Abstract
Alternations in the glycosylation of proteins have been described in connection with several cancers, including hepatocellular carcinoma (HCC) and colorectal cancer. Analytical tools, which use combination of liquid chromatography and mass spectrometry, allow precise and sensitive description of these changes. In this study, we use MRM and FT-ICR operating in full-MS scan, to determine ratios of intensities of specific glycopeptides in HCC, colorectal cancer, and liver metastasis of colorectal cancer. Haptoglobin, hemopexin and complement factor H were detected after albumin depletion and the N-linked glycopeptides with fucosylated glycans were compared with their non-fucosylated forms. In addition, sialylated forms of an O-linked glycopeptide of hemopexin were quantified in the same samples. We observe significant increase in fucosylation of all three proteins and increase in bisialylated O-glycopeptide of hemopexin in HCC of hepatitis C viral (HCV) etiology by both LC-MS methods. The results of the MRM and full-MS scan FT-ICR analyses provide comparable quantitative readouts in spite of chromatographic, mass spectrometric and data analysis differences. Our results suggest that both workflows allow adequate relative quantification of glycopeptides and suggest that HCC of HCV etiology differs in glycosylation from colorectal cancer and liver metastasis of colorectal cancer.
Significance
The article compares N- and O-glycosylation of several serum proteins in different diseases by a fast and easy sample preparation procedure in combination with high resolution Fourier transform ion cyclotron resonance mass spectrometry. The results show successful glycopeptides relative quantification in a complex peptide mixture by the high resolution instrument and the detection of glycan differences between the different types of cancer diseases. The presented method is comparable to conventional targeted MRM approach but allows additional curation of the data.
Keywords: Mass spectrometry, Glycomics, FT-ICR, MRM, Hepatocellular carcinoma, Quantification, N-glycopeptides, O-glycopeptides, Haptoglobin, Hemopexin
1. Introduction
Protein glycosylation fulfills many biochemical and physiological functions including antigen recognition, cell-cell interaction, cell signal regulation, and protein stability [1–4]. It is known that under pathological conditions such as inflammation, the glycan composition is altered mostly due to changes in the activity of enzymes responsible for building the oligosaccharide components [5–8]. Several types of glycan modifications are commonly detected in cancer diseases depending on the type, activity, and localization of specific glycosyltransferases [9,10]. We and others have described changes in the glycosylation of liver secreted glycoproteins in hepatocellular carcinoma [11–16]. The best known examples in this context represent changes in glycan structures related to the increased expression of fucosyltransferases FUT-8 or FUT- 6 which result in elevated core or outer arm fucosylation of complex glycans, respectively. Formations of Lewis type epitopes such as SLeX, LeX, or LeY were recently observed on liver secreted glycoproteins [17–21], in addition to the well-known increase in core fucosylated α- fetoprotein in hepatocellular carcinoma [22–24]. These alterations in the glycosylation of proteins were extensively studied in human tissues and especially in serum [13,15,25–27]. Other described alterations of glycans include increased branching, sialylation, agalactosylated glycans or polylactosamine chains [16,28,29].
Recent developments in mass spectrometry and separation techniques have enabled characterization and quantification of glycopeptides in complex biological matrixes by the detection of released glycans [30–34], detection of glycopeptides [35–38], or top-down mass spectrometry of intact proteins [39]. Each of the approaches has its benefits and the methods offer complementary view of protein glycosylation. Major benefit of the quantification of glycopeptides is its ability to quantify site-specific glycoforms because the knowledge of glycan distribution within a protein is important for its function [40–42]. We therefore describe two LC-MS workflows for quantitative analysis of glycopeptides and document their applicability to serologic analysis of glycoproteins in cancer diseases.
Quantitative analysis by multiple reaction monitoring (MRM) is typically performed on triple quadrupole instruments in combination with liquid chromatography (LC). We and others have recently shown that this workflow can be used for quantification of glycopeptides [13,25, 43–46]. Collision induced dissociation of glycopeptides (CID) yields high intensity oxonium ions that originate from the glycans and less intense peptide-glycan or peptide fragments. The highly sensitive but less specific oxonium ions are typically used as quantitative transitions in the MRM mode [13,25,47]. In complex sample mixture, the most intense peptide-glycan fragments are often selected as specific transitions used for glycopeptide confirmation.
Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry provides the highest mass resolution and accuracy measurements of all analyzers. The high resolving power (>100,000 at m/z 400) and accuracy bellow 1 ppm in combination with high separation efficiency of HPLC allow accurate mass detection of analytes in complex sample matrices. We have used the 12T FT-ICR with ParaCell (solariX XR, Bruker Daltonics) to evaluate high resolution quantification of the glycopeptides. The 12T magnet provides a high magnetic field, which improves the dynamic range, mass accuracy, mass selectivity and signal-to- noise level [48,49]. The ultra-high resolution MS survey scan without data dependent acquisition (DDA) performed on such a high magnetic field instrument enables precise mass determination and quantification of peptides or glycopeptides [50]. Here, we demonstrate, for the first time, quantification of glycopeptides from partially depleted human sera by ultra-high resolution full-MS scan analysis performed by nanocapillary reversed phase chromatography coupled with the FT-ICR mass spectrometer and compare these data with results obtained by our previously optimized LC-MS/MS-MRM workflow [11,13,27].
2. Experimental section
2.1. Chemicals and materials
Unless stated otherwise, all chemicals were purchased from Sigma- Aldrich (St. Louis, MO). Acetonitrile and water for chromatography were from Merck (Darmstadt, Germany) and from Fisher Scientific (Fair Lawn, NJ). Trypsin was obtained from Promega (Madison, WI). Neuraminidase was from New England Biolabs (Ipswich, MA).
2.2. Study population
All participants were enrolled under the protocols approved by the Georgetown University Institutional Review Board and by the Pilsen Faculty Hospital Board. The HCC/HCV patients (n = 10) and healthy individuals (n = 10) were enrolled into the study in collaboration with the Department of Hepatology and Liver Transplantation, Georgetown University Hospital, Washington D.C., USA. The diagnosis of HCC was made by the attending physician based on liver imaging and/or liver biopsy. All The HCC/HCV patients had early stage disease according to the 7th Edition of the American Joint Committee on Cancer Staging manual and had chronic hepatitis C virus infection as the primary diagnosis. The HCC patients (n = 10), colorectal cancer patients (n = 10), and colorectal cancer/liver metastasis patients (n = 10) were enrolled into the study with the Laboratory of Immunoanalysis, Faculty Hospital in Pilsen, Pilsen, Czech Republic. The basic characteristics of the study population are summarized in Supplemental Table 1.
2.3. Albumin depletion
Albumin was depleted according to a previously described method with some modifications [51]. Ten microliters of human serum was diluted with 290 μL of water and centrifuged at RT for 15 min at 15,000 ×g. Lipids floating on the top were removed; the delipidated serum (150 μL) was mixed with 300 μL of 150 mM NaCl followed by addition of 350 μL of ice cold 100% ethanol. After 1 h incubation at 4 °C, samples were centrifuged for 45 min at 4 °C and 16,000 ×g. Supernatant was removed and the pellet was re-suspended in 200 μL of ice cold 42% ethanol and centrifuged for 15 min at 4 °C and 16,000 ×g. Sample pellet was dried and stored at −80 °C until further usage.
