Abstract
Congenital adrenal hyperplasia, due to 21-hydroxylase deficiency (21-OHD) is an autosomal recessive disorder of adrenal steroidogenesis caused by mutations in the CYP21A2 gene. Direct comparison of established and novel methodologies of CYP21A2 genetic analysis in a large cohort representing a wide range of genotypes has not been previously reported. We genotyped a cohort of 129 unrelated patients with 21-OHD, along with 145 available parents, using Southern blot (SB) analysis, multiplex ligation-dependent probe amplification (MLPA), PCR-based restriction fragment length polymorphism (RFLP) analysis, multiplex minisequencing and conversion-specific PCR, duplication-specific amplification, and DNA sequencing. CYP21A2 genotyping identified four duplicated CYP21A2 genes (1.53%) and 79 chimeric CYP21A1P/CYP21A2 genes (30.15%). Parental SB data were essential for determining the CYP21 haplotype in three cases, whereas PCR-based RFLP analysis was necessary for MLPA results to be accurately interpreted in the majority of cases. The comparison of different methods in detecting deletion and duplication showed that MLPA with PCR-based RFLP was comparable with SB analysis, with parental data of 100% sensitivity and specificity. DNA sequencing was required for the identification of 16 (6.1%) rare point mutations and determination of clinically significant chimera junction sites. MLPA with PCR-based RFLP analysis is an excellent substitute for SB analysis in detecting CYP21A2 deletion and duplication and a combination of MLPA, PCR-based RFLP, duplication-specific amplification, and DNA sequencing is a convenient and comprehensive strategy for mutation analysis of the CYP21A2 gene in patients with 21-OHD.
CME Accreditation Statement: This activity (“JMD 2013 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint sponsorship of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2013 CME Program in Molecular Diagnostics”) for a maximum of 48 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
Congenital adrenal hyperplasia [(CAH) OMIM 201910] is a group of autosomal recessive disorders characterized by impairment of cortisol biosynthesis, with or without impairment of aldosterone biosynthesis,1 and approximately 95% of cases are due to 21-hydroxylase deficiency (21-OHD).2 CAH is manifested in a variety of clinical severities comprised of three subtypes: i) classic salt wasting, ii) classic simple virilizing, and iii) nonclassic (mild or late onset) forms. The incidence of classic CAH worldwide ranges from 1 in 10,000 to 1 in 20,000 births.3
The steroid 21-hydroxylase gene, CYP21A2, is located in the human leukocyte antigen class III region on chromosome 6p21.3, closely adjacent in tandem with three other genes (serine/threonine kinase RP, complement C4, and tenascin TNX), forming a genetic module termed RCCX (RP-C4-CYP21-TNX).4 The RCCX module shows a high homology between the functional genes (RP1, CYP21A2, and TNXB) and the corresponding pseudogenes (RP2, CYP21A1P, and TNXA), leading to gene conversions5 and gene deletions due to homologous recombination, which inactivate the functional genes.
Molecular analysis of CYP21A2 gene is of great importance to understanding the etiology of 21-OHD, both in basic science and in clinical diagnosis.1 Approximately 95% of defective CYP21A2 genes seen in CAH fall into three categories6: i) approximately 65% to 70% are deleterious mutations derived from pseudogene CYP21A1P due to small gene conversions, including In2G [IVS-13 A/C->G (28%)], p.I172N (9%), p.V281L (9%), p.Q318X (4%), p.R356W (4%), E6 cluster [p.I235N, p.V236E, p.M238K (4%)], p.G110fx21 (3%), p.P30L (2%), and p.L307fx15 (1%)2, 7; ii) approximately 5% are spontaneous point mutations; and iii) approximately 25% to 30% are large gene rearrangements generated by unequal meiotic crossing over.8, 9, 10, 11, 12, 13
Large CYP21A2 gene rearrangements have been traditionally detected by Southern blot (SB) analysis.10, 14, 15 Other strategies including PCR-based restriction fragment length polymorphism (RFLP)16, 17, 18 and the real-time quantitative PCR–based method for CYP21A2 copy number detection9, 19 have also been described; however, only 82% of studied subjects showed agreement between SB analysis and the quantitative PCR-based method.9 Recently, multiplex ligation-dependent probe amplification (MLPA) has been increasingly used for identification of CYP21A2 gene deletion and duplication,11, 20 suggesting that MLPA may replace SB analysis as an efficient strategy to identify gene copy number. Coeli et al21 combined MLPA and SB in the genetic analysis of 20 Brazilian patients with 21-OHD. However, to our knowledge, a study comparing SB and MLPA in the evaluation of CYP21A2 gene rearrangements in a large cohort of patients representing diverse genotypes with 21-OHD has not been performed. Moreover, distinction among CYP21A2 deletions is clinically significant. Certain junction sites of CYP21A1P/CYP21A2 chimera have been associated with a milder phenotype, the so-called attenuated chimera.8 In addition, contiguous deletion of CYP21A2 and TNXB has been associated with Ehlers Danlos syndrome in a significant subset of CAH patients.22 Identification of these clinically significant chimeras has been incorporated into our mutation analysis strategy.
An additional challenge in the molecular evaluation of CYP21A2 results from the proximity and homology (98% in exons and 96% in introns15) of a pseudogene, CYP21A1P. Impure PCR products can be potentially generated by amplifying the pseudogene, with detection of pseudogene mutations rather than functional gene mutations. To address this issue, several methods have been developed in the past three decades, such as high-resolution melting curve,23 locus-specific PCR,9, 13, 24, 25 denaturing high-performance liquid chromatography analysis,26 and multiplex minisequencing.25 These methods adopt a common core strategy based on the known mutations and have some limitations with the maximum overall sensitivity of mutation detection achieved, which is reported to range from 90%9, 27 to 95%.28
We sought to establish a multistep definitive strategy to perform a convenient and comprehensive mutation analysis of the CYP21A2 gene for patients with 21-OHD. Thus, we compared data from SB, MLPA, and multiplex minisequencing and conversion-specific PCR (MMCP) in 129 unrelated patients with 21-OHD, and we included the identification of clinically significant chimeras.
The strategy presented here provides an accurate and inclusive genetic picture of the CYP21A genes in each patient with 21-OHD, and thus represents a useful tool in the diagnosis and management of CAH.
Materials and Methods
Characterization of the Patients
Patients with 21-OHD underwent comprehensive clinical29 and genetic30 evaluations as part of an ongoing observational study at the National Institutes of Health Clinical Center (http://clinicaltrials.gov/ct2/show/NCT00250159?term=NCT00250159&rank=1, last accessed February 1, 2013). DNA was obtained from peripheral blood leukocytes using standard methodology. All studies were approved by the Eunice Kennedy Shriver National Institute of Child Health and Human Development Institutional Review Board and the MedStar Health Institutional Review Board. All patients and their family members gave informed consent for DNA analysis, and all minors ≥8 years old gave their assent.
