Abstract
Satellite DNAs (satDNAs) are major components of eukaryote genomes. However, because of their quick divergence, the evolutionary origin of a given satDNA family can rarely be determined. Herein we took advantage of available primate sequenced genomes to determine the origin of the CapA satDNA (approx. 1500 bp long monomers), first described in the tufted capuchin monkey Sapajus apella. We show that CapA is an abundant satDNA in Platyrrhini, whereas in the genomes of most eutherian mammals, including humans, this sequence is present only as a single copy located within a large intron of the NOS1AP (nitric oxide synthase 1 adaptor protein) gene. Our data suggest that this intronic CapA-like sequence gave rise to the CapA satDNA and we discuss possible mechanisms implicated in this event. This is the first report to our knowledge of a single copy intronic sequence giving origin to a satDNA that reaches up to 100 000 copies in some genomes.
Keywords: repetitive DNA, satellite DNA origin, Platyrrhini
1. Introduction
Eukaryote genomes are replete with repetitive DNA sequences among which satellite DNAs (satDNAs) are prominent components [1]. These tandem repeats (TRs) are commonly found as very long arrays located in heterochromatic regions of chromosomes, such as pericentric and centromeric heterochromatin, and subtelomeric regions [2]. Because satDNAs are fast-evolving, their origin is often hard to determine [1,2]. The most recurrent evolutionary path suggested for satDNA origin is the tandem amplification of internal segments of transposable elements (TEs), with many cases described so far in animals and plants [3].
The study of primate genomes is paramount for medical genetics and comparative evolutionary studies of the human genome. Nevertheless, as for most eukaryotes, the satDNA fraction of these genomes is often overlooked. Herein, we took advantage of the available primate sequenced genomes to study the evolution of a satDNA called CapA. This satDNA was first described in the New World monkey (NWM) Sapajus apella (previously classified as Cebus apella, Cebidae, Platyrrhini). It has large monomers (approx. 1500 bp), and DNA–DNA hybridization experiments revealed its presence in other NWMs [4,5]. However, given the paucity of DNA sequence data available at the time of its description, CapA origin and evolution remained elusive.
A similarity search against the NCBI nucleotide collection using BLASTn and the CapA sequence described in Malfoy et al. [4] returned highly similar sequences from several mammals (electronic supplementary material, table S1). Interestingly, one hit (77% coverage with 79% identity) was annotated as the Homo sapiens nitric oxid synthase 1 adaptor protein (NOS1AP), located at the proximal region of the chromosome 1 long arm (1q23.3). An inspection of this gene revealed that the region with similarity to the deposited CapA monomer is approximately 1180 bp long and is situated in the second intron of NOS1AP. Subsequent BLAT searches using this intronic CapA segment against the whole human genome (hg38) retrieved no additional matches, indicating that this sequence is not repetitive in humans.
We used this human CapA-like intronic sequence as query against all vertebrate genomes available in NCBI. This search retrieved similar sequences mostly as single matches in each genome, with CapA-like sequences being present in most eutherians. Data from a few species with partial chromosome assembly and chromosome painting data with human probes confirm that the CapA-like sequence remains in the same locus, probably associated with the NOS1AP gene (electronic supplementary material, table S2). Additionally, to confirm the association of the CapA-like sequence with NOS1AP, we analysed the CapA-like surrounding sequence of one representative of each order (electronic supplementary material, table S3). Some eutherian clades appear to lack this CapA-like