2.4. Sample preparation for MS analysis
The pellet was re-suspended in 100 μL of buffer containing 4 M Guanidine-HCl in 50 mM ammonium bicarbonate, pH 7.8. After the pellet was dissolved, the sample was further diluted with 50 mM ammonium bicarbonate to final concentration of 1 M Guanidine-HCl and the protein concentration was measured by BCA assay using the NanoDrop system (Thermo Scientific, Wilmington, DE). The protein concentration of each sample was adjusted to 2 μg/μL by addition of 50 mM ammonium bicarbonate. Ten microliters (20 μg) of total protein mixture was reduced by dithiothreitol (DTT), final concentration 10 mM, and incubated for 45 min at 60 °C. Free –SH groups were modified by iodoacetamide, final concentration 30 mM, for 30 min in the dark. The alkylation reaction was quenched by addition of DTT, final concentration 30 mM and the proteins were digested by trypsin (0.4 μg per sample) over night at 37 °C. Trypsin was inactivated by heating the samples for 5 min at 90 °C and half of each sample was transferred to new tube for treatment with 100 units of α-2/3,6,8 neuraminidase from Clostridium perfringens overexpressed in Escherichia coli (New England Biolabs) for 24 h at 37 °C. Both neuraminidase-treated and non-treated samples were desalted on a MicroTrap peptide cartridge (Michrom Bioresources, Auburn, CA) as follows: cartridge was equilibrated with 2 × 250 μL of 0.1% trifluoroacetic acid (TFA). Samples were diluted with 150 μL of 0.1% TFA and gently loaded on the cartridge and desalted with 4 × 250 μL of 0.1% TFA. Peptides were eluted with 80% acetonitrile (ACN) containing 0.1% TFA, and dried using a SpeedVac concentrator. To document robustness and reproducibility of the sample preparation procedure, three technical replicates of a control serum sample were processed as described above and analyzed by FT-ICR.
2.5. Detection and quantification of glycopeptides by FT-ICR
One microgram of tryptic digest was injected on a nano-capillary system (Ultimate 3000 RSLC, Thermo Scientific, Waltham, MA) coupled with 12T solariX XR FT-ICR mass spectrometer equipped with Captive Spray ion source (Bruker Daltonics, Billerica, MA). For identification of glycoproteins in depleted serum, 1 μg of protein digest was injected on reversed phase C18 column (Acclaim PepMap 100, 100 μm × 2 cm, nanoViper C18, 5 μm, 100 Å, Thermo Scientific) and trapped for 3 min at a flow rate of 10 μL/min using an aqueous loading solution of 0.1% formic acid (FA). Peptides were further separated on an analytical reversed phase C18 column (Acclaim PepMap RSLC, 75 μm × 15 cm, nanoViper C18, 2 μm, 100 Å, Thermo Scientific) at a flow rate 0.5 μL/ min and ACN gradient: 3–45% ACN, 0.1% FA for 5–60 min; 50–98% ACN, 0.1% FA for 60–65 min; 98% ACN, 0.1% FA for 65–70 min. Solvent A consisted of 2% ACN + 0.1% FA and solvent B of 98% ACN + 0.1% FA. Both trap and analytical columns were heated to 60 °C. The FT-ICR instrument was operated in data dependent mode. All spectra were acquired in broadband detection mode, number of data points were set to 512 k at m/z 250, ion accumulation to 0.4 s and average scan to 1. The capillary voltage was set at −1500 V, the dry heat gas temperature at 180 °C, dry gas flow at 6.0 L/min and the nebulizer gas at 1.0 bar. The three most intense precursor ions were selected for CID fragmentation with active exclusion set to 0.5 min and with exclusion after 2 spectra with the minimum threshold intensity higher than 1 × 107. The sweep excitation power was set to 30%.
The relative quantification of glycopeptides was performed on the same HPLC system, columns and solvents, but using a shorter ACN gradient: 3–50% ACN, 0.1% FA for 5–30 min; 50–98% ACN, 0.1% FA for 30–35 min; 98% ACN, 0.1% FA for 35–40 min. The FT-ICR mass spectrometer was operated in MS survey scan and broadband detection mode, number of data points was set at 2 M at m/z 250, ion accumulation at 0.4 s, and average scan at 4. The processing mode was set at Magnitude and Full-Sine window mode. The ion source parameters were set up as described above. The instrument was calibrated on-line using the lock masses 322.048121 Da, 622.02896 Da and 922.009798 Da (Agilent, Santa Clara, CA). The calibration solution was applied to Captive Spray filter as recommended by the manufacturer. The CID spectra for the comparative detection of core fucosylated glycopeptide of complement factor H were acquired by the FT-ICR operating in selective fragmentation mode with fixed collisional energy set at 30 eV, which was optimized for this glycopeptide.
2.6. Quantification of glycopeptides by MRM
One microgram of the tryptic digest was separated by reversed phase chromatography (NanoAcquity, Waters Associates, Milford, MA) on a Symmetry C18 (3 μm × 180 μm × 20 mm, Waters Associates, Milford, MA) trap column and capillary column (BEH 300 Å, 1.7 μm × 150 mm × 75 μm, Waters Associates, Milford, MA) coupled with 6500 Q-TRAP mass spectrometer (AB Sciex, Framingham, MA). Peptides were first trapped for 3 min using 2% ACN, 0.1% FA solvent at a flow rate of 10 μL/min. Separation of peptides was achieved by the following gradient at a flow rate 0.4 μL/min: 3–50% ACN, 0.1% FA for 5–30 min; 50–98% ACN, 0.1% FA for 30–35 min; 98% ACN, 0.1% FA for 35–40 min. The analytical column was heated to 60 °C. The source parameters of the Q-TRAP instrument were set as follows: curtain gas flow was set at 13 PSI, ion spray voltage at 2400 V, ion source gas at 130 PSI, interface heater temperature at 180 °C, entrance potential at 10 V, and collision exit potential at 10 V. Optimal collision energies for fragmentation of glycopeptide precursors were calculated for each oxonium ion based on the following equations: for oxonium ion 204: y = 0.0349x + 14.877, for oxonium ion 366: y = 0.0688x − 37.825 and for oxonium ion 512: y = 0.0303x − 0.8182, where y represented collision energy and x represented m/z of the precursor ion [13].