SB Analysis and MMCP
To genotype the copy number variation of RCCX modules, SB was performed for unrelated patients with 21-OHD and their parents using TaqI and PshAI (both from New England BioLabs, Ipswich, MA) as previously described.30 MMCP analysis was conducted to identify the 12 most common CYP21A2 mutations, 12 single nucleotide polymorphic markers, and the 30-kb deletion, as previously described.30
MLPA
MLPA, a rapid and high-throughput technique for copy number quantification31 was performed using the SALSA MLPA Kit P050-B2 CAH (MRS Holland, the Netherlands) according to the manufacturer’s recommendations. Amplification products were run on ABI Genetic Analyzer 3130xl (Applied Biosystems, Foster City, CA). The kit is designed to detect deletions and duplications of one or more exons of CYP21A and TNX genes, and contains five probes for CYP21A2 (p.P30L, p.G110fsX21, p.I172N, E6cluster, and p.Q318X mutations) and three CYP21A1P-specific probes.
PCR-Based RFLP
To genotype large gene deletions at the centromeric tail of the RCCX module, primers CYP779f and Tena32F18 (Table 1) were used to amplify a fragment of 8515-bp by using the Expand Long-Range dNTPack (Roche Applied Science, Indianapolis, IN). The reaction condition followed the manufacturer’s recommendations, except for using an annealing temperature of 62°C. The 8.5 kb PCR products were purified with QIAquick PCR Purification Kit (Qiagen, Valencia, CA) and were then digested with 20 U TaqαI (New England BioLabs) for 2.5 hours at 37°C followed by 15 minutes at 85°C. Digested products were electrophoresed for 16 hours on a 0.8% agarose gel.
Table 1.
Primers Used for PCR and Sequencing
| Primer | Sequence | Location | Genes | Purpose | Reference |
|---|---|---|---|---|---|
| seq1 | 5′-CCCCAAAACAGTCTACACAG-3′ | 5′UTR | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq2 | 5′-GGCCCGAATTTCTGAGTCA-3′ | Exon1 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq3 | 5′-TGTGGTGGTGCTGAACTC-3′ | Exon2 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq4 | 5′-AGCAGGGAGTAGTCTCCC-3′ | Exon3 | CYP21A2 (G110)∗ | Sequencing | Present study |
| seq5 | 5′-GCTTTCCAGAGCAGAGACC-3′ | Exon3 | CYP21A1P (G110fx21)∗ | Sequencing | Present study |
| seq6 | 5′-TTCTGTGAGGTAAGGCTGG-3′ | Exon3/Intron3 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq7 | 5′-TACTGTGAGAGGCGAGGCTGA-3′ | Intron3 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq8 | 5′-TTTCTCAGGGTGAGGACCTG-3′ | Exon5/Intron5 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq9 | 5′-CCTCACTCAGCTCTGAGCACT-3′ | Intron7 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq10 | 5′-TTGTGCCCTTAGCCTTGC-3′ | Exon8 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq11 | 5′-TTTCCTCACTCATCCCCAAC-3′ | Intron8 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq12 | 5′-AAGAACTCCAGAGCTCTGGC-3′ | Exon10 | CYP21A2 and CYP21A21P | Sequencing | Present study |
| seq13 | 5′-TCTCAGCTTCATTTCCGTGA-3′ | 3′UTR | CYP21A2 and CYP21A21P | Sequencing | Present study |
| CYP779f | 5′-AGGTGGGCTGTTTTCCTTTCA-3′ | 5′ end | CYP21A2 and CYP21A21P | PCR (sense) | 18 |
| Tena32F | 5′-CTGTGCCTGGCTATAGCAAGC-3′ | Intron32 | TNXB | PCR (antisense) | 18 |
| 21BF1 | 5′-CCCAGGTGGGGGCGGACACTA-3′ | 5′ end | CYP21A2 | PCR (sense) | 32 |
| XA† | 5′-ACTCCAGGTGGGAGTCAAGAA-3′ | 120-bp del | TNXA | PCR (antisense) | Present study |
Primer seq4 and seq5 are specific to wild-type and mutant of p.G110, respectively.
XA is reverse complementary to the XA-specific antisense primer32.
Sequencing CYP21A2 and Chimeric CYP21A1P/CYP21A2 Genes
To identify point mutations in the CYP21A2 and chimeric CYP21A1P/CYP21A2 genes, another aliquot (50 μL PCR product [CYP779f/Tena32F]) was obtained for each sample. After agarose gel discrimination, the PCR product was purified with 1 U of shrimp alkaline phosphatase (USB, Cleveland, OH) and 3.5 U of exonuclease I (USB), followed by sequencing using Big Dye Terminator V3.1 (Applied Biosystems) and inner sequencing primers (Table 1) on ABI Genetic Analyzer 3100xl (Applied Biosystems). Results were aligned with the reference CYP21A2 (ENSG00000206338) and CYP21A1P (ENSG00000204338) sequences.
21BF1/XA Amplification to Identify Duplicated CYP21A2 Gene
Primers 21BF1 and XA (Table 1) were used to amplify a 5.6-kb fragment by Expand Long-Range dNTPack for identification of duplicated CYP21A2 genes telomeric to the TNXA gene. The reaction conditions followed the manufacturer recommendations except for using an annealing temperature of 58°C and addition of 2% DMSO. Primer 21BF1 matched the 5′ end of the CYP21A2 gene exclusively,33 and primer XA was located in the TNXA-specific region containing a 121-bp deletion closing the reading frame.34 The 5.6-kb PCR products were then purified and sequenced as described above.
Results
Methods Comparison
SB analysis, MLPA, PCR-based RFLP, DNA sequencing, and MMCP were performed on 129 unrelated patients with 21-OHD and 145 available parents. Based on our analysis of the cohort, each method had advantages and disadvantages (Table 2). Parental SB data were essential for determining the CYP21 haplotype in three cases (2.32%), two of which involved a combination of duplication and deletion.
Table 2.
