present with haemolytic uremic syndrome and or bloody diarrhoea or not). Unlike the studies in Europe, this study did not focus on only patients with haemolytic uremic syndrome or bloody diarrhoea. Additionally, the distribution of tellurite resistant STEC (which is targeted by CHROMagarTMSTEC) in this setting is not known.
The prevalence of stx genes in stool samples (6.2%) was lower than the 9% previously reported by Kullin et al., 2015.24 Among the 53 isolates that formed mauve colonies on CHROMagarTMSTEC, five were STEC (two serotype O26 and others non-typeable), four were enteroaggregative E. coli (serotype O104), two were atypical EPEC (serotype O101 and non-typeable) and two were typical EPEC (serotype O55). Serotypes O26 and O104 are among the top six STEC serotypes globally.25,26 CHROMagarTMSTEC is intended for STEC culture; however, we detected other diarrhoeic E. coli pathotypes using this medium. The other pathotypes detected might have been hybrid strains that lost the stx genes or hybrid strains whose stx genes could not be detected using the primers employed in this study. Notably, bacteriophages carrying the stx genes are very quickly lost both in vivo and in vitro,27 and not all stx primers can detect all the stx gene variants.28
Limitations
Not all STEC are tellurite resistant and may have been missed on CHROMagarTMSTEC culture. This study only focused on the strains that formed mauve colonies. The stool samples in this study were collected between September 2014 and May 2015, and therefore the results of this study as regards prevalence of STEC may not reflect the entire year or areas of South Africa other than Cape Town.
Conclusions
The in-house developed real-time PCR assay is a sensitive and specific alternative to the currently used diagnostic strategy. Due to the high false positivity rate, CHROMagarTMSTEC can only be used as an adjunt to a more sensitive and specific assay such as real-time PCR.
Trustworthiness
To the best of our knowledge, the findings of this study can be used as per the scope of the study and in light of the study limitations as clearly pointed out.
Reliability
We confirm that the experiments conducted in this study will yield the same results during repeated trials using the same reagents and detection platforms.
Validity
To the best of our knowledge, the findings of this study, as obtained using the methods we employed, are valid for the study area and season. The in-house developed real-time PCR may, however, be adopted in other laboratories in developing countries.
Acknowledgements
We acknowledge the Center for Enteric Disease Research Unit, National Institute of Communicable Disease (Sandringham, Johannesburg, South Africa) for serotyping of the E. coli isolates. We also acknowledge the National Research Foundation, South Africa, the ACP-RISE scholarship fund and the ADB-HEST fund.
Competing interests
The authors declare that there are no financial or personal relationships that may have had influence in the writing of this article.
Sources of support
Funds for this study were provided by the National Research Foundation, South Africa, the ADB-HEST scholarship fund and the ACP-RISE scholarship fund.
Authors’ contributions
J.B.K. participated in processing the samples, data analysis and writing of the manuscript. K.H.K. participated in the supervision of the work, development of the idea and writing of the manuscript. A.S. participated in the writing of the manuscript and supervision of the work. M.P.N. participated in the development of the idea, supervision of the work and writing of the manuscript. L.R. developed the idea, participated in supervision and in writing of the manuscript.
Footnotes
How to cite this article: Kalule JB, Keddy KH, Smith A, Nicol MP, Robberts L. Development of a real-time PCR assay and comparison to CHROMagarTM STEC to screen for Shiga toxin-producing Escherichia coli in stool, Cape Town, South Africa. Afr J Lab Med. 2017;6(1), a609. https://doi.org/10.4102/ajlm.v6i1.609
References
- 1.Slayton RB, Turabelidze G, Bennett SD, et al. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O157:H7 associated with romaine lettuce consumption, 2011. PLoS One. 2013;8(2):e55300 https://doi.org/10.1371/journal.pone.0055300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friesema I, Schimmer B, Stenvers O, et al. STEC O157 outbreak in the Netherlands, September–October 2007. Euro Surveill. 2007;12(11):E071101 1. [DOI] [PubMed] [Google Scholar]
- 3.Heiman KE, Mody RK, Johnson SD, et al. Escherichia coli O157 outbreaks in the United States, 2003–2012. Emerg Infect Dis. 2015;21(8):1293–1301. https://doi.org/10.3201/eid2108.141364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luna-Gierke RE, Griffin PM, Gould LH, et al. Outbreaks of non-O157 Shiga toxin-producing Escherichia coli infection: USA. Epidemiol Infect. 2014;142(11):2270–2280. https://doi.org/10.1017/s0950268813003233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould LH. Recommendations for diagnosis of Shiga toxin producing Escherichia coli infections by clinical laboratories. Wkly Rep. 2009;58:1e14 https://doi.org/10.1016/j.clinmicnews.2012.04.004 [PubMed] [Google Scholar]
- 6.Germani Y, Soro B, Vohito M, et al. Enterohaemorrhagic Escherichia coli in Central African Republic. Lancet. 1997;349(9066):1670 https://doi.org/10.1016/S0140-6736(05)62636-0 [DOI] [PubMed] [Google Scholar]
