Abstract
We report the clinical and analytical performance of an isothermal thermophilic helicase-dependent amplification assay for blood Plasmodium parasite detection and species-level identification. The assay amplifies the 18S rRNA gene fragment of all Plasmodium species and uses a species-specific probe and a pan-malarial probe to definitively identify Plasmodium falciparum from other infectious Plasmodium species. Amplicon-probe hybridization products are detected with a disposable dipstick enclosed in a cassette. With a pan-malarial–positive and P. falciparum–negative result, an additional test is performed to detect if the pan-malarial–positive band was the result of the presence of Plasmodium vivax. The assay uses only 2 μL of human whole blood directly for a 50-μL amplification reaction, without any pre-amplification processing. The clinical performance of the assay was validated using 88 samples from New York patients suspected of malaria or babesiosis. The overall sensitivity of the assay was 96.6% (95% CI, 87.3% to 99.4%), and the specificity was 100% (95% CI, 85.4% to 100%), compared with gold standard microscopy and a laboratory-developed molecular assay, respectively. The analytical sensitivity was 50 copies of DNA per assay or 200 parasites per microliter of blood, and the assay can detect samples with parasitemia levels <1%. This novel molecular diagnostic assay requires minimal laboratory instrumentation and uses un-processed blood as input; it can be readily performed in the field.
Malaria is a mosquito-borne disease caused by intraerythrocytic protozoa of the genus Plasmodium. Five Plasmodium species are known to infect human beings: Plasmodium falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi.1 People infected by the malaria parasite experience fever, chills, and a flu-like illness that, if left untreated, may develop into severe complications, potentially leading to death. The World Health Organization reports that, in 2010, approximately 216 million people were infected with malaria, and 655,000 died as a result of their infection (World Malaria Report 2011, http://www.who.int/malaria/world_malaria_report_2011/en, last accessed June 24, 2013). Most fatalities (91%) occur in Africa, and are due to infection by P. falciparum. Although relatively less severe than P. falciparum, P. vivax and P. ovale can enter a dormant stage as hypnozoites in the liver of the host and cause relapses by invading the bloodstream weeks, or even years, later (Centers for Disease Control and Prevention, http://www.cdc.gov/malaria/about/facts.html, last accessed June 24, 2013). Because P. vivax and P. ovale present a health risk for travelers to the Middle East, Asia, South America, and Central America, P. falciparum and P. vivax have different drug-resistance profiles2; thus, species-specific identification is prudent if patients come from regions of the globe where more than one species is present.
Although microscopy of stained thick or thin blood film smears remains the gold standard for diagnosis of malaria infection, properly identifying the species of Plasmodium in a patient's blood is difficult for western clinical laboratories.3 The rapid lateral-flow antigen detection tests target the histidine-rich protein of P. falciparum and a pan-malarial lactose dehydrogenase protein; however, the persistence of histidine-rich protein of P. falciparum in patients receiving therapy renders these rapid lateral-flow antigen detection tests unsuitable for monitoring clearance of the pathogen.4 Moreover, recent reports on the deletion of the HRP2 gene from the Plasmodium population in certain endemic areas have demonstrated that this deletion results in significantly higher false-negative diagnoses.5, 6 Rapid lateral-flow antigen detection tests have good sensitivity for P. falciparum but lack sensitivity for the other species using the pan-malarial test line.7, 8, 9 Therefore, western laboratories often perform a laboratory-developed nucleic acid amplification test to detect and further identify the species of Plasmodium present in a patient's blood.10, 11
Several isothermal nucleic acid amplification tests have been developed to detect Plasmodium using loop-mediated isothermal amplification technology (LAMP).12, 13, 14, 15, 16, 17, 18 Some of these tests do not offer species discrimination,16 whereas others do distinguish between the species.15, 18 Some target the multicopy rRNA genes,12, 17 whereas others target other sequences also present in multiple copies in each cell.13, 16 LAMP is viewed as a good platform for the detection of malaria in the field because it can be performed using a constant temperature incubator or water bath,19 and it allows for the detection of amplification products using the naked eye.20 However, the complexity of the LAMP reaction, and its reliance on the use of six primer pairs, makes LAMP assays more difficult to manufacture than PCR tests, and complicates assay design.21 In addition, the LAMP-based assays still require dilution and heating of the blood sample before isothermal amplification,16 which potentially increases the complexity of assay workflow.
In this study, we report the development of an alternative isothermal nucleic acid amplification platform that can detect all five species of Plasmodium, and can specifically differentiate P. falciparum and P. vivax parasites using a simple, handheld, disposable detection device. The assay uses thermophilic helicase–dependent amplification (tHDA)22, 23 chemistry to amplify the 18S rRNA genes24 of all Plasmodium species, followed by a species-specific, probe-based detection of P. falciparum versus the other Plasmodium species. A reflex test, using one species-specific primer, is performed to resolve P. vivax from P. ovale, P. malariae, and P. knowlesi. Because tHDA can be performed using a small amount (2 μL) of unprocessed blood and a dry heat block, this assay has the potential of being used as a point-of-care device. The sensitivity and specificity of this assay were evaluated with a panel of 88 blood samples collected from either symptomatic travelers returning from malaria-endemic countries to the New York City area or patients from eastern Long Island with a diagnosis of possible Babesia infection.