Summary of Different Methods for Genotyping CYP21A2 Genes
| Method | Targeted mutations | Advantage | Disadvantage |
|---|---|---|---|
| SB | 1. Deletion and duplication | 1. Informative for the whole RCCX module | 1. Laborious |
| 2. Radioactive biohazard | |||
| 3. Requires high quantity and good quality DNA | |||
| 4. RCCX trimodule causes densitometric error | |||
| 5. Cannot determine junction sites of chimeras | |||
| 6. Parental data often required | |||
| MLPA | 1. Deletion and duplication | 1. Convenient | 1. Duplication masks deletion. |
| 2. Point mutations | 2. Rapid | 2. Signals are sensitive to quality of DNA and probe binding. | |
| 3. High-throughput | 3. Requires extensive knowledge of RCCX module | ||
| 4. Low DNA quantity required | 4. Difficult to determine phase of deletion/duplication | ||
| 5. Indicates junction sites of chimeras | 5. Results must be confirmed by other methods | ||
| PCR-RFLP | 1. Deletion | 1. Convenient | 1. Cannot determine junction sites of chimeras |
| 2. Low DNA quantity and quality required | |||
| MMCP | 1. Deletion and duplication | 1. Convenient | 1. Cannot detect novel mutations |
| 2. Known point mutations | 2. Rapid | 2. Cannot determine phase of deletion/duplication | |
| 3. High-throughput | 3. Some results are equivocal | ||
| Sequencing | 1. Point mutations | 1. Detects point mutations | 1. Risk of false negative/positive results |
| 2. Determines junction sites of chimeras | 2. Primers must be carefully designed |
MLPA, multiplex ligation-dependent probe amplification; MMCP, multiplex mini-sequencing and conversion-specific PCR; PCR-RFLP, PCR-based restriction fragment length polymorphism analysis; RCCX, RP-C4-CYP21-TNX; SB, Southern blot.
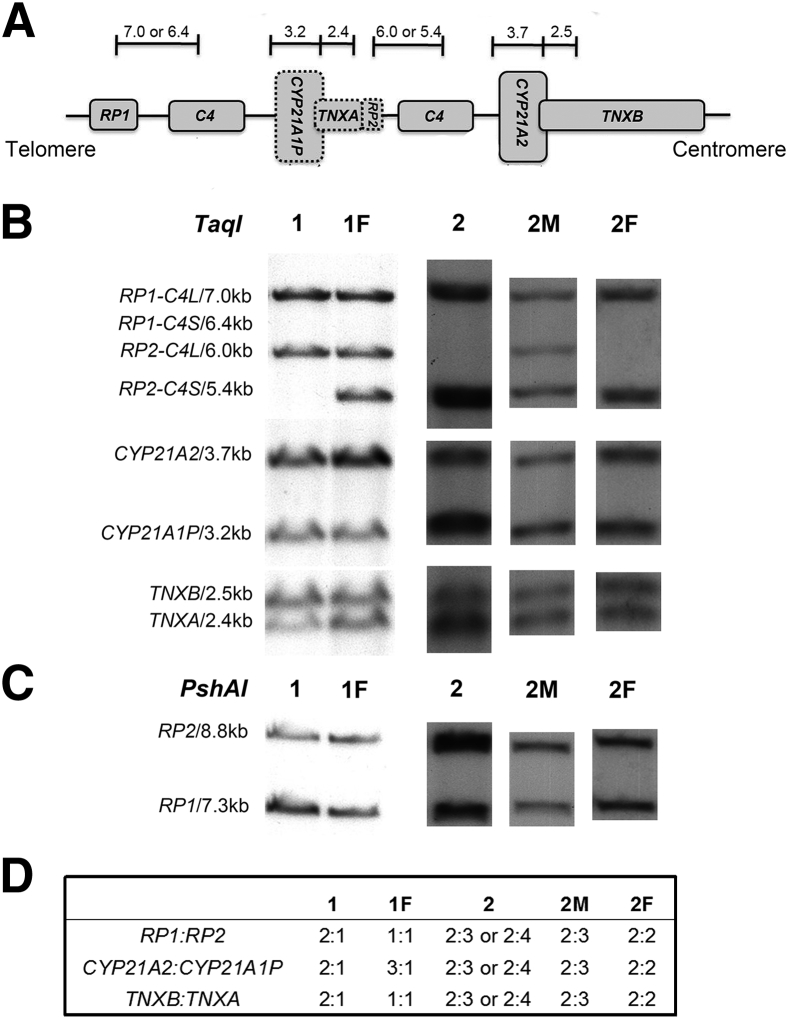
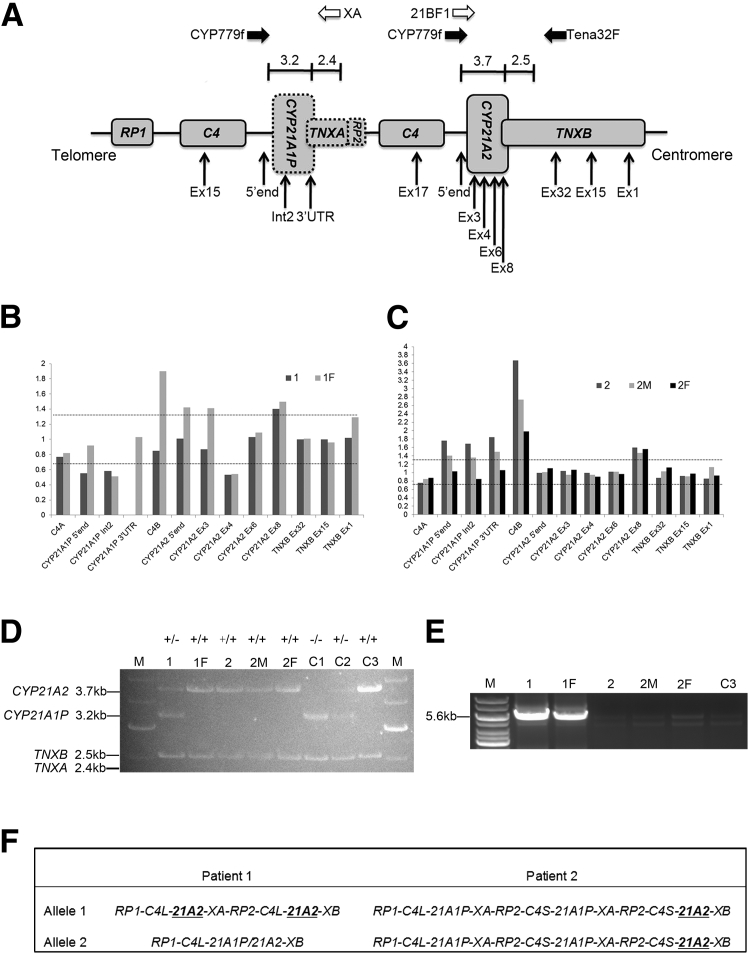
A multistep strategy was necessary to accurately define genotype, illustrated by two representative families (Figure 1). The SB densitometric ratio of CYP21A2 versus CYP21A1P in patient 1 was 2:1 (Figure 1, B and D), consistent with MLPA data (Figure 2B). This observation thus led to an interpretation of no CYP21A2 gene deletion/duplication in patient 1. However, a 3.2-kb CYP21A1P-specific band was observed in PCR-based RFLP, indicating CYP21A2 gene deletion (Figure 2D) and we obtained a positive amplification using primers 21BF1/XA (Figure 2E), suggesting CYP21A2 gene duplication. DNA sequencing identified p.P30L, p.G110fsX21, and p.I172N mutations in heterozygosity in CYP779f/Tena32F amplicon and p.I172N, E6cluster, p.V281L, and p.L307fx15 mutations in 21BF1/XA amplicon from patient 1. Therefore, as shown in Figure 2F, the genotype of patient 1 was two TNXB-adjacent CYP21A2 genes (one CH-1 chimera8 and one with p.I172N), and one TNXA-adjacent CYP21A2 gene (carrying p.I172N, E6cluster, p.V281L, and p.L307fx15 mutations), consistent with MMCP data.