2.7. Data processing and evaluation
The MS/MS data files acquired by FT-ICR mass spectrometer were processed in DataAnalysis 4.2 (Bruker Daltonics, Billerica, MA), exported to mgf format and searched by using Mascot search engine (version 2.2.07) against the SwissProt human database (SwissProt 2014x) with following searching parameters: digestion by trypsin, 1 allowed missed cleavage, carbamidomethylation as fixed modification, and acetyl (N-term) and methionine oxidation as variable modification. The minimum ion score was set at 15 and significant threshold at p < 0.05. The accept Mascot score for protein assessment was higher than 80 and the accept Mascot score for peptide assessment was higher than 20. The minimum number of peptides per protein was set at 1. The MS data for quantification of glycopeptides were processed in DataAnalysis 4.2 (Bruker Daltonics, Billerica, MA) and the intensities of the third isotope of the extracted ion chromatograms (XIC) of particular glycopeptides from the FT-ICR data were used for calculation of the glycopeptide ratios. The MRM data files were processed with MultiQuant 2.0 (AB Sciex, Framingham, MA) and the results were exported as text files for further data evaluation. Peak intensities of the 366 transition were used for calculation of the ratios of the glycopeptides. Other transitions were used for confirmation of the specificity of detection of individual glycopeptides. Both results from MRM and FT- ICR-MS were statistically analyzed by Kruskal-Wallis test. The p-values were calculated for each patient group and compared with control. We adopt the N-glycan nomenclature from the NIBRT GlycoBase databases [52].
3. Results and discussion
It has been recently reported that fucosylated glycopeptides in several serum glycoproteins are elevated compared to non-fucosylated glycopeptides in colon cancer, HCC and cirrhotic patients [7,11,15,27,53, 54]. The most common analytical strategies for quantification of glycopeptides in these studies include enrichment of glycoproteins or glycopeptides by affinity chromatography [55–57] (lectins or antibodies) or by chromatographic enrichment of glycopeptides on HILIC [12,58] or graphitized carbon [59,60] columns. Final step for the detection of glycopeptides is typically a reversed phase liquid chromatography separation coupled to a mass spectrometer operating in data dependent mode [12,13,27,61,62]. In our previous studies, we have observed glycopeptides that bear up to six fucoses forming different variations of Lewis antigens in cirrhotic HCC patients of HCV etiology [11]. Significantly elevated concentrations of singly fucosylated glycopeptides were observed on majority of the glycoproteins that we examined, in this disease context, so far. In this report, we simplified the detection of the liver secreted glycoproteins from serum, improved robustness of the quantitative analysis, and documented feasibility of glycopeptide quantification by a full-MS scan FT-ICR mass spectrometric workflow. We applied the optimized workflow to patient populations with hepatocellular carcinoma, colorectal cancer, and metastases of colorectal cancer to the liver to improve our understanding of the elevation of fucosylated glycoproteins in the context of cancer diseases. Patients with colorectal cancer and colorectal cancer with liver metastases were selected to evaluate whether metastatic liver disease leads to similar increase in glycopeptides as primary liver cancer. All the HCC/HCV patients had cirrhotic liver.
3.1. Sample preparation
To reduce complexity of the serum samples, we chose ethanol precipitation as a fast efficient alternative to affinity enrichment [51]. The method uses ethanol in final concentration of 42% to precipitate proteins of interest while leaving albumin in the supernatant. Protein of all the albumin-depleted samples was adjusted to the same concentration for further processing and identification by MS/MS analysis was performed on a solariX FT-ICR instrument (Supplemental Table 2). Although the FT-ICR instrument was set to scan as fast as possible, the relatively low speed and fragmentation efficiency of precursor ions of low intensity had limited coverage to the 45 most abundant proteins in the depleted serum samples. This was, however, sufficient to assure that several glycoproteins of interest were detected in the depleted serum and further used for quantification, the main goal of our analysis. We have recently characterized in detail site specific glycopeptides of haptoglobin, hemopexin, and complement factor H [11,13,25,27]. All of these proteins were detected in the albumin-depleted serum samples and were selected for the FT-ICR quantification.
3.2. Full-MS scan FT-ICR quantification
The same serum sample set used for MRM quantification was analyzed by full-MS scan performed on a 12T solariX-XR-FT-ICR instrument (Fig. 1A and B). Our main goal was to document that the ultra-high resolution power and accuracy of FT-ICR allows quantification of glycopeptides in a complex mixture comparable to the established MRM workflow. The intensities of the third isotope of the XICs of a particular glycopeptides from the FT-ICR data were used for calculation of the glycopeptide ratios. This allows acquisition of at least 5 data points per XIC peak which is the minimum number of points per peak allowed in our quantification. The glycopeptides were searched with the mass error <1 ppm (Table 1). Glycopeptides of the same peptide co-eluted in our chromatography within a one-minute retention time window and confirmed the correctness of glycopeptide matching. An inclusion list of selected glycopeptide masses was created for further validation by CID fragmentation. Although some of the low intensity glycopeptides were not selected by the data acquisition software for fragmentation, the CID spectra of their major glycopeptides confirmed the correctness of assignment (Supplemental Table 4). FT-ICR allowed us to quantify even low abundant glycoforms of several peptides of haptoglobin, hemopexin, and complement factor H in the sample. Three replicates of control serum measurement are shown in Supplemental Fig. 2 to demonstrate the robustness of the method. Extracted ion chromatograms of the low abundant tri-antennary fucosylated VVLHPNYSQVDIGLIK glycopeptide from haptoglobin overlap in retention times and intensities. Table 1 summarizes observed and quantified glycopeptides. The high accuracy and resolution achieved by FT-ICR allows quantification of haptoglobin glycopeptide VVLHPNYSQVDIGLIK bearing the tri-antennary fucosylated glycan observed as triply charged ion at m/z 1310.26626. This ion co-elutes with other unknown analytes whose isotopes overlap with the isotopic envelope of the glycopeptide of interest. The resolving power 45,000 (at m/z 1310) separated glycopeptide isotopes from the unknown analytes sufficiently and enabled its quantification. The observed pattern of the triply charged fucosylated glycopeptide correlates well with the simulated isotopic envelope which supports the correctness of our assignment (Supplemental Fig. 1A).
Fig. 1.
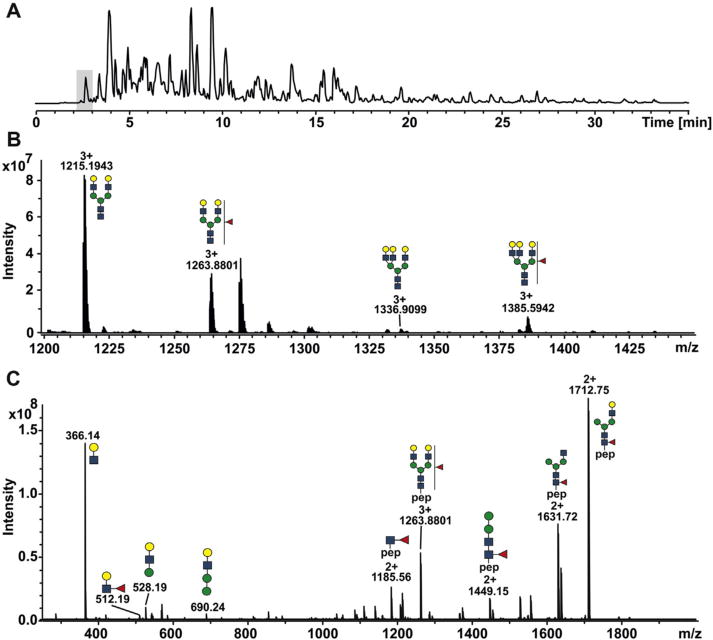
A) Total ion chromatogram of tryptic digest of HCC/HCV serum separated on a reversed phase C18 column coupled with solariX XR FT-ICR mass spectrometer. The highlighted region represents elution window of complement factor H glycopeptides. B) MS spectrum represents the sum of scans within a 0.5 min retention window. Only bi- and tri-antennary glycopeptides of complement factor H glycopeptide IPCSQPPQIEHGTINSSR and their singly fucosylated forms were detected. C) CID spectrum of a fucosylated bi-antennary glycopeptide. The peptide-glycan fragments bearing fucose identify the core fucosylated glycopeptides.