Materials and Methods
Control DNA
Genomic DNA of P. falciparum (3D7 strain) was purified from laboratory culture–derived parasites by the Chelex method.25, 26 Briefly, cultured 3D7 parasites were mixed with red blood cells and normal human serum to achieve a final concentration of 50% hematocrit and 50% normal human serum; then, they were spotted on Whatman filter paper and allowed to dry at room temperature overnight. The dried filter paper was soaked in sterile 0.5% saponin in PBS for 30 minutes at 37°C. The filter paper was washed with sterile PBS and treated with 200 μL of 5% Chelex 100 (Bio-Rad, Hercules, CA) (preheated at 65°C for 10 minutes) at 100°C. The supernatant obtained after centrifugation (2 minutes at 10,000 × g) was respun to ensure complete removal of Chelex. The resulting supernatant was stored at −20°C until further use. Dried blood spots of P. vivax, P. ovale, and P. malariae on filter papers were obtained from the Malaria Research and Reference Reagent Resource Center (Manassas, VA). DNA was extracted by the Chelex method, as previously described. The total DNA concentration was assessed by OD260 measurement. The OD260/OD280 ratio was also measured, and the values were approximately 2.0, indicating acceptable purity of the extracted DNA.
Plasmids carrying partial sequences of the 18S rRNA of P. malariae, P. knowlesi, P. ovale, P. vivax, or the internal control (IC) sequences were synthesized by BlueHeron (Bothell, WA) by cloning the desired fragment of 200 to 400 bp into pUC vector.
Clinical Samples
Fresh negative human whole blood used in spiking experiments was purchased from Research Blood Components (Boston, MA). EDTA whole blood samples (n = 88) collected from 55 patients between October 2009 and January 2012 for routine diagnostic purposes, from either symptomatic travelers returning from malaria-endemic countries to the New York City area or patients from eastern Long Island with a diagnosis of possible Babesia infection, were used for this clinical evaluation. At sample collection, EDTA whole blood samples were tested by thin-and-thick blood film microscopy and a nucleic acid sequence–based amplification malaria test (NASBAMT) developed by a laboratory10, 11 (North Shore–LIJ Health System Laboratories, Lake Success, NY). Thirty-nine samples from 20 patients contained P. falciparum, 16 samples from nine patients contained P. vivax, three samples from two patients contained P. malariae, one sample from one patient contained P. ovale, 15 samples from 10 patients contained Babesia species, and 14 samples from 13 patients were negative for both Plasmodium and Babesia. Samples from the same patients were collected on different days or at different times on the same day. Samples were stored frozen at −80°C until shipment to BioHelix Corporation (Beverly, MA) for the tDHA-based molecular tests. Whole blood samples were de-identified in accordance with a protocol approved by the Feinstein Institute for Medical Research Institutional Review Board (Manhasset, NY). All samples provided to BioHelix Corporation were stripped of all patient identifiers and provided with a reference number. Samples used for initial assay verification contained Plasmodium or Babesia or were negative for both. The initial panel of samples (n = 20) was provided with microscopy and NASBAMT results. A second panel of samples (n = 68) containing various types of Plasmodium and Babesia and samples negative for both blood parasites were provided blinded (without microscopy and NASBAMT results) to BioHelix Corporation.
Design of Primers, Probes, and ICs for tHDA Amplification
The tHDA-based malaria detection and typing assay consisted of two separate tests. The first test (test 1) detects all five Plasmodium species, P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi, known to infect humans. In addition, it selectively identifies the presence of P. falciparum. Primers for tHDA amplification in test 1 were designed using Primer3 software version 0.4.0,27 following the specifications described in the IsoAmp III tHDA amplification kit (BioHelix Corporation). Primer pairs were evaluated by real-time tHDA amplification in the presence of EvaGreen fluorescent dye (Biotium, Hayward, CA). The tHDA reaction conditions were then optimized using the best-performing primer pairs. Asymmetric tHDA amplification conditions were evaluated with one of the primers labeled with biotin at 5′ and in excess to the other unlabeled primer.
A digoxigenin (DIG)–labeled probe was designed to anneal to a conserved region in the tHDA amplification product among all five Plasmodium species. A second probe labeled with FAM was designed to anneal specifically only to P. falciparum but not the other four species. After amplification, the reactions were applied to BESt cassettes (BioHelix Corporation). The probes annealed to the DNA strand were generated by the biotin-labeled primer at room temperature and were captured by anti-DIG or anti-FAM antibody lines on a lateral-flow test strip in the BESt cassettes. Detection of the probe-amplicon complex was achieved using streptavidin-coated color latex particles.28
An IC sequence was designed to be amplified by the same primer pairs as the target amplicon, with a random sequence in the middle between the primers. The IC sequence was cloned into a pUC plasmid and included in the tHDA amplification reaction. A probe labeled with 2,4-dinitrophenyl (DNP) was designed to anneal to the IC sequence. The amplification products were detected on the lateral-flow strip by an anti-DNP antibody.
The second test (test 2), which serves as a reflex test, detects the presence of P. vivax. Amplification of P. vivax in test 2 used the same reverse primer as in test 1. The forward primer was designed to be specific for P. vivax and would not amplify other Plasmodium species. The same DIG-labeled detection probe in test 1 was also used in test 2. After tHDA amplification, the product was detected on the lateral-flow strip through anti-DIG antibodies. An IC sequence for this test was designed similar to test 1. The IC sequence for test 2 was the same as that for test 1, except for the forward primer-binding region, which only binds the forward primer for P. vivax. This IC sequence was also cloned into a pUC plasmid and included in the reaction, and its amplification product was detected on the lateral-flow strip through binding to the same DNP-labeled probe as in test 1.
The DNP-labeled probe for the ICs was synthesized and high-performance liquid chromatography purified by TriLink (San Diego, CA). All other primers and probes were synthesized and high-performance liquid chromatography purified by Eurofins MWG Operon (Huntsville, AL).