Figure 1.
Southern blot analysis of RCCX module in two representative patients and their respective parents. A: Schematic of a typical RCCX bimodule allele. Dashed boxes denote pseudogenes; short vertical lines, TaqI restriction sites; numbers, fragment sizes (in kb). B and C: TaqI and PshAI Southern blots. Enzyme digested fragments with size (in kb) correspond to target gene(s). D: Densitometric ratios were measured in Southern blots. The ratios indicate gene dosages, which can be used to infer gene arrangement. Columns 1 and 1F represent patient 1 and the father, respectively; columns 2, 2M, and 2F represent patient 2, the mother, and the father, respectively.
Figure 2.
MLPA, PCR-based RFLP, and PCR-based duplication analysis of two representative patients and their respective parents. A: Scheme shows MLPA probes and PCR primers. Dashed boxes indicate pseudogenes; white arrows, primers used to amplify duplicated CYP21A2 gene; black arrows, PCR-based RFLP primers; short vertical lines, TaqαI restriction site of CYP779f/Tena32F amplified products, with numbers showing the fragment size (in kb); vertical arrows, hybridization sites of the MLPA probes. ex, exon; int, intron; UTR, untranslated region. B and C: Normalized MLPA data of families of patient 1 and patient 2 with probes on x axis. Columns correspond to normalized electropherogram peak areas calculated using Coffalyser software. Diamonds denote results of point mutations; circles denote results of chimeric genes. Ratio values (y axis) between 0.7 and 1.3 (dashed lines) indicate two copies, bottom dashed lines (one copy) and top dashed lines (two copies). 1 and 1F represent patient 1 and the father, respectively. 2, 2M, and 2F represent patient 2, the mother, and the father, respectively. D: Electropherogram of TaqαI digested CYP779f/Tena32F amplified products. Target fragments are labeled with gene names and size. −, CYP21A2 deletion on one allele; +, no deletion; M, 1 kb DNA ladder; C1–C3, controls with two, one, and no CYP21A2 deletions, respectively. The 3.2 kb band for the CYP21A1P gene was not detected in samples 1F, 2, 2M, and 2F, which indicated no CYP21A2 deletion; sample 1 shows one 3.2 kb band for CYP21A1P gene, confirming CYP21A2 deletion. E: Electropherogram of products of duplication-specific amplification using primer 21BF1 and XA; no amplification is observed without CYP21A2 duplication. A positive 5.6 kb band for samples 1 and 1F indicates CYP21A2 duplication. The faint bands shown in samples 2, 2M, 2F, and C3 result from unspecific amplification. F: Genotyping results of patient 1 and 2. The following 21A2, 21A1P, XA, and XB stand for CYP21A2, CYP21A1P, TNXA, and TNXB, respectively. CYP21A2 genes identified with point mutations are set bold and underlined.
Patient 2 SB showed a weaker band of CYP21A2 gene than CYP21A1P gene indicating a possible CYP21A2 gene deletion, yet the bands were too dense to measure the exact ratios, rendering an ambiguous interpretation (Figure 1). The SB on the parents of patient 2 revealed the ratios of CYP21A2 versus CYP21A1P 2:3 in the mother and 2:2 in the father, illustrating that parental SB data were essential for determining the CYP21 haplotype (Figure 1). MLPA showed that patient 2 had two dosages of CYP21A2 gene (Figure 2C). No CYP21A2 gene deletion and duplication was observed in the PCR-based RFLP (Figure 2D) or 21BF1/XA amplification (Figure 2E). DNA sequencing of the CYP779f/Tena32F amplicon of patient 2 identified the p.V281L mutation in homozygosity, consistent with the MMCP data. Thus, we inferred that patient 2 carried two dosages of the CYP21A2 gene, both with the p.V281L mutation, and four dosages of CYP21A1P gene. In both cases, accurate genotyping required PCR-based RFLP in combination with MLPA and parental data for SB due to duplication and/or deletion involving the CYP21 genes.
We compared data from SB analysis of probands and available parents, MLPA with PCR-based RFLP and MMCP in detecting CYP21A2 gene deletion and duplication (Table 3). We found 100% agreement between SB with parental data and MLPA with PCR-based RFLP, both yielding authentic results, whereas MMCP showed 91.6% sensitivity (76 of 83 alleles; 95% confidence interval [CI] 86 to 98) and 88.8% specificity (159 of 179 alleles; 95% CI 84 to 93) because it missed one duplicated CYP21A2 gene, and the equivocal results were also observed in 13 individuals. Indeed, both SB analysis and MLPA with PCR-based RFLP identified CYP21A2 gene deletion on one allele in six of these 13 individuals.
Table 3.
Comparison of Different Methods Used to Detect Deletion and Duplication of the CYP21A2 Gene
| Detection | Combined methods∗ | SB† | MLPA andPCR-RFLP | MMCP |
|---|---|---|---|---|
| Deletion on one allele | 49 | 49 | 49 | 43 |
| Deletion on two alleles | 15 | 15 | 15 | 15 |
| Duplication | 4 | 4 | 4 | 3 |
| No deletion and duplication | 65 | 65 | 65 | 58 |
| Status unknown | 0 | 0 | 0 | 13 |
Numbers of detected samples are shown.
Final results were given based on all assays used.
Based on proband and parental data.
Mutation Analysis and Genotyping Strategy Proposal
The CYP21A2 genotyping results for all 129 individuals revealed a wide spectrum of mutations (Table 4), and each mutation or deletion was confirmed by at least two methods. Of 262 CYP21A2 genes, four (1.53%) were duplicated CYP21A2 genes and 79 (30.15%) were chimeric CYP21A1P/CYP21A2 genes. Of the 79 chimeras, 76 truncated the functional CYP21A2 gene, whereas the other three were of attenuated form associated with less severe phenotype.8 Among the nine most common mutations, In2G (23.57%), p.V281L (17.87%), and p.I172N (13.69%) were most frequent. Sixteen uncommon mutations were identified by sequencing, accounting for 6.11% allele frequency.