- 7.Germani Y, Cunin P, Tedjouka E, et al. Enterohaemorrhagic Escherichia coli in Ngoila (Cameroon) during an outbreak of bloody diarrhoea. Lancet. 1998;352(9128):625–626. https://doi.org/10.1016/s0140-6736(05)62636-0 [DOI] [PubMed] [Google Scholar]
- 8.Koyange L, Ollivier G, Muyembe JJ, et al. Enterohemorrhagic Escherichia coli O157, Kinshasa. Emerg Infect Dis. 2004;10(5):968–969. https://doi.org/10.3201/eid1005.031034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soares BE. Quality of medical laboratory services in resource-limited settings. Afr J Infect Dis. 2012;6(1):10–11. https://doi.org/10.4314/ajid.v6i1.77736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loefler IJ. Surgical wound infection in the Third World: The African experience. J Med Microbiol. 1998;47(6):471–473. https://doi.org/10.1099/00222615-47-6-471 [DOI] [PubMed] [Google Scholar]
- 11.Van Biljon G. Causes, prognostic factors and treatment results of acute renal failure in children treated in a tertiary hospital in South Africa. J Trop Pediatr. 2008;54(4):233–237. https://doi.org/10.1093/tropej/fmm079 [DOI] [PubMed] [Google Scholar]
- 12.Gouali M, Ruckly C, Carle I, et al. Evaluation of CHROMagarSTEC and STEC O104 chromogenic agar media for detection of Shiga Toxin-producing Escherichia coli in stool specimens. J Clin Microbiol. 2013;51(3):894–900. https://doi.org/10.1128/jcm.03121-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirvonen JJ, Siitonen A, Kaukoranta SS. Usability and performance of CHROMagarSTEC medium in detection of Shiga toxin-producing Escherichia coli strains. J Clin Microbiol. 2012;50(11):3586–3590. https://doi.org/10.1128/jcm.01754-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson KE, Thorpe CM, Sears CL. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis: An official publication of the Infectious Diseases Society of America. 2006;43(12):1587–1595. https://doi.org/10.1086/509573 [DOI] [PubMed] [Google Scholar]
- 15.Karch H, Bielaszewska M, Bitzan M, et al. Epidemiology and diagnosis of Shiga toxin-producing Escherichia coli infections. Diagn Microbiol Infect Dis. 1999;34(3):229–243. https://doi.org/10.1016/s0732-8893(99)00031- [DOI] [PubMed] [Google Scholar]
- 16.Grys TE, Sloan LM, Rosenblatt JE, et al. Rapid and sensitive detection of Shiga toxin-producing Escherichia coli from nonenriched stool specimens by real-time PCR in comparison to enzyme immunoassay and culture. J Clin Microbiol. 2009;47(7):2008–2012. https://doi.org/10.1128/jcm.02013-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perna NT, Plunkett G, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409(6819):529–533. https://doi.org/10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- 18.Paton AW, Paton JC. Detection and characterization of Shigatoxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36(2):598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bettelheim KA, Thompson CJ. New method of serotyping Escherichia coli: Implementation and verification. J Clin Microbiol. 1987;25(5):781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura K, Sakazaki R, Murase M, Kosako Y. Serotyping and categorisation of Escherichia coli strains isolated between 1958 and 1992 from diarrhoeal diseases in Asia. J Med Microbiol. 1996;45(5):353–358. https://doi.org/10.1099/00222615-45-5-353 [DOI] [PubMed] [Google Scholar]
- 21.Olesen B, Jensen C, Olsen K, et al. VTEC O117:K1:H7. A new clonal group of E. coli associated with persistent diarrhoea in Danish travellers. Scand J Infect Dis. 2005;37(4):288–294. https://doi.org/10.1080/00365540410021090 [DOI] [PubMed] [Google Scholar]
- 22.Wylie JL, Van Caeseele P, Gilmour MW, et al. Evaluation of a new chromogenic agar medium for detection of Shiga toxin-producing Escherichia coli (STEC) and relative prevalences of O157 and non-O157 STEC in Manitoba, Canada. J Clin Microbiol. 2013;51(2):466–471. https://doi.org/10.1128/jcm.02329-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tzschoppe M, Martin A, Beutin L. A rapid procedure for the detection and isolation of enterohaemorrhagic Escherichia coli (EHEC) serogroup O26, O103, O111, O118, O121, O145 and O157 strains and the aggregative EHEC O104:H4 strain from ready-to-eat vegetables. Int J Food Microbiol. 2012;152(1–2):19–30. https://doi.org/10.1016/j.ijfoodmicro.2011.10.009 [DOI] [PubMed] [Google Scholar]
- 24.Kullin B, Meggersee R, D’Alton J, et al. Prevalence of gastrointestinal pathogenic bacteria in patients with diarrhoea attending Groote Schuur Hospital, Cape Town, South Africa. SAMJ. 2015;105:121–125. https://doi.org/10.7196/samj.8654 [DOI] [PubMed] [Google Scholar]
- 25.Hegde NV, Cote R, Jayarao BM, et al. Detection of the top six non-O157 Shiga toxin-producing Escherichia coli O groups by ELISA. Foodborne Pathog Dis. 2012;9(11):1044–1048. https://doi.org/10.1089/fpd.2012.1231 [DOI] [PubMed] [Google Scholar]
- 26.Buchholz U, Bernard H, Werber D, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med. 2011;365(19):1763–1770. https://doi.org/10.1016/j.yped.2011.12.017 [DOI] [PubMed] [Google Scholar]
- 27.Bielaszewska M, Kock R, Friedrich AW, et al. Shiga toxin-mediated hemolytic uremic syndrome: Time to change the diagnostic paradigm? PLoS One. 2007;2(10):e1024 https://doi.org/10.1371/journal.pone.0001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins C, Lawson AJ, Cheasty T, Willshaw GA. Assessment of a real-time PCR for the detection and characterization of verocytotoxigenic Escherichia coli. J Med Microbiol. 2012;61(Pt 8):1082–1085. https://doi.org/10.1099/jmm.0.041517-0 [DOI] [PubMed] [Google Scholar]