Assay Procedure
Both tHDA tests 1 and 2 were performed in a 50-μL reaction volume. Test 1 contained 1× annealing buffer II (BioHelix Corporation), 4 mmol/L MgSO4, 40 mmol/L NaCl, 3.6 μL IsoAmp II dNTP mix (BioHelix Corporation), 40 nmol/L forward primer, 120 nmol/L reverse primer, 50 nmol/L DIG-labeled probe for all Plasmodium species, 30 nmol/L FAM-labeled P. falciparum–specific probe, 30 nmol/L DNP-labeled IC probe, 5 U of DdeI (New England Biolabs, Ipswitch, MA), 150 ng extreme thermostable single-stranded DNA binding protein (BioHelix Corporation), 500 copies of the IC 1 IC plasmid, 2 μL IsoAmp III Enzyme Mix (BioHelix Corporation), and input template, as indicated in the figures and text. Test 2 was performed in similar conditions as test 1, except for the primers/probes and IC, which contained 50 nmol/L of the P. vivax–specific forward primer, 100 nmol/L reverse primer, 30 nmol/L DIG-labeled probe, 30 nmol/L DNP-labeled IC probe, and 2000 copies of IC 2 IC plasmid. The reaction mixture was allowed to incubate at 64°C for 90 (without blood) or 120 (with blood) minutes. The reaction tubes were applied to BESt cassettes after incubation, and images were obtained 15 minutes later.
Determination of Analytical Sensitivity and Specificity of the tHDA Assay
Genomic DNA from P. falciparum was extracted from laboratory culture–derived parasites. The concentration of the DNA was determined by OD260 measurement. Tenfold serial dilutions (from 104 to 10 copies per μL) of the P. falciparum genomic DNA were used to determine the limit of detection for the tHDA-based malaria test 1. Because parasite cultures of the other four Plasmodium species could not be obtained, plasmids containing a fragment of the 18S rRNA region for P. vivax, P. ovale, P. malariae, and P. knowlesi were constructed, serially diluted (from 104 to 10 copies per μL), and used as templates to determine the analytical sensitivity of test 1 for each of these four species and that of test 2 for P. vivax. Each dilution was tested in triplicate. High copy numbers (50,000) of either genomic DNA from P. falciparum or plasmid DNA containing a partial sequence of P. ovale, P. malariae, and P. knowlesi were used to assess the specificities of test 2.
Amplification by tHDA in the Presence of Unprocessed Human Whole Blood
To test the effects of human blood on tHDA amplification, 2 to 5 μL of fresh human whole blood (clinical test input volume) was added to each 50-μL tHDA reaction test 1 containing 50 or 500 copies of P. falciparum genomic DNA. Amplification in the presence of human blood was detected using BESt cassettes (BioHelix Corporation).
Determination of Clinical Sample Sensitivity and Specificity
The 88 frozen EDTA whole blood samples were thawed at room temperature just before the tHDA tests. To each of the 50-μL tHDA reactions in test 1, 2 μL of the blood sample was added directly without heat denaturation or nucleic acid extraction. Samples being identified by the tHDA test as positive for Plasmodium (DIG-line positive) but negative for P. falciparum (FAM-line negative) were subjected to test 2 to identify the presence or absence of P. vivax. The samples were tested in tHDA assays either not blinded (n = 20) or blinded (n = 68). Results were compared with microscopy and NASBAMT. The analytical sensitivity of the tHDA assay from human whole blood samples was evaluated by serial dilutions of the frozen blood samples with known parasitemia levels into fresh blood. Four P. falciparum samples with parasitemia levels ranging from 0.3% to 8.8% were diluted up to 2000-fold. The final concentrations of the diluted samples were between 100 and 250 parasites per μL of blood. Each dilution was tested by tHDA test 1 in duplicates.
Results
Assay Overview
The tHDA-based malaria detection and typing assay consisted of two separate tests. The first test (test 1) detects all five Plasmodium species, P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi, known to infect humans. In addition, it selectively identifies the presence of P. falciparum. The second test (test 2), which serves as a reflex test, detects the presence of P. vivax. Both tests 1 and 2 are based on isothermal amplification of the 18S rRNA gene (Figure 1) by tHDA platform.22, 23 In both tests, 2 μL of human whole blood is added directly to the tHDA reaction mix for DNA amplification. No nucleic acid extraction or any other type of sample preparation is necessary. The reactions are incubated at a constant temperature of 64°C for 2 hours. During incubation, thermal stable UvrD helicase unwinds double-stranded DNA template for hybridization to sequence specific primers, which are subsequently extended by thermal stable DNA polymerase.22, 23 During amplification, probes labeled with either FAM or DIG hybridize specifically to the target amplicon (Table 1 and Figure 1). The end product is then detected using a disposable lateral-flow strip by binding to anti-FAM or anti-DIG antibodies.28 In both tests, an IC plasmid was included in the reaction mix to monitor the amplification and was detected through a DNP-labeled probe on the lateral-flow strip (Table 1 and Figure 2).
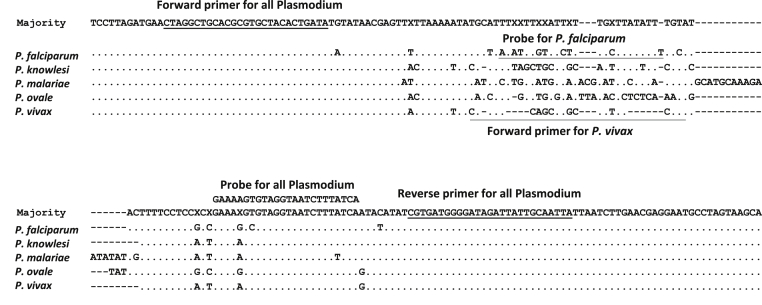
Figure 1.