Table 4.
CYP21A2 Mutations Found in 129 Unrelated Patients with 21-OHD
| Type | Allele No. | Frequency in 262 alleles (%) |
|---|---|---|
| Classic chimeric CYP21A1P/CYP21A2∗ | ||
| CH-1 | 30 | 11.45 |
| CH-2 | 0 | 0.00 |
| CH-3 | 4 | 1.53 |
| CH-5 | 29 | 11.07 |
| CH-6 | 0 | 0.00 |
| CH-7 | 0 | 0.00 |
| CH-8 | 13 | 4.96 |
| Total | 76 | 29.01 |
| Attenuated chimeric CYP21A1P/CYP21A2∗ | ||
| CH-4 | 3 | 1.15 |
| CH-9 | 0 | 0.00 |
| Total | 3 | 1.15 |
| Total chimeric CYP21A1P/CYP21A2 | ||
| 79 | 30.15 | |
| Most common mutations† | ||
| In2G | 62 | 23.66 |
| V281L | 47 | 17.94 |
| I172N | 36 | 13.74 |
| Q318X | 11 | 4.20 |
| R356W | 9 | 3.44 |
| L307fx15 | 6 | 2.29 |
| E6cluster | 5 | 1.91 |
| G110fsX21 | 2 | 0.76 |
| P30L | 1 | 0.38 |
| Other point mutations | ||
| 16 | 6.11 | |
| CYP21A2 duplications | ||
| 4 | 1.53 | |
Chimeric types were assigned according to Chen et al.8
Nomenclature at the protein level is based on conventional codon numbering. Nomenclature at the cDNA level, based on ENST00000418967, is as follows: P30L (c.92C>T), In2G (c.293-13A/C>G), G110fsX21 (c.332_339del), I172N (c.518T>A), D183E (c.552C>G), D234D (c.705T>C), E6cluster [I236N (c.710T>A), V237E (c.713T>A), M239K (c.719T>A)], V281L (c.844G>T), L307fx15 (c.923_924insT), Q318X (c.955C>T), and R356W (c.1069C>T); 21-OHD, 21-hydroxylase deficiency.
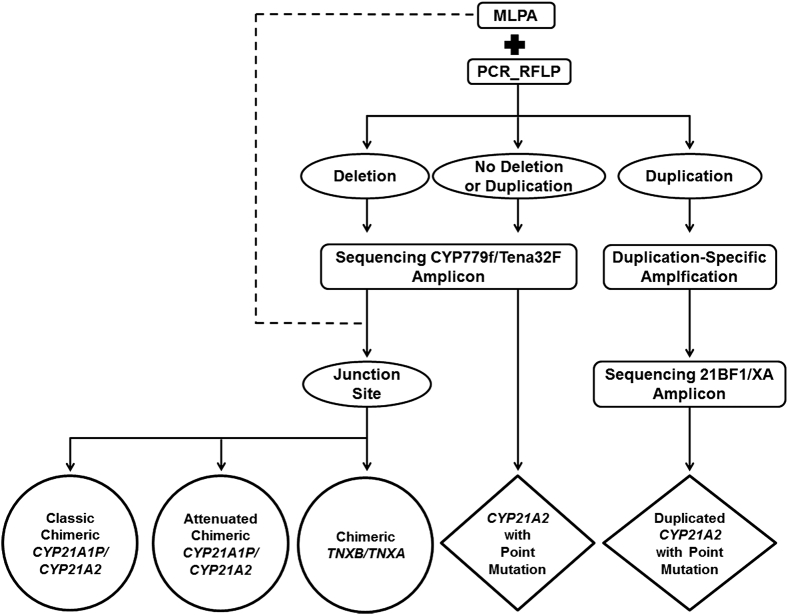
Based on our results, we developed a comprehensive strategy for the genetic analysis of the CYP21A2 gene in CAH patients (Figure 3). First, MLPA with PCR-based RFLP should be performed on each patient to identify gene deletion and/or duplication. Sequencing CYP779f/Tena32F and/or 21BF1/XA amplicon is then conducted to identify point mutations. The junction site of the chimeric CYP21A1P/CYP21A2 gene is defined by the identified point mutations and the MLPA data.
Figure 3.
Molecular analysis strategy of the CYP21A2 gene in CAH patients. The target of this strategy is to identify mutations in deleted, duplicated, and normal CYP21A2 genes. Final genotyping result of each patient is given in the bottom row, including chimera type of CYP21A1P/CYP21A2 genes and point mutations in normal and duplicated CYP21A2 genes. Rectangles denote the methods; ovals, results from the upstream methods; diamonds, results of point mutations; circles, results of chimeric genes. The junction site of each chimeric gene was identified based on data from MLPA (dashed lines) and sequencing CYP779f/TenaF amplicon.
Discussion
CAH due to 21-OHD is a life-threatening disease, and neonatal screening for CAH is performed in the United States. The molecular diagnosis is useful in evaluating borderline cases3 but is fraught with challenges. Our study is the first comprehensive comparison of various existing methodologies that has been proposed for CYP21A2 mutation analysis.
In our study, we conducted extensive mutation analysis of the CYP21A2 gene for 129 unrelated patients with 21-OHD, using SB analysis, MLPA, PCR-based RFLP, MMCP, and DNA sequencing. We found that MLPA with PCR-based RFLP when combined with duplication-specific amplification and DNA sequencing is the preferred methodology, and this provides an accurate and comprehensive approach to the molecular diagnosis of CAH due to 21-OHD.
Our extensive comparison of various methodologies revealed that MLPA in combination with PCR-based RFLP is comparable with SB in detecting the CYP21A2 gene deletion and duplication. The sole use of MLPA methodology has limitations and it requires a deep knowledge of CYP21A2 gene arrangements. It should be noted that a mutation or polymorphism in a probe-binding region could cause a reduction in the relative peak area, even when it is not located exactly on the ligation site. Another major limitation of MLPA is that the probes do not determine the phase of the deletion and duplication. Thus, it is possible that adjacent probes with apparently reduced signals are on different chromosomes or represent noncontiguous point mutations. Additionally, based on our experience, some probes of the MLPA kit P050-B2 CAH (MRC-Holland, Holland, the Netherlands) are extremely sensitive to DNA quality and small changes in experimental manipulation. Our analysis of patient 1 provides evidence that MLPA is prone to producing an inconclusive result without PCR-based RFLP data when CYP21A2 deletion is masked by duplication. Thus, as suggested by Concolino et al,20 for most if not all cases, MLPA alone is not able to yield accurate results, and thus a combination of MLPA and PCR-based RFLP should be used for the analysis of CYP21A2 gene deletion and duplication in patients with 21-OHD.