Table 1.
List of identified and quantified glycopeptides; glycopeptides marked with a star were quantified.
| Protein | Peptide | Glycan | m/z observed | m/z calculated | Error (ppm) |
|---|---|---|---|---|---|
| Serotransferrin | CGLVPVLAENYNK | A2G2 | 1033.7833 (3+) | 1033.7825 (3+) | 0.7 |
| QQQHLFGSNVTDCSGNFCLFR | A2G2 | 1379.9010 (3+) | 1379.9067 (3+) | 1 | |
| Hemopexin | SWPAVGNCSSALR | A2G2* | 1514.1293 (2+) | 1514.1289 (2+) | 0.4 |
| A2G2F1* | 1587.1567 (2+) | 1587.1578 (2+) | 0.7 | ||
| A3G3* | 1696.6942 (2+) | 1696.6950 (2+) | 0.5 | ||
| A3G3F1* | 1769.7223 (2+) | 1769.7239 (2+) | 0.9 | ||
| TPLPPTSAHGNVAEGETKPDPDVTER | 1N1H1NA* | 1124.5225 (3+) | 1124.5214 (3+) | 1 | |
| 1N1H2NA* | 1221.5528 (3+) | 1221.5532 (3+) | 0.3 | ||
| Haptoglobin | VVLHPNYSQVDIGLIK | A2G2* | 1139.8693 (3+) | 1139.8691 (3+) | 0.1 |
| A2G2F1* | 1188.5553 (3+) | 1188.5551 (3+) | 0.2 | ||
| A3G3* | 1261.5792 (3+) | 1261.5799 (3+) | 0.5 | ||
| A3G3F1* | 1310.2663 (3+) | 1310.2658 (3+) | 0.3 | ||
| A4G4 | 1383.2909 (3+) | 1383.2906 (3+) | 0.2 | ||
| A4G4F1 | 1431.9771 (3+) | 1431.9765 (3+) | 0.4 | ||
| MVSHHNLTTGATLINEQWLLTTAK | A2G2* | 1434.6615 (3+) | 1434.6628 (3+) | 0.9 | |
| A2G2F1* | 1483.3479 (3+) | 1483.3488 (3+) | 0.6 | ||
| A3G3* | 1556.3728 (3+) | 1556.3736 (3+) | 0.5 | ||
| A3G3F1* | 1605.0605 (3+) | 1605.0596 (3+) | 0.6 | ||
| Complement factor H | IPCSQPPQIEHGTINSSR | A2G2* | 1215.1938 (3+) | 1215.1944 (3+) | 0.5 |
| A2G2F1* | 1263.8807 (3+) | 1263.8804 (3+) | 0.2 | ||
| A3G3* | 1336.9059 (3+) | 1336.9051 (3+) | 0.6 | ||
| A3G3F1* | 1385.5918 (3+) | 1385.5911 (3+) | 0.5 | ||
| Clusterin 1 | HNSTGCLR | A2G2 | 1284.0117 (2+) | 1284.0128 (2+) | 0.9 |
Core fucosylation was previously shown to be increased in cancers [25,27,63]. Here, we monitored core fucosylation of the bi-antennary singly fucosylated glycopeptide IPCSQPPQIEHGTINSSR from complement factor H by selective CID fragmentation of the precursor ion at m/z 1263.8801. We compared only control and HCC/HCV group, because we observed major alteration in the fucosylation of glycans in these groups. Core fucosylation of the bi-antennary glycopeptide was confirmed by the presence of high intensity peptide-glycan fragments (peptide + GlcNAc, peptide + 2GlcNAc + 2Man) bearing a fucose, in agreement with our recent findings [25,27]. A low intensity oxonium ion at m/z 512.1895 was also observed in the fragmentation spectrum suggesting the presence of outer arm fucose (Fig. 1C). This is likely due to a fucose rearrangement observed in glycopeptide analysis by mass spectrometry [64]. We have not observed any difference in the intensities of the 512.1895 Da fucose-glycan fragment between the groups. These findings indicate that the bi-antennary glycopeptide of complement factor H with core fucosylation and not outer-arm fucosylation is increased in HCC/HCV patients. It is likely that the tri- antennary glycoforms of complement factor H and hemopexin contain the core fucose. However, the quality of the CID spectra of the tri- antennary fucosylated glycoforms of complement factor H and hemopexin were not sufficient to confirm it. Tetra-antennary and multiply fucosylated glycopeptides, which were recently observed on enriched glycoproteins including haptoglobin, hemopexin, complement factor H and kininogen-127, were not detected in this study or did not pass the quantification criteria.
3.3. Verification of FT-ICR quantification by MRM
We created a list of known and predicted glycopeptides of the tryptic digest of serum and used these as a template for calculation of precursor ion masses and transitions for the targeted quantification assays (Supplemental Table 3). The collision energies for each glycopeptide were calculated by equations created based on collision energies optimized in our previous study [13]. CID spectra of glycopeptides provide abundant oxonium ions at m/z 204 (N-acetylglucosamin/N-acetylgalactosamin), 366 (lactosamine), or 512 (fucosylated lactosamine); CID also generates specific peptide-glycan fragments at lower intensity. In our MRM analysis, we used the oxonium ions for quantification and a specific peptide-N-acetylglucosamine (N-linked glycopeptides) or peptide-N-acetylgalactosamine (O-linked glycopeptides) fragments, and their retention times, to assure specificity of detection. The peak areas were calculated for each glycopeptide from the oxonium ion m/z 366. The validation was done manually in MultiQuant 2.0 software and appropriate peaks were individually matched to each glycopeptide. Although we used in this study quite complex fraction of serum and fast chromatographic separations, we were able to quantify in one run the selected N-glycopeptides of haptoglobin and two O-glycopeptides of hemopexin. For example, the three transitions (366, 204 and 512) of the glycopeptide VVLHPNYSQVDIGLIK bearing the tri-antennary fucosylated glycan have two retention times at 12.5 min and 16.5 min in the chromatogram (Supplemental Fig. 1B). However, all four transitions (366, 204, 512 and 999.5), including the specific transition (pepGlcNAc) at m/z 999.5, were observed only at RT 12.5 min. Overall, the intensity of specific transitions for less abundant glycopeptides was usually low or under quantification and detection limit. This prohibited definitive assignment of the low abundant glycopeptides and limited coverage of the quantification process.