Alignment of P. falciparum, P. knowlesi, P. malariae, P. ovale, and P. vivax 18S rRNA sequences. The sequences were obtained from GenBank (http://www.ncbi.nlm.nih.gov/nucleotide; accession numbers AL010278, L07560, AB489196, AJ001527, and U93233) for each species, respectively. Only the region surrounding the tHDA amplification product is shown. The primers and probes used in the two tHDA tests are indicated on the alignment.
Table 1.
Primer, Probe, and IC Sequences
| Test no. | Primer/probe | Sequence | Label | Concentration (nmol/L) |
|---|---|---|---|---|
| 1 | Forward primer | 5′-CTAGGCTGCACGCGTGCTACACTGATA-3′ | None | 40 |
| Reverse primer | 5′-TAATTGCAATAATCTATCCCCATCACG-3′ | 5′ Biotin | 120 | |
| Probe for all 5 Plasmodium species | 5′-GAAAAGTGTAGGTAATCTTTATCA-3′ | 3′ DIG | 50 | |
| Probe for P. falciparum | 5′-ATATTTGTATCTTTGCTTATATT-3′ | 3′ FAM | 30 | |
| Probe for IC | 5′-CCGCTATTTCATAGTAAGTC-3′ | 3′ DNP | 30 | |
| IC 1 | 5′-CTAGGCTGCACGCGTGCTACACTGATACAACATTGACAACTAAAACCGCTATTTCATAGTAAGTCCTTCGTCTATGACTATCTGTGCCAAATCATCTAGCAAACTGCTAATCCAATTTATTTCATCAAATCGTGATGGGGATAGATTATTGCAATTA-3′ | None | 500 Copies | |
| 2 | Forward primer | 5′-CGATTCAGCTTGCTGTTTCGTA-3′ | None | 50 |
| Reverse primer | Same as in test 1 | None | 100 | |
| Probe for P. vivax | Same as in test 1 | 3′ DIG | 30 | |
| Probe for IC | Same as in test 1 | 3′ DNP | 30 | |
| IC 2 | 5′-CGATTCAGCTTGCTGTTTCGTATTTTTCCTCAACATTGACAACTAAAACCGCTATTTCATAGTAAGTCCTTCGTCTATGACTATCTGTGCCAAATCATCTAGCAAACTGCTAATCCAATTTATTTCATCAAATCGTGATGGGGATAGATTATTGCAATTA-3′ | None | 2000 Copies |
Forward and reverse primer-binding regions are underlined.
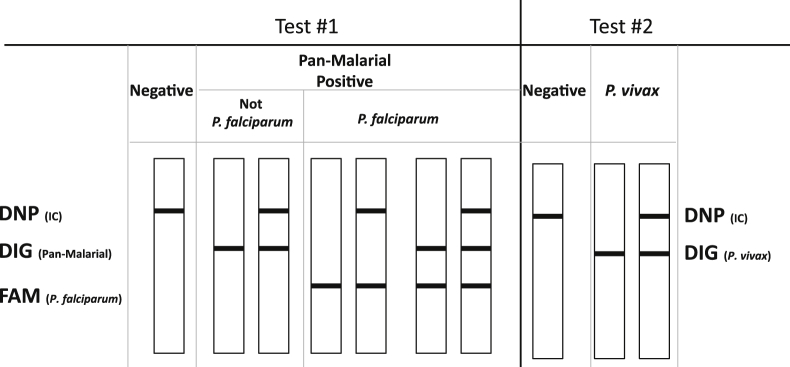
Figure 2.
Schematic diagram of the tHDA-based malaria assay results. There are three antibody lines on the lateral-flow test strip: anti-DNP, anti-DIG, and anti-FAM (from top to bottom). In test 1, if only the DNP line is present, the test result is negative for Plasmodium. If the FAM line is present (the other two lines can be present or absent), the test result is positive for P. falciparum. If the FAM line is absent, but the DIG line is present (the DNP line can be either present or absent), the result is positive for Plasmodium but negative for P. falciparum. In test 2, if only the DNP line is present, the test result is negative for P. vivax; and if the DIG line is present (the DNP line can be either present or absent), the test is positive for P. vivax. The FAM line should always be absent for test 2.
Analytical Sensitivity and Specificity
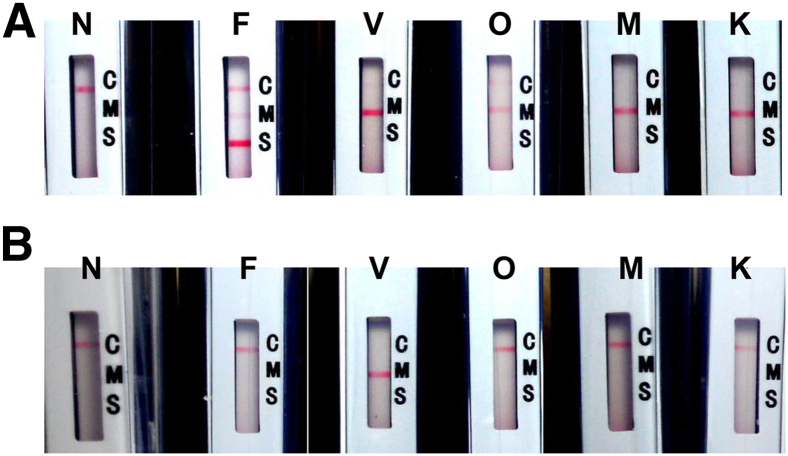
For all five species, the tHDA-based malaria assay test 1 was able to detect as low as 50 copies of input template DNA (genomic DNA for P. falciparum and constructed plasmid DNA containing sequences of the other four species) per reaction (Figure 3A). This test also successfully identified the presence of as low as 50 copies of P. falciparum per reaction (Figure 3A). The specificity of the P. falciparum probe was demonstrated by the absence of the FAM line in the tests using any of the other four Plasmodium species as input template (Figure 3A).