SB analysis is the reference standard in detection of large gene arrangement in the RCCX module.10, 14, 15 However, densitometric screening of fragments is prone to error for an allele with a trimodular RCCX, which is estimated to occur in 2.7% to 7% of patients.30, 35 Our analysis of SB data showed that in three out of 129 cases (2.32%), two of which involved duplications and deletions, and parental data were necessary for accurate interpretation. Because SB is laborious, time-consuming, involves the use of radioisotopes, and requires a relatively large quantity of DNA sample, alternative methodologies are preferable. In our comparison of SB, MLPA with PCR-based RFLP, and MMCP, we found that SB and MLPA with PCR-based RFLP are impressively equivalent and more sensitive than MMCP in detecting CYP21A2 gene deletion and duplication. The combined MLPA with PCR-based RFLP method shows great advantage in comparison with SB because it requires a smaller amount of genomic DNA (25 to 250 ng) and no use of radioisotopes. None of our cases required parental samples for interpretation using the combined MLPA with PCR-based RFLP method. Moreover, MLPA also indicates particular point mutations (c.1-126C>T, c.1-112G>A, c.1-110T>C, c.1-103A>G, p.G110fsX21, p.I172N, p.I236N, p.V237E, p.M239L, and p.Q318X), as these mutations are located in the probe-binding regions. MLPA with PCR-based RFLP, and is, therefore, is an informative, convenient, and sensitive tool for detection of deletion and duplication, and the junction site of each chimera could be preliminarily predicted based on the contiguous probes.8
DNA sequencing is often needed in CYP21A2 genotyping, especially for identification of uncommon mutations. Previous studies8, 13 show that analysis of the junction sites of chimeric CYP21A1P/CYP21A2 genes may be useful in the clinical diagnosis and genetic counseling of patients with 21-OHD because some chimeras attenuate the severity of the CAH phenotype. Sequencing of the CYP21A2 gene, especially in cases with a chimeric CYP21A1P/CYP21A2 gene, may be necessary to localize the junction site. Moreover, three of the common mutations (p.P30L, p.V281L, and p.R356W) cannot be detected by the MLPA kit P050-B2 CAH (MRC-Holland), underlining the utility of sequencing.
Duplicated CYP21A2 genes in RCCX modules have been found in previous studies.30, 35, 36 In the present study, primers 21BF1 and XA were used for the detection of duplication, and the amplicon was then sequenced to identify point mutation in the duplicated CYP21A2 gene. The mutation carrier ship in the duplicated CYP21A2 gene is of clinical importance through modulation of phenotypes. An alternative PCR method to detect a duplicated CYP21A2 gene was previously proposed.35 However, the sense primer used for the CYP21A2 gene in this methodology is located at the sequence of wild-type p.G110fsX21, suggesting that these primers will not work if the duplicated CYP21A2 gene carries a p.G110fsX21 mutation.
In our study, we propose a comprehensive strategy for accurate mutation analysis of the CYP21A2 gene: MLPA, PCR-based RFLP, duplication-specific amplification, and DNA sequencing. Vrzalová et al11 used a similar strategy to study chimeric CYP21A1P/CYP21A2 genes in a CAH cohort. However, to amplify the CYP21A1P/CYP21A2 genes, they used CYP21A1P-specific AF1 and CYP21A2-specific 21BR primers, located in the 5′ end of CYP21A1P gene and the 3′ untranslated region of CYP21A2 gene, respectively. Our recent study,37 uncovers that 21BR primer is not CYP21A2 specific because CYP21A1P-specific polymorphisms were observed in high frequency (5 of 28 alleles) in the primer-binding region. Thus, amplification of primers AF1/21BR will undoubtedly produce false-negative results. Other most commonly used PCR primers are two overlapping CYP21A2-specific primer pairs, one located in the 5′ end and exon 3 (wild-type of p.G110fx21) and the other in exon 6 (wild-type of E6cluster) in the 3′ end.13, 25 This methodology would fail to amplify the CYP21A2 gene carrying both p.G110fx21 and E6culster mutations, although this CYP21A2 gene has not been found yet in Argentinean,38 Italian,39 Chinese,13 or American30 populations. Day et al40 suggest that using internal mutation-specific primers might yield false-negative results due to preferential amplification or allele dropout; thus, a PCR product with a full CYP21A2 gene containing the 5′ end of the CYP21A2 gene and the downstream sequence of the TNXB gene is preferable in CYP21A2 mutation analysis.
We recently described that a significant subset (13%) of patients with 21-OHD CAH suffer from clinical features of Ehlers-Danlos syndrome due to TNXB haploinsufficiency, and these patients carrying a CYP21A2 deletion had a clinically significant contiguous CYP21A2 deletion that extended into the TNXB gene.22 Most strategies for CYP21A2 genetic analysis12, 13, 21, 23 are not able to identify this clinically important chimera. However, in our strategy, the primer pair, CYP779f and Tena32F (Figure 2A), is capable of amplification of any CYP21A2, CYP21A1P, or chimeric CYP21A1P/CYP21A2 gene that is closely adjacent to the TNXB gene. Additionally, by using our strategy, we are able to identify a chimeric TNXA/TNXB gene,16 which is expected to be dysfunctional due to a 121-bp deletion shifting the reading frame from the TNXA gene and resulting in TNXB haploinsufficiency.
To our knowledge, this is the first direct comparison of various commonly used methods to analyze the CYP21A2 gene in a large cohort of CAH patients. By combining existing methodologies and optimizing primer pair choice, we have established a convenient and comprehensive approach to the molecular analysis of CAH. We have also addressed the clinically important issue of identifying attenuated and tenascin-X affected chimera. This strategy will provide genetic information that surpasses previously described strategies and will inevitably aid in clinical care and genetic counseling of patients affected by CAH due to 21-OHD.
Acknowledgments
We thank the patients and their parents for their participation in this study, Dr. Frank Fujimura and Esoterix Laboratories for their excellent technical support, and the NIA Core Laboratory staff for DNA extraction and sample processing.
Footnotes
Supported by the Intramural Research Programs of the NIH, National Institute on Aging, the Clinical Center, and the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
D.P.M. serves as a Commissioned Officer in the United States Public Health Service.
Z.X. and W.C. contributed equally to this work.