We focused on the quantification of fucosylated N-glycopeptides following enzymatic removal of sialic acid from the glycans to increase the signal of fucosylated forms of the microheterogeneous sialylated glycopeptides. The list of all selected glycoproteins for MRM analysis with predicted glycopeptides is shown in Supplemental Table 2. We were limited to 300 transitions and selected only the most abundant glycoproteins. The box plots in Fig. 2 represent quantitative comparison of two haptoglobin glycopeptides bearing bi- and tri-antennary glycopeptides and their singly fucosylated forms. Panel A documents the results of MRM quantification; panel B shows results of the full-MS scan quantification performed by FT-ICR. Statistical evaluation of the haptoglobin data identified significant increase in the fucosylated tri-antennary glycopeptides VVLHPNYSQVDIGLIK in the HCC/HCV group; the increase was consistently detected by both approaches. The p-values shown in MRM box plots (panel A) look more consistent than these shown in FT-ICR box plots (panel B). Differences between both approaches were observed in the ratios of tri- vs bi-antennary glycopeptide VVLHPNYSQVDIGLIK. Here, the error bars for the MRM approach are higher compared to FT-ICR method. On the other hand, FT-ICR data shows significant difference (p-value 0.002) between control and colorectal/liver metastasis groups and between control and HCC group (p- value 0.001), which we do not expect. Other differences in the p-values obtained by the two MS approaches were observed for the fucosylated tri-antennary glycopeptide MVSHHNLTTGATLINEQWLLTTAK; the MRM approach detected significant difference (p-value 0.0008) in the HCC/HCV group while FT-ICR measurement did not reach significance (p-value 0.18). This data inconsistency could be caused by different fragmentation behavior of fucosylated vs non-fucosylated glycopeptides, which may result in different yield of oxonium ions used for quantification by MRM. Interestingly, the colorectal cancer and its liver metastasis are not separated from the control group. Similar ratios of non-fucosylated tri- vs bi-antennary glycopeptides of the MVSHHNLTTGATLINEQWLLTTAK peptide between patients groups nicely shows that fucosylation itself and not increased branching cause the increase in fucosylated glycopeptides. Although the p-values show differences between both mass spectrometric methods, collectively, the results document consistently a significant increase of the fucosylated forms of all glycopeptides in the patients with HCC/HCV compared to all other patient groups. Supplemental Table 5 summarizes all quantified glycopeptides using both methods. The other predicted glycopeptides selected for MRM analysis were not quantified, because we did not observe the specific transitions (peptide + HexNAc) and could not assign the correct glycopeptide. The results show that the sensitivity of both methods for relative quantification of glycopeptides is comparable.
Fig. 2.

Box plots represent relative quantification of two haptoglobin peptides bearing tri-antennary glycopeptides and their fucosylated forms for each patient group. Peak intensities of the 366 transition were used for calculation of the ratio of glycopeptides for the MRM quantification (shown on the left). Intensities of the XIC of the third isotope of each glycopeptide were used for calculation of the ratios of fucosylated and non-fucosylated glycopeptides or the ratio of tri-antennary and bi-antennary glycopeptide for the FT-ICR quantification (shown on the right). The numbers above each box plot represent p-values.
Park et al. have recently observed alterations of haptoglobin fucosylation in patients with colorectal cancer studied by a combination of lectins recognizing fucosylated glycopeptides and monoclonal antibodies against the Lewis epitopes [54]. AAL lectin, which has the affinity to several Lewis type antigens [54], had increased blotting index in colorectal cancer. Park et al. have confirmed increased outer arm fucosylation of haptoglobin in colon cancer compared to normal and inflammatory bowel disease in a follow up publication [53]. Moreover, they have found higher fucosylation of glycans at Asn 241, which is represented in our study by the VVLHPNYSQVDIGLIK peptide [53]. They have used nano liquid chromatography coupled with FT-ICR for site specific characterization of haptoglobins glycopeptides. We cannot confirm these results on our set of colorectal patients and observe an insignificant increase in the fucosylation of haptoglobin in colorectal cancer even in case of its metastasis to the liver. Another study has detected higher concentration of bi-fucosylated glycans in HCC compared to cirrhotic patients by the analysis of glycans released from haptoglobin isolated from patients with HCC and cirrhosis of different etiologies, including HCV, HBV and heavy alcohol consumption. The authors have observed differences in the expression of singly fucosylated glycopeptides between healthy controls and all patients groups, but not between the patient groups themselves [14]. In our study, mono fucosylated glycopeptides increase the HCC/HCV group compared to healthy controls. Further, we observed significantly elevated fucosylated glycopeptides of hemopexin and complement factor H in HCC/HCV patients compared to other groups (Fig. 4).
Fig. 4.
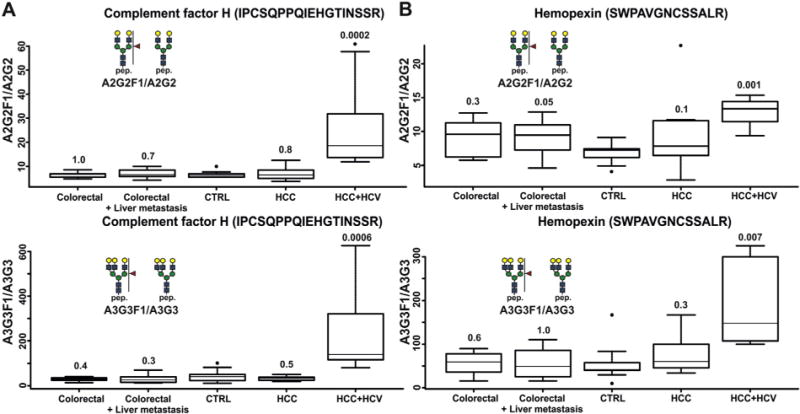
Relative quantification of complement factor H and hemopexin N-glycopeptides performed by the FT-ICR instrument. The glycopeptides were identified by exact mass search. The numbers above each box plot represent p-values.
3.4. Quantification of the O-glycopeptides of hemopexin
In our recent paper, we have described an LC-MS3 method for the quantification of O-glycopeptides of hemopexin in serum [65]. Here, we compared mono- and bi-sialylated glycopeptides of T-antigen of the N-terminal TPLPPTSAHGNVAEGETKPDPDVTER peptide of hemopexin by full-MS scan FT-ICR and confirmed the results by LC- MS/MS-MRM (Fig. 3). Results of our quantification show a significant elevation of bi-sialylated T-antigen in patients with HCC/HCV compared to all groups using both quantification methods. In contrast to the fucosylated glycopeptides, we observed increase in the bi-sialylated T- antigen in colorectal cancer (p-value: 0.08 for MRM and 0.01 for FT- ICR) and colorectal cancer with liver metastasis (p-value: 0.007 for MRM and 0.03 for FT-ICR) compared to the control group. Changes of expression of O-linked glycopeptides of hemopexin in patients with colorectal cancer are to our knowledge reported for the first time and should be further explored in addition to their increase in HCC. The results clearly show that not only fucosylation of N-glycopeptides, but also sialylation of O-glycopeptides can be reliably quantified by full-MS scan FT-ICR analysis of proteins after ethanol precipitation of patient serum. This simple protocol opens new methods for quantification of glycopeptides in complex samples with specificity of detection assured by standardized retention times of the analytes and high resolution and mass accuracy of the MS method.