Figure 3.
Amplification and detection of all five Plasmodium species and identification of P. falciparum and P. vivax by tHDA and lateral-flow cassettes. A: Test 1. Genomic DNA of P. falciparum (lane F) or plasmids containing the 18S rRNA fragment of the other Plasmodium species (lanes V, O, M, and K) were added to 50 μL of tHDA test 1 amplification reaction to a final amount of 50 or 500 copies. B: Test 2. Plasmid DNA containing the 18S rRNA fragment of P. vivax was added to tHDA test 2 at 50 copies per reaction (lane V). Plasmid containing P. ovale, P. malariae, or P. knowlesi DNA fragment or genomic DNA of P. falciparum was added to tHDA test 2 at 1000× limit of detection (final 50,000 copies) per reaction (lanes F, O, M, and K). In both A and B, the reactions were incubated at 64°C for 90 minutes before the tubes were applied to BESt cassettes. Negative reactions without input template (NTC) were also performed (lane N). Each reaction was performed in triplicate. In A, only NTC and reactions containing 50 copies of the input DNA template were shown. Only one of the triplicates was shown for each reaction. Label C on cassettes indicates the DNP line for IC; M, DIG line for Plasmodium; and S, FAM line for P. falciparum.
Similarly, test 2 detected 50 copies of the plasmid containing the P. vivax 18S rRNA fragment at 50 copies per reaction (Figure 3B). The amplification was specific for P. vivax. The test result was negative (Figure 3B) when 50,000 copies, which was 1000× the limit of detection of this test, of P. falciparum, P. ovale, P. malariae, or P. knowlesi were used as input template.
Although purified genomic DNA from Plasmodium species other than P. falciparum could not be obtained, total DNA from dried blood papers containing P. falciparum, P. vivax, P. ovale, and P. malariae was used to further verify the ability of test 1 to detect genomic DNA from these species. Different amounts (from 1 to 0.05 μL) of each extracted sample were added to the 50-μL test 1 reaction mix. The test was able to detect all four species using as low as 0.05 μL of sample input (data not shown).
Effect of Human Whole Blood on tHDA Amplification
The effect of human whole blood on tHDA amplification was evaluated using test 1. Our results demonstrated that the presence of 2 μL of human whole blood did not inhibit tHDA amplification. When 2 μL of whole blood was added to the 50-μL reaction mix of test 1, the analytical sensitivity of 50 copies per reaction was not compromised (data not shown). When the volume of the blood increased to 5 μL per reaction, we started to observe failure of amplification and detection when the target input was close to the limit of detection of 50 copies (data not shown).
Clinical Sensitivity and Specificity
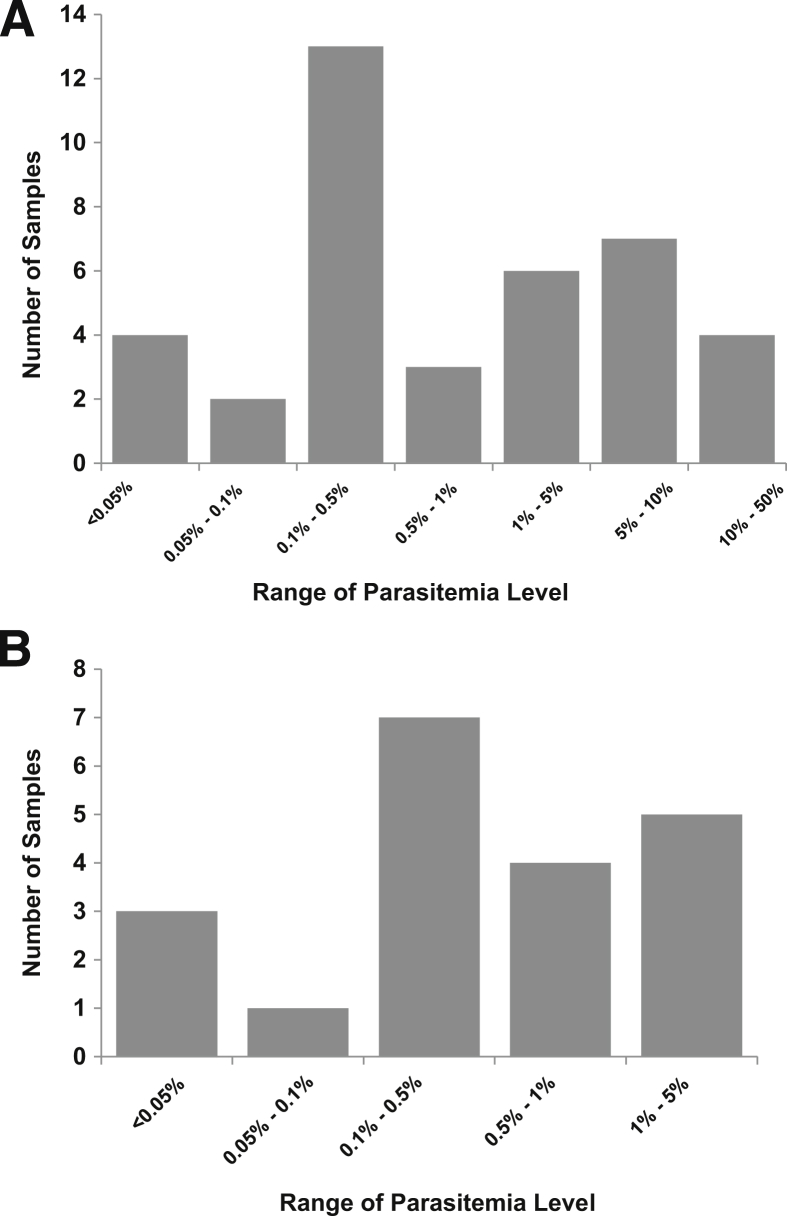
Among the 88 EDTA whole blood samples, 59 were identified as positive for malaria by microscopy/NASBAMT. These 59 samples contained various types of Plasmodium, with 39 P. falciparum, 16 P. vivax, 3 P. malariae, and 1 P. ovale. The parasitemia levels range from 0.01% to 36.9% (median, 0.5%), with most (n = 37) of the 59 positive samples being <1%. Ten of them were <0.1% (Table 2 and Figure 4). The five lowest parasitemia levels were 0.01%, 0.03%, 0.03%, 0.05%, and 0.05%.