References
- 1.Merke D.P., Bornstein S.R. Congenital adrenal hyperplasia. Lancet. 2005;365:2125–2136. doi: 10.1016/S0140-6736(05)66736-0. [DOI] [PubMed] [Google Scholar]
- 2.White P.C., Speiser P.W. Congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Endocr Rev. 2000;21:245–291. doi: 10.1210/edrv.21.3.0398. [DOI] [PubMed] [Google Scholar]
- 3.Speiser P.W., Azziz R., Baskin L.S., Ghizzoni L., Hensle T.W., Merke D.P., Meyer-Bahlburg H.F., Miller W.L., Montori V.M., Oberfield S.E., Ritzen M., White P.C. Congenital adrenal hyperplasia due to steroid 21-hydroxylase deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2010;95:4133–4160. doi: 10.1210/jc.2009-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Z., Mendoza A.R., Welch T.R., Zipf W.B., Yu C.Y. Modular variations of the human major histocompatibility complex class III genes for serine/threonine kinase RP, complement component C4, steroid 21-hydroxylase CYP21, and tenascin TNX (the RCCX module). A mechanism for gene deletions and disease associations. J Biol Chem. 1999;274:12147–12156. doi: 10.1074/jbc.274.17.12147. [DOI] [PubMed] [Google Scholar]
- 5.White P.C., Tusie-Luna M.T., New M.I., Speiser P.W. Mutations in steroid 21-hydroxylase (CYP21) Hum Mutat. 1994;3:373–378. doi: 10.1002/humu.1380030408. [DOI] [PubMed] [Google Scholar]
- 6.Speiser P.W., White P.C. Congenital adrenal hyperplasia. New Engl J Med. 2003;349:776–788. doi: 10.1056/NEJMra021561. [DOI] [PubMed] [Google Scholar]
- 7.Baumgartner-Parzer S.M., Schulze E., Waldhäusl W., Pauschenwein S., Rondot S., Nowotny P., Meyer K., Frisch H., Waldhauser F., Vierhapper H. Mutational spectrum of the steroid 21-hydroxylase gene in Austria: identification of a novel missense mutation. J Clin Endocrinol Metab. 2001;86:4771–4775. doi: 10.1210/jcem.86.10.7898. [DOI] [PubMed] [Google Scholar]
- 8.Chen W., Xu Z., Sullivan A., Finkielstain G.P., Van Ryzin C., Merke D.P., McDonnell N.B. Junction site analysis of chimeric CYP21A1P/CYP21A2 genes in 21-hydroxylase deficiency. Clin Chem. 2012;58:421–430. doi: 10.1373/clinchem.2011.174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olney R.C., Mougey E.B., Wang J., Shulman D.I., Sylvester J.E. Using real-time, quantitative PCR for rapid genotyping of the steroid 21-hydroxylase gene in a north Florida population. J Clin Endocrinol Metab. 2002;87:735–741. doi: 10.1210/jcem.87.2.8273. [DOI] [PubMed] [Google Scholar]
- 10.White P.C., Vitek A., Dupont B., New M.I. Characterization of frequent deletions causing steroid 21-hydroxylase deficiency. Proc Natl Acad Sci U S A. 1988;85:4436–4440. doi: 10.1073/pnas.85.12.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vrzalová Z., Hrubá Z., Hrabincová E.S., Vrábelová S., Votava F., Kolouŝková S., Fajkusová L. Chimeric CYP21A1P/CYP21A2 genes identified in Czech patients with congenital adrenal hyperplasia. Eur J Med Genet. 2011;54:112–117. doi: 10.1016/j.ejmg.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Choi J.H., Jin H.Y., Lee B.H., Ko J.M., Lee J.J., Kim G.H., Jung C.W., Lee J., Yoo H.W. Clinical phenotype and mutation spectrum of the CYP21A2 gene in patients with steroid 21-hydroxylase deficiency. Exp Clin Endocrinol Diabetes. 2012;120:23–27. doi: 10.1055/s-0031-1287789. [DOI] [PubMed] [Google Scholar]
- 13.Chan A.O., But W.M., Ng K.L., Wong L.M., Lam Y.Y., Tiu S.C., Lee K.F., Lee C.Y., Loung P.Y., Berry I.R., Brown R., Charlton R., Cheng C.W., Ho Y.C., Tse W.Y., Shek C.C. Molecular analysis of congenital adrenal hyperplasia due to 21-hydroxylase deficiency in Hong Kong Chinese patients. Steroids. 2011;76:1057–1062. doi: 10.1016/j.steroids.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Lobato M.N., Aledo R., Meseguer A. High variability of CYP21 gene rearrangements in Spanish patients with classic form of congenital adrenal hyperplasia. Hum Hered. 1998;48:216–225. doi: 10.1159/000022804. [DOI] [PubMed] [Google Scholar]
- 15.White P.C., New M.I., Dupont B. Structure of human steroid 21-hydroxylase genes. Proc Natl Acad Sci U S A. 1986;83:5111–5115. doi: 10.1073/pnas.83.14.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H.H., Lee Y.J., Lin C.Y. PCR-based detection of the CYP21 deletion and TNXA/TNXB hybrid in the RCCX module. Genomics. 2004;83:944–950. doi: 10.1016/j.ygeno.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Lee H.H., Lee Y.J., Chan P., Lin C.Y. Use of PCR-based amplification analysis as a substitute for the southern blot method for CYP21 deletion detection in congenital adrenal hyperplasia. Clin Chem. 2004;50:1074–1076. doi: 10.1373/clinchem.2003.028597. [DOI] [PubMed] [Google Scholar]
- 18.Lee H.H., Chang S.F., Lee Y.J., Raskin S., Lin S.J., Chao M.C., Lo F.S., Lin C.Y. Deletion of the C4-CYP21 repeat module leading to the formation of a chimeric CYP21P/CYP21 gene in a 9.3-kb fragment as a cause of steroid 21-hydroxylase deficiency. Clin Chem. 2003;49:319–322. doi: 10.1373/49.2.319. [DOI] [PubMed] [Google Scholar]
- 19.Parajes S., Quinterio C., Dominguez F., Loidi L. A simple and robust quantitative PCR assay to determine CYP21A2 gene dose in the diagnosis of 21-hydroxylase deficiency. Clin Chem. 2007;53:1577–1584. doi: 10.1373/clinchem.2007.087361. [DOI] [PubMed] [Google Scholar]