Fig. 3.
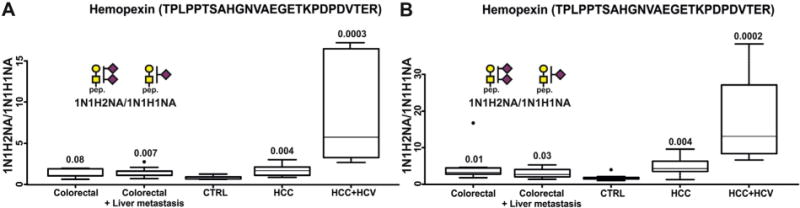
Box plots showing MRM quantification (left) and FT-ICR quantification (right) of differentially sialylated T antigen of the TPLPPTSAHGNVAEGETKPDPDVTER O-glycopeptide of hemopexin. The bi-sialylated glycopeptide is significantly elevated in HCC/HCV patients. The numbers above each box plot represent p-values.
4. Conclusions
The data presented in this paper illustrate the utility of ultra-high resolution full-MS scan analysis performed by FT-ICR for glycopeptide quantification. Detection of the low-abundant glycopeptides in the digest of albumin-depleted serum requires sensitive instrumentation. We show that the FT-ICR workflow for quantification of glycopeptides achieves sensitivity comparable to our MRM method. Resolution and quantification of the bi- and tri-antennary N-glycopeptides of three proteins (haptoglobin, hemopexin, and complement factor H) and sialylated O-glycopeptides of hemopexin was achieved by a thirty minute reversed phase chromatographic analysis. The high resolution and accuracy measurements performed by the FT-ICR allow detection of other glycopeptides than the glycopeptides selected for targeted MRM quantification. The possibility of additional data curation is the major benefit of full-MS scan FT-ICR approach compared to MRM. This is demonstrated by the detection and quantification of the N-glycopeptides of complement factor H (IPCSQPPQIEHGTINSSR) and hemopexin (SWPAVGNCSSALR) which were not selected for our MRM quantification. There is also no need for the FT-ICR approach to design glycopeptides transitions. On the other hand, data processing in the MRM approach is faster and easier using the available software. The glycopeptides quantification by FT-ICR requires manual data processing and calculation of peak intensities.
The significant increase in fucosylation of N-glycopeptides was observed in patients with HCC previously diagnosed with HCV compared to colorectal cancers and HCC without HCV etiology. The core fucosylation detected in the bi-antennary glycopeptide of complement factor H was also elevated only in the HCC/HCV patients. Interestingly, the same situation was observed in bi-sialylated O-linked glycopeptide of hemopexin.
Supplementary Material
Acknowledgments
Ministry of Education, Youth and Sports of the Czech Republic (AMVIS Czech Republic-U.S. exchange program LH13051), Project UNCE204025/2012, Operational Program Prague – Competitiveness project CZ.2.16/3.1.00/24023, IMIC institutional research concept RVO61388971. This work was supported in part by National Institutes of Health Grants UO1 CA168926 and RO1 CA135069 (to R.G.) and CCSG Grant P30 CA51008 (to Lombardi Comprehensive Cancer Center supporting the Proteomics and Metabolomics Shared Resource).
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2016.09.004.
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.van Kooyk Y, Rabinovich GA. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol. 2008;9(6):593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 2.Taylor ME, Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr Opin Cell Biol. 2007;19(5):572–577. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Sharon N. Lectins: carbohydrate-specific reagents and biological recognition molecules. J Biol Chem. 2007;282(5):2753–2764. doi: 10.1074/jbc.X600004200. [DOI] [PubMed] [Google Scholar]
- 4.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell. 2006;126(5):855–867. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Nakano M, Nakagawa T, Ito T, Kitada T, Hijioka T, Kasahara A, Tajiri M, Wada Y, Taniguchi N, Miyoshi E. Site-specific analysis of N-glycans on haptoglobin in sera of patients with pancreatic cancer: a novel approach for the development of tumor markers. Int J Cancer. 2008;122(10):2301–2309. doi: 10.1002/ijc.23364. [DOI] [PubMed] [Google Scholar]
- 6.Blomme B, Van Steenkiste C, Callewaert N, van Vlierberghe H. Alteration of protein glycosylation in liver diseases. J Hepatol. 2009;50(3):592–603. doi: 10.1016/j.jhep.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang M, Long RE, Comunale MA, Junaidi O, Marrero J, Di Bisceglie AM, Block TM, Mehta AS. Novel fucosylated biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol Biomark Prev. 2009;18(6):1914–1921. doi: 10.1158/1055-9965.EPI-08-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saldova R, Royle L, Radcliffe CM, Abd hamid UM, Evans R, Arnold JN, Banks RE, Hutson R, Harvey DJ, Antrobus R, et al. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17(12):1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- 9.Aebi M. N-linked protein glycosylation in the ER, Biochim. Biophys Acta Mol Cell Res. 2013;1833(11):2430–2437. doi: 10.1016/j.bbamcr.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Schwarz F, Aebi M. Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. 2011;21(5):576–582. doi: 10.1016/j.sbi.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site-specific glycopeptides of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics. 2013;12(5):1281–1293. doi: 10.1074/mcp.M112.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pompach P, Chandler KB, Lan R, Edwards N, Goldman R. Semi-automated identification of N-glycopeptides by hydrophilic interaction chromatography, nano-reverse-phase LC-MS/MS, and glycan database search. J Proteome Res. 2012;11(3):1728–1740. doi: 10.1021/pr201183w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanda M, Pompach P, Brnakova Z, Wu J, Makambi K, Goldman R. Quantitative liquid chromatography-mass spectrometry-multiple reaction monitoring (LC-MS- MRM) analysis of site-specific glycopeptides of haptoglobin in liver disease. Mol Cell Proteomics. 2013;12(5):1294–1305. doi: 10.1074/mcp.M112.023325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J, Lin Z, Wu J, Yin H, Dai J, Feng Z, Marrero J, Lubman DM. Analysis of serum haptoglobin fucosylation in hepatocellular carcinoma and liver cirrhosis of different etiologies. J Proteome Res. 2014;13(6):2986–2997. doi: 10.1021/pr500128t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaji H, Ocho M, Togayachi A, Kuno A, Sogabe M, Ohkura T, Nozaki H, Angata T, Chiba Y, Ozaki H, et al. Glycoproteomic discovery of serological biomarker candidates for HCV/HBV infection-associated liver fibrosis and hepatocellular carcinoma. J Proteome Res. 2013;12(6):2630–2640. doi: 10.1021/pr301217b. [DOI] [PubMed] [Google Scholar]