Table 2.
Parasitemia Level of the Malaria-Positive Clinical Samples
| Sample | No. of samples | Range of parasitemia (%) | Mean parasitemia (%) | Median parasitemia (%) |
|---|---|---|---|---|
| P. falciparum | 39 | 0.01–36.9 | 4.17 | 0.7 |
| Others | 20 | 0.03–2 | 0.55 | 0.39 |
| All positives | 59 | 0.01–36.9 | 2.98 | 0.5 |
Figure 4.
Histogram of the parasitemia levels of the positive clinical samples. A: P. falciparum–positive samples. B: All other Plasmodium-positive samples.
The tHDA malaria assay test 1 identified 57 of the 59 microscopy/NASBAMT-positive malaria samples as positive for Plasmodium, of which 38 were identified as P. falciparum by tHDA. One sample, identified as P. falciparum by microscopy/NASBAMT with a parasitemia level of 0.05%, was negative by tHDA. Among the 20 non–P. falciparum malaria samples, 19 were identified by tHDA as Plasmodium positive but P. falciparum negative (16 P. vivax, 2 P. malariae, and 1 P. ovale by microscopy/NASBAMT). These samples were subjected to test 2, which identified the 16 P. vivax samples as positive for P. vivax. The two P. malariae samples and one P. ovale sample were identified by tHDA test 2 as negative for P. vivax. One sample was identified by tHDA as malaria negative but by NASBAMT as P. malariae. This sample was also negative by microscopy. The 14 negative and 15 Babesia samples were all identified by tHDA assay as negative for Plasmodium. Overall, compared with combined microscopy and NASBAMT results, the tHDA malaria assay resulted in a sensitivity of 96.6% (95% CI, 87.3% to 99.4%) and specificity of 100% (95% CI, 85.4% to 100%) (Table 3).
Table 3.
Clinical Performance of the tHDA Assay Compared with Microscopy/NASBAMT
| Microscopy/NASBAMA |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| P.falciparum | ||||
| HDA | Positive | 38 | 0 | 38 |
| Negative | 1 | 49 | 50 | |
| Total | 39 | 49 | 88 | |
| 95% CI | ||||
| Sensitivity (%) | 97.4 | 84.9–99.9 | ||
| Specificity (%) | 100.0 | 90.9–100.0 | ||
| P.vivax | ||||
|---|---|---|---|---|
| HDA | Positive | 16 | 0 | 16 |
| Negative | 0 | 72 | 72 | |
| Total | 16 | 71 | 88 | |
| 95% CI | ||||
| Sensitivity (%) | 100.0 | 75.9–100.0 | ||
| Specificity (%) | 100.0 | 93.7–100.0 | ||
| Other Plasmodium | ||||
|---|---|---|---|---|
| HDA | Positive | 3 | 0 | 3 |
| Negative | 1 | 84 | 85 | |
| Total | 4 | 84 | 88 | |
| 95% CI | ||||
| Sensitivity (%) | 75.0 | 21.9–98.7 | ||
| Specificity (%) | 100.0 | 94.6–100.0 | ||
| Overall | ||||
|---|---|---|---|---|
| HDA | Positive | 57 | 0 | 57 |
| Negative | 2 | 29 | 31 | |
| Total | 59 | 29 | 88 | |
| 95% CI | ||||
| Sensitivity (%) | 96.6 | 87.3–99.4 | ||
| Specificity (%) | 100.0 | 85.4–100.0 | ||
The analytical sensitivity of the tHDA assay from human whole blood samples was evaluated by serial dilutions of the frozen blood samples with a known parasitemia level into fresh blood. Four P. falciparum samples with parasitemia levels ranging from 0.3% to 8.8% were diluted up to 2000-fold. The final concentrations of the diluted samples were between 100 and 250 parasites per μL of blood. Each dilution was tested by tHDA test 1 in duplicate. All four samples were positive by tHDA test for at least one of the duplicates (Table 4). These results suggested that the sensitivity of the tHDA assay was approximately 200 parasites per μL of blood.
Table 4.