- 20.Concolino P., Mello E., Toscano V., Ameglio F., Zuppi C., Capoluongo E. Multiplex ligation-dependent probe amplification (MLPA) assay for the detection of CYP21A2 gene deletions/duplications in congenital adrenal hyperplasia: first technical report. Clin Chim Acta. 2009;402:164–170. doi: 10.1016/j.cca.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 21.Coeli F.B., Soardi F.C., Bernardi R.D., de Araújo M., Paulino L.C., Lau I.F., Petroli R.J., de Lemos-Marini S.H., Baptista M.T., Guerra-Junior G., de-Mello M.P. Novel deletion alleles carrying CYP21A1P/A2 chimeric genes in Brazilian patients with 21-hydroxylase deficiency. BMC Med Genet. 2010;11:104. doi: 10.1186/1471-2350-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merke D.P., Chen W., Morissette R., Xu Z., Van Ryzin C., Sachdev V., Hannoush H., Shanbhag S.M., Acevedo A.T., Nishitani M., Arai A.E., McDonnell N.B. Tenascin-X haploinsufficiency associated with Ehlers-Danlos Syndrome in patients with congenital adrenal hyperplasia. J Clin Endocrinol Metab. 2013;98:E379–E387. doi: 10.1210/jc.2012-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y.C., Liu T.C., Chang J.G., Lee H.H. High-resolution melting curve (HRM) analysis to establish CYP21A2 mutations converted from the CYP21A1P in congenital adrenal hyperplasia. Clin Chim Acta. 2011;412:1918–1923. doi: 10.1016/j.cca.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 24.Wilson R.C., Wei J.Q., Cheng K.C., Mercado A.B., New M.I. Rapid deoxyribonucleic acid analysis by allele-specific polymerase chain reaction for detection of mutations in the steroid 21-hydroxylase gene. J Clin Endocrinol Metab. 1995;80:1635–1640. doi: 10.1210/jcem.80.5.7745011. [DOI] [PubMed] [Google Scholar]
- 25.Keen-Kim D., Redman J.B., Alanes R.U., Eachus M.M., Wilson R.C., New M.I., Nakamoto J.M., Fenwick R.G. Validation and clinical application of a locus-specific polymerase chain reaction- and minisequencing-based assay for congenital adrenal hyperplasia (21-hydroxylase deficiency) J Mol Diagn. 2005;7:236–246. doi: 10.1016/S1525-1578(10)60550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai L.P., Cheng C.F., Hsieh J.P., Teng M.S., Lee H.H. Application of the DHPLC method for mutational detection of the CYP21A2 gene in congenital adrenal hyperplasia. Clin Chim Acta. 2009;410:48–53. doi: 10.1016/j.cca.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Ezquieta B., Oliver A., Gracia R., Gancedo P.G. Analysis of steroid 21-hydroxylase gene mutations in the Spanish population. Hum Genet. 1995;96:198–204. doi: 10.1007/BF00207379. [DOI] [PubMed] [Google Scholar]
- 28.Speiser P.W., Dupont J., Zhu D., Serrat J., Buegeleisen M., Tusie-Luna M.T., Lesser M., New M.I., White P.C. Disease expression and molecular genotype in congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Invest. 1992;90:584–595. doi: 10.1172/JCI115897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finkielstain G.P., Kim M.S., Sinaii N., Nishitani M., Van Ryzin C., Hill S.C., Reynolds J.C., Hanna R.M., Merke D.P. Clinical characteristics of a cohort of 244 patients with congenital adrenal hyperplasia. J Clin Endocrinol Metab. 2012;97:4429–4438. doi: 10.1210/jc.2012-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finkielstain G.P., Chen W., Mehta S.P., Fujimura F.K., Hanna R.M., Van Ryzin C., McDonnell N.B., Merke D.P. Comprehensive genetic analysis of 182 unrelated families with congenital adrenal hyperplasia due to 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2011;96:E161–E172. doi: 10.1210/jc.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burch G.H., Gong Y., Liu W., Dettman R.W., Curry C.J., Smith L., Miller W.L., Bristow J. Tenascin-X deficiency is associated with Ehlers-Danlos syndrome. Nat Genet. 1997;17:104–108. doi: 10.1038/ng0997-104. [DOI] [PubMed] [Google Scholar]
- 33.Lee H.H. The chimeric CYP21P/CYP21 gene and 21-hydroxylase deficiency. J Hum Genet. 2004;49:65–72. doi: 10.1007/s10038-003-0115-2. [DOI] [PubMed] [Google Scholar]
- 34.Gitelman S.E., Bristow J., Miller W.L. Mechanism and consequences of the duplication of the human C4/P450c21/gene X locus. Mol Cell Biol. 1992;12:3313–3314. doi: 10.1128/mcb.12.7.3313-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parajes S., Quinteiro C., Dominguez F., Loidi L. High frequency of copy number variations and sequence variants at CYP21A2 locus: implication for the genetic diagnosis of 21-hydroxylase deficiency. PloS One. 2008;3:e2138. doi: 10.1371/journal.pone.0002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koppens P.F., Hoogenboezem T., Degenhart H.J. Duplication of the CYP21A2 gene complicates mutation analysis of steroid 21-hydroxylase deficiency: characteristics of three unusual haplotypes. Hum Genet. 2002;111:405–410. doi: 10.1007/s00439-002-0810-7. [DOI] [PubMed] [Google Scholar]
- 37.Chen W., Xu Z., Merke D.P., McDonnell N.B. Reply to CLINCHEM/2012/190769 CH-8 phenotype in congenital adrenal hyperplasia: fact or fancy? Clin Chem. 2012;58:1601–1603. [Google Scholar]
- 38.Marino R., Ramirez P., Galeano J., Garrido N.P., Rocco C., Ciaccio M., Warman D.M., Guercio G., Chaler E., Maceiras M., Bergada I., Gryngarten M., Balbi V., Pardes E., Rivarola M.A., Belgorosky A. Steroid 21-hydroxylase gene mutational spectrum in 454 argentinean patients: genotype-phenotype correlation in a large cohort of patients with congenital adrenal hyperplasia. Clin Endocrinol (Oxf) 2011;75:427–435. doi: 10.1111/j.1365-2265.2011.04123.x. [DOI] [PubMed] [Google Scholar]
- 39.Ghizzoni L., Cappa M., Vottero A., Ubertini G., Carta D., Di Iorgi N., Gasco V., Marchesi M., Raggi V., Ibba A., Napoli F., Massimi A., Maghnie M., Loche S., Porzio O. Relationship of Cyp21a2 genotype and serum 17-hydroxyprogesterone and cortisol levels in a large cohort of Italian children with premature pubarche. Eur J Endocrinol. 2011;135:307–314. doi: 10.1530/EJE-11-0119. [DOI] [PubMed] [Google Scholar]
- 40.Day D.J., Speiser P.W., Schulze E., Bettendorf M., Fitness J., Barany F., White P.C. Identification of non-amplifying CYP21 genes when using PCR-based diagnosis of 21-hydroxylase deficiency in congenital adrenal hyperplasia (CAH) affected pedigrees. Hum Mol Genet. 1996;5:2039–2048. doi: 10.1093/hmg/5.12.2039. [DOI] [PubMed] [Google Scholar]