- 16.Campion B, Léger D, Wieruszeski JM, Montreuil J, Spik G. Presence of fucosylated triantennary, tetraantennary and pentaantennary glycans in transferrin synthesized by the human hepatocarcinoma cell line Hep G2. Eur J Biochem. 1989;184(2):405–413. doi: 10.1111/j.1432-1033.1989.tb15032.x. [DOI] [PubMed] [Google Scholar]
- 17.Barthel SR, Wiese GK, Cho J, Opperman MJ, Hays DL, Siddiqui J, Pienta KJ, Furie B, Dimitroff CJ. Alpha 1,3 fucosyltransferases are master regulators of prostate cancer cell trafficking. Proc Natl Acad Sci U S A. 2009;106(46):19491–19496. doi: 10.1073/pnas.0906074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higai K, Miyazaki N, Azuma Y, Matsumoto K. Transcriptional regulation of the fucosyltransferase VI gene in hepatocellular carcinoma cells. Glycoconj J. 2008;25(3):225–235. doi: 10.1007/s10719-008-9114-z. [DOI] [PubMed] [Google Scholar]
- 19.Mathieu S, Gerolami R, Luis J, Carmona S, Kol O, Crescence L, Garcia S, Borentain P, El-Battari A. Introducing alpha(1,2)-linked fucose into hepatocarcinoma cells inhibits vasculogenesis and tumor growth. Int J Cancer. 2007;121(8):1680–1689. doi: 10.1002/ijc.22797. [DOI] [PubMed] [Google Scholar]
- 20.Guo Q, Guo B, Wang Y, Wu J, Jiang W, Zhao S, Qiao S, Wu Y. Functional analysis of α1,3/4-fucosyltransferase VI in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;417(1):311–317. doi: 10.1016/j.bbrc.2011.11.106. [DOI] [PubMed] [Google Scholar]
- 21.Wei T, Liu Q, He F, Zhu W, Hu L, Guo L, Zhang J. The role of Nacetylglucosaminyltransferases V in the malignancy of human hepatocellular carcinoma. Exp Mol Pathol. 2012;93(1):8–17. doi: 10.1016/j.yexmp.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Du MQ, Hutchinson WL, Johnson PJ, Williams R. Differential alpha-fetoprotein lectin binding in hepatocellular carcinoma. Diagnostic utility at low serum levels. Cancer. 1991;67(2):476–480. doi: 10.1002/1097-0142(19910115)67:2<476::aid-cncr2820670226>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 23.Taketa K, Sekiya C, Namiki M, Akamatsu K, Ohta Y, Endo Y, Kosaka K. Lectin-reactive profiles of alpha-fetoprotein characterizing hepatocellular carcinoma and related conditions. Gastroenterology. 1990;99(2):508–518. doi: 10.1016/0016-5085(90)91034-4. [DOI] [PubMed] [Google Scholar]
- 24.Sterling RK, Jeffers L, Gordon F, Sherman M, Venook AP, Reddy KR, Satomura S, Schwartz ME. Clinical utility of AFP-L3% measurement in North American patients with HCV-related cirrhosis. Am J Gastroenterol. 2007;102(10):2196–2205. doi: 10.1111/j.1572-0241.2007.01405.x. [DOI] [PubMed] [Google Scholar]
- 25.Benicky J, Sanda M, Pompach P, Wu J, Goldman R. Quantification of fucosylated hemopexin and complement factor H in plasma of patients with liver disease. Anal Chem. 2014;86(21):10716–10723. doi: 10.1021/ac502727s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsutsumi M, Wang JS, Takada A. Microheterogeneity of serum glycoproteins in alcoholics: is desialo-transferrin the marker of chronic alcohol drinking or alcoholic liver injury? Alcohol Clin Exp Res. 1994;18(2):392–397. doi: 10.1111/j.1530-0277.1994.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 27.Pompach P, Ashline DJ, Brnakova Z, Benicky J, Sanda M, Goldman R. Protein and site specificity of fucosylation in liver-secreted glycoproteins. J Proteome Res. 2014;13(12):5561–5569. doi: 10.1021/pr5005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mori S, Aoyagi Y, Yanagi M, Suzuki Y, Asakura H. Serum Nacetylglucosaminyltransferase I11 activities in hepatocellular carcinoma. J Gastroenterol Hepatol. 1998;13(6):610–619. doi: 10.1111/j.1440-1746.1998.tb00699.x. [DOI] [PubMed] [Google Scholar]
- 29.Vanderschaeghe D, Laroy W, Sablon E, Halfon P, Van Hecke A, Delanghe J, Callewaert N. GlycoFibroTest is a highly performant liver fibrosis biomarker derived from DNA sequencer-based serum protein glycomics. Mol Cell Proteomics. 2009;8(5):986–994. doi: 10.1074/mcp.M800470-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stumpo KA, Reinhold VN. The N-glycome of human plasma research articles. J Proteome Res. 2010;9(9):4823–4830. doi: 10.1021/pr100528k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekesova S, Kosti O, Chandler KB, Wu J, Madej HL, Brown KC, Simonyan V, Goldman R. N-glycans in liver-secreted and immunoglogulin-derived protein fractions. J Proteome. 2012;75(7):2216–2224. doi: 10.1016/j.jprot.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis, Rapid Commun. Mass Spectrom. 2005;19(23):3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mechref Y, Novotny MV. Structural characterization of oligosaccharides using MALDI/TOF/TOF tandem mass spectrometry. Anal Chem. 2003;75(18):4895–4903. doi: 10.1021/ac0341968. [DOI] [PubMed] [Google Scholar]
- 34.Mechref Y, Novotny MV. Mass spectrometric mapping and sequencing of N-linked oligosaccharides derived from submicrogram amounts of glycoproteins. Anal Chem. 1998;70(3):455–463. doi: 10.1021/ac970947s. [DOI] [PubMed] [Google Scholar]
- 35.Christiansen MN, Kolarich D, Nevalainen H, Packer NH, Jensen PH. Challenges of determining o-glycopeptide heterogeneity: a fungal glucanase model system. Anal Chem. 2010;82(9):3500–3509. doi: 10.1021/ac901717n. [DOI] [PubMed] [Google Scholar]
- 36.Deshpande N, Jensen PH, Packer NH, Kolarich D. GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J Proteome Res. 2010;9(2):1063–1075. doi: 10.1021/pr900956x. [DOI] [PubMed] [Google Scholar]
- 37.Mysling S, Palmisano G, Hojrup P, Thaysen-Andersen M. Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal Chem. 2010;82(13):5598–5609. doi: 10.1021/ac100530w. [DOI] [PubMed] [Google Scholar]
- 38.Nilsson J, Rüetschi U, Halim A, Hesse C, Carlsohn E, Brinkmalm G, Larson G. Enrichment of glycopeptides for glycan structure and attachment site identification. Nat Methods. 2009;6(11):809–811. doi: 10.1038/nmeth.1392. [DOI] [PubMed] [Google Scholar]