Analytical Sensitivity of the tHDA Malaria Assay
| Sample no. | Parasitemia (%) | Fold of dilution | Final concentration (parasites/μL)∗ | tHDA test 1 results (positive/total) |
|---|---|---|---|---|
| NS-046 | 2.3 | 1000 | 115 | 1/2 |
| NS-060 | 8.8 | 2000 | 220 | 1/2 |
| NS-109 | 0.3 | 100 | 150 | 2/2 |
| NS-110 | 1.1 | 333.3 | 165 | 1/2 |
The parasite concentration in blood was calculated using the following formulation: Number of Parasites/μL = % Parasitemia × (5 × 106 Red Blood Cells/mL)/(100 × Fold of Dilution).
Discussion
The tHDA-based malaria assay demonstrated the capability of detecting the five malaria causing Plasmodium species using only 2 μL of unprocessed human whole blood without the need for nucleic acid extraction. The tHDA amplifications were performed under isothermal conditions at a constant temperature of 64°C, which can be performed in a simple dry heat block or water bath. The detection of the target-specific amplification products was achieved by single-use disposable lateral-flow–based cassettes.
The tHDA-based malaria assay was able to identify the two most common Plasmodium species, P. falciparum and P. vivax. P. falciparum causes approximately 81% of total malaria worldwide (World Malaria Report 2011), with P. vivax making up the rest.29 P. falciparum is the most dangerous cause of malaria, with the highest mortality rate (World Malaria Report 2011). P. vivax is often nonfatal, but more widespread, than P. falciparum (World Malaria Report 2011). It is believed to account for 70 to 80 million cases annually (ie, 34% of the global incidence and 56% of malaria cases outside of Africa).29 Moreover, P. vivax can cause relapses years after the initial infection if not completely eradicated (Centers for Disease Control and Prevention, http://www.cdc.gov/malaria/about/facts.html). In contrast, the incidence of the other species of Plasmodium is seldom discussed in the literature. P. falciparum and P. vivax also have different drug-resistance profiles.2 Therefore, the ability of the tHDA assay for species-specific identification would help determine the proper treatment in a timely manner, especially in regions where more than one species is present.
The analytical sensitivity of the tHDA-based assay was 50 copies of Plasmodium genomic DNA or plasmid, in the presence of 2 μL of human whole blood spiked into the 50-μL reaction. Compared with the analytical sensitivity using pure DNA with spiked blood, the analytical sensitivity using patient samples was approximately 5 to 10 times lower (approximately 200 parasites per μL of blood). This could be because of the fact that, after long-term storage and repeated freeze and thaw, the parasite DNA might have partially degraded or the quality of the blood may have caused some inhibition to tHDA reactions. These possibilities can be tested in future studies by using fresh blood samples as a template for tHDA amplification.
In summary, the combination of the two tHDA tests offers a simple, cost-effective way to detect Plasmodium species and identify P. falciparum and P. vivax in unpressed blood without a sample preparation step. The work flow renders the assay essentially instrument free, eliminating the need for costly thermocyclers required for molecular diagnostic assays based on PCR. The tests can also be performed in a random access manner, allowing for on-demand testing that would be particularly beneficial for the rapid diagnosis of malaria and other blood-borne parasites, such as Babesia, in persons seen in both emergency departments and urgent care centers. Potentially, this assay can be developed as a low-cost and fast turnaround point-of-care assay performed in the field by people with minimal training.
Acknowledgment
We thank the Malaria Research and Reference Reagent Resource Center for providing dry blood filter papers containing P. vivax, P. ovale, and P. malariae blood samples.
Footnotes
Supported by National Institute of Allergy and Infectious Diseases award R43AI088787.
Disclosures: Y.L., B.L., and H.K. are employees of BioHelix Corporation, which supplied the tHDA amplification reagents.
References
- 1.Hay S.I., Guerra C.A., Gething P.W., Patil A.P., Tatem A.J., Noor A.M., Kabaria C.W., Manh B.H., Elyazar I.R., Brooker S., Smith D.L., Moyeed R.A., Snow R.W. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parija S.C., Praharaj I. Drug resistance in malaria. Indian J Med Microbiol. 2011;29:243–248. doi: 10.4103/0255-0857.83906. [DOI] [PubMed] [Google Scholar]
- 3.Abanyie F.A., Arguin P.M., Gutman J. State of malaria diagnostic testing at clinical laboratories in the United States, 2010: a nationwide survey. Malar J. 2011;10:340. doi: 10.1186/1475-2875-10-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houze S., Boly M.D., Le Bras J., Deloron P., Faucher J.F. PfHRP2 and PfLDH antigen detection for monitoring the efficacy of artemisinin-based combination therapy (ACT) in the treatment of uncomplicated falciparum malaria. Malar J. 2009;8:211. doi: 10.1186/1475-2875-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koita O.A., Doumbo O.K., Ouattara A., Tall L.K., Konare A., Diakite M., Diallo M., Sagara I., Masinde G.L., Doumbo S.N., Dolo A., Tounkara A., Traore I., Krogstad D.J. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg. 2012;86:194–198. doi: 10.4269/ajtmh.2012.10-0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar N., Pande V., Bhatt R.M., Shah N.K., Mishra N., Srivastava B., Valecha N., Anvikar A.R. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop. 2013;125:119–121. doi: 10.1016/j.actatropica.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Ashton R.A., Kefyalew T., Tesfaye G., Counihan H., Yadeta D., Cundill B., Reithinger R., Kolaczinski J.H. Performance of three multi-species rapid diagnostic tests for diagnosis of Plasmodium falciparum and Plasmodium vivax malaria in Oromia Regional State, Ethiopia. Malar J. 2010;9:297. doi: 10.1186/1475-2875-9-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubio J.M., Buhigas I., Subirats M., Baquero M., Puente S., Benito A. Limited level of accuracy provided by available rapid diagnosis tests for malaria enhances the need for PCR-based reference laboratories. J Clin Microbiol. 2001;39:2736–2737. doi: 10.1128/JCM.39.7.2736-2737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjitra E., Suprianto S., McBroom J., Currie B.J., Anstey N.M. Persistent ICT malaria P.f/P.v panmalarial and HRP2 antigen reactivity after treatment of Plasmodium falciparum malaria is associated with gametocytemia and results in false-positive diagnoses of Plasmodium vivax in convalescence. J Clin Microbiol. 2001;39:1025–1031. doi: 10.1128/JCM.39.3.1025-1031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoone G.J., Oskam L., Kroon N.C., Schallig H.D., Omar S.A. Detection and quantification of Plasmodium falciparum in blood samples using quantitative nucleic acid sequence-based amplification. J Clin Microbiol. 2000;38:4072–4075. doi: 10.1128/jcm.38.11.4072-4075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang F, Patel U, Manji R, Ebrahimzadeh A, Ennis JG, Teal AE, Ginocchio CC: Rapid real time detection and speciation of Plasmodium spp. using Nucleic Acid Sequence Based Amplification (NASBA) and molecular beacon detection. Presented at the ASM General Meeting, May 23-27, 2004, New Orleans, LA. Abstract number C368
- 12.Han E.T., Watanabe R., Sattabongkot J., Khuntirat B., Sirichaisinthop J., Iriko H., Jin L., Takeo S., Tsuboi T. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol. 2007;45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lau Y.L., Fong M.Y., Mahmud R., Chang P.Y., Palaeya V., Cheong F.W., Chin L.C., Anthony C.N., Al-Mekhlafi A.M., Chen Y. Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J. 2011;10:197. doi: 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu F., Gao Q., Zhou H., Cao J., Wang W., Lim C.S., Na S., Tsuboi T., Han E.T. Molecular test for vivax malaria with loop-mediated isothermal amplification method in central China. Parasitol Res. 2012;110:2439–2444. doi: 10.1007/s00436-011-2783-8. [DOI] [PubMed] [Google Scholar]
- 15.Lucchi N.W., Demas A., Narayanan J., Sumari D., Kabanywanyi A., Kachur S.P., Barnwell J.W., Udhayakumar V. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS One. 2010;5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polley S.D., Mori Y., Watson J., Perkins M.D., González I.J., Notomi T., Chiodini P.L., Sutherland C.J. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol. 2010;48:2866–2871. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pöschl B., Waneesorn J., Thekisoe O., Chutipongvivate S., Karanis P. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am J Trop Med Hyg. 2010;83:56–60. doi: 10.4269/ajtmh.2010.09-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamamura M., Makimura K., Ota Y. Evaluation of a new rapid molecular diagnostic system for Plasmodium falciparum combined with DNA filter paper, loop-mediated isothermal amplification, and melting curve analysis. Jpn J Infect Dis. 2009;62:20–25. [PubMed] [Google Scholar]
- 19.LaBarre P., Hawkins K.R., Gerlach J., Wilmoth J., Beddoe A., Singleton J., Boyle D., Weigl B. A simple, inexpensive device for nucleic acid amplification without electricity-toward instrument-free molecular diagnostics in low-resource settings. PLoS One. 2011;6:e19738. doi: 10.1371/journal.pone.0019738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tao Z.Y., Zhou H.Y., Xia H., Xu S., Zhu H.W., Culleton R.L., Han E.T., Lu F., Fang Q., Gu Y.P., Liu Y.B., Zhu G.D., Wang W.M., Li J.L., Cao J., Gao Q. Adaptation of a visualized loop-mediated isothermal amplification technique for field detection of Plasmodium vivax infection. Parasit Vectors. 2011;4:115. doi: 10.1186/1756-3305-4-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gandelman O., Jackson R., Kiddle G., Tisi L. Loop-mediated amplification accelerated by stem primers. Int J Mol Sci. 2011;12:9108–9124. doi: 10.3390/ijms12129108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An L., Tang W., Ranalli T.A., Kim H.J., Wytiaz J., Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J Biol Chem. 2005;280:28952–28958. doi: 10.1074/jbc.M503096200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent M., Xu Y., Kong H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004;5:795–800. doi: 10.1038/sj.embor.7400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Safeukui I., Millet P., Boucher S., Melinard L., Fregeville F., Receveur M.C., Pistone T., Fialon P., Vincendeau P., Fleury H., Malvy D. Evaluation of FRET real-time PCR assay for rapid detection and differentiation of Plasmodium species in returning travellers and migrants. Malar J. 2008;7:70. doi: 10.1186/1475-2875-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plowe C.V., Djimde A., Bouare M., Doumbo O., Wellems T.E. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg. 1995;52:565–568. doi: 10.4269/ajtmh.1995.52.565. [DOI] [PubMed] [Google Scholar]
- 26.Wooden J., Kyes S., Sibley C.H. PCR and strain identification in Plasmodium falciparum. Parasitol Today. 1993;9:303–305. doi: 10.1016/0169-4758(93)90131-x. [DOI] [PubMed] [Google Scholar]
- 27.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 28.Goldmeyer J., Kong H., Tang W. Development of a novel one-tube isothermal reverse transcription thermophilic helicase-dependent amplification platform for rapid RNA detection. J Mol Diagn. 2007;9:639–644. doi: 10.2353/jmoldx.2007.070012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendis K., Sina B.J., Marchesini P., Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]