- 39.Guerrero A, Lerno L, Barile D, Lebrilla CB. Top-down analysis of highly post- translationally modified peptides by Fourier transform ion cyclotron resonance mass spectrometry. J Am Soc Mass Spectrom. 2014;26(3):453–459. doi: 10.1007/s13361-014-1034-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee LY, Lin CH, Fanayan S, Packer NH, Thaysen-Andersen M. Differential site accessibility mechanistically explains subcellular-specific N-glycosylation determinants. Front Immunol. 2014 Aug;5:1–13. doi: 10.3389/fimmu.2014.00404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pompach P, Brnakova Z, Sanda M, Wu J, Edwards N, Goldman R. Site-specific glycopeptides of haptoglobin in liver cirrhosis and hepatocellular carcinoma. Mol Cell Proteomics. 2013;12(5):1281–1293. doi: 10.1074/mcp.M112.023259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pompach P, Ashline DJ, Brnakova Z, Benicky J, Sanda M, Goldman R. Protein and site specificity of fucosylation in liver-secreted glycoproteins. 2014;13(12):5561–5569. doi: 10.1021/pr5005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song E, Pyreddy S, Mechref Y. Quantification of glycopeptides by multiple reaction monitoring liquid chromatography/tandem mass spectrometry, Rapid Commun. Mass Spectrom. 2012;26(17):1941–1954. doi: 10.1002/rcm.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute quantitation of immunoglobulin G and its glycopeptides using multiple reaction monitoring. Anal Chem. 2013;85(18):8585–8593. doi: 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruhaak LR, Lebrilla CB. Applications of multiple reaction monitoring to clinical glycomics. Chromatographia. 2014;78(5–6):335–342. doi: 10.1007/s10337-014-2783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurogochi M, Matsushista T, Amano M, Furukawa J, Shinohara Y, Aoshima M, Nishimura SI. Sialic acid-focused quantitative mouse serum glycoproteomics by multiple reaction monitoring assay. Mol Cell Proteomics. 2010;9(11):2354–2368. doi: 10.1074/mcp.M110.000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldman R, Sanda M. Targeted methods for quantitative analysis of protein glycosylation, PROTEOMICS - Clin. Appl. 2015;9(1–2):17–32. doi: 10.1002/prca.201400152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amster IJ. Fourier transform mass spectrometry. J Mass Spectrom. 1996;31(12):1325–1337. [Google Scholar]
- 49.Marshall AG, Hendrickson CL, Jackson GS. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom Rev. 1998;17(1):1–35. doi: 10.1002/(SICI)1098-2787(1998)17:1<1::AID-MAS1>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 50.Fenyo D, Chait BT. Protein identification using mass spectrometric information. Electrophoresis. 1998;19:998–1005. doi: 10.1002/elps.1150190615. [DOI] [PubMed] [Google Scholar]
- 51.Fu Q, Garnham CP, Elliott ST, Bovenkamp DE, Van Eyk JE. A robust, streamlined, and reproducible method for proteomic analysis of serum by delipidation, albumin and IgG depletion, and two-dimensional gel electrophoresis. Proteomics. 2005;5(10):2656–2664. doi: 10.1002/pmic.200402048. [DOI] [PubMed] [Google Scholar]
- 52.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, et al. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376(1):1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Park SY, Lee SH, Kawasaki N, Itoh S, Kang K, Ryu SHee, Hashii N, Kim JM, Kim JY, Kim JHoe. α1-3/4 fucosylation at Asn 241 of β-haptoglobin is a novel marker for colon cancer: a combinatorial approach for development of glycan biomarkers. Int J Cancer. 2012;130(10):2366–2376. doi: 10.1002/ijc.26288. [DOI] [PubMed] [Google Scholar]
- 54.Park SY, Yoon SJ, Jeong YT, Kim JM, Kim JY, Bernert B, Ullman T, Itzkowitz SH, Kim JH, Hakomori S. N-glycosylation status of beta-haptoglobin in sera of patients with colon cancer, chronic inflammatory diseases and normal subjects. Int J Cancer. 2010;126(1):142–155. doi: 10.1002/ijc.24685. [DOI] [PubMed] [Google Scholar]
- 55.Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography-mass spectrometry. J Chromatogr B Anal Technol Biomed Life Sci. 2005;825(2):124–133. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 56.Zielinska DF, Gnad F, Wiśniewski JR, Mann M. Precision mapping of an in vivo Nglycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141(5):897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z, Hancock WS. Approach to the comprehensive analysis of glycoproteins isolated from human serum using a multi-lectin affinity column. J Chromatogr A. 2004;1053(1–2):79–88. [PubMed] [Google Scholar]
- 58.Zauner G, Deelder AM, Wuhrer M. Recent advances in hydrophilic interaction liquid chromatography (HILIC) for structural glycomics. Electrophoresis. 2011;32(24):3456–3466. doi: 10.1002/elps.201100247. [DOI] [PubMed] [Google Scholar]
- 59.Karlsson NG, Wilson NL, Wirth HJ, Dawes P, Joshi H, Packer NH. Negative ion graphitised carbon nano-liquid chromatography/mass spectrometry increases sensitivity for glycoprotein oligosaccharide analysis. Rapid Commun Mass Spectrom. 2004;18(19):2282–2292. doi: 10.1002/rcm.1626. [DOI] [PubMed] [Google Scholar]
- 60.Pabst M, Bondili JS, Stadlmann J, Mach L, Altmann F. Mass + retention time = structure: a strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal Chem. 2007;79(13):5051–5057. doi: 10.1021/ac070363i. [DOI] [PubMed] [Google Scholar]
- 61.Lam MPY, Siu SO, Lau E, Mao X, Sun HZ, Chiu PCN, Yeung WSB, Cox DM, Chu IK. Online coupling of reverse-phase and hydrophilic interaction liquid chromatography for protein and glycoprotein characterization. Anal Bioanal Chem. 2010;398(2):791–804. doi: 10.1007/s00216-010-3991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zauner G, Koeleman CAM, Deelder AM, Wuhrer M. Nano-HPLC-MS of glycopeptides obtained after nonspecific proteolysis. Methods Mol Biol. 2013;951:113–127. doi: 10.1007/978-1-62703-146-2_9. [DOI] [PubMed] [Google Scholar]
- 63.Miyoshi E, Moriwaki K, Nakagawa T. Biological function of fucosylation in cancer biology. J Biochem. 2008;143(6):725–729. doi: 10.1093/jb/mvn011. [DOI] [PubMed] [Google Scholar]
- 64.Wuhrer M, Deelder AM, van der Burgt YEM. Mass spectrometric glycan rearrangements. Mass Spectrom Rev. 2011;30(4):664–680. doi: 10.1002/mas.20337. [DOI] [PubMed] [Google Scholar]
- 65.Sanda M, Pompach P, Benicky J, Goldman R. LC-MS3 quantification of O-glycopeptides in human serum. Electrophoresis. 2013;34(16):2342–4349. doi: 10.1002/elps.201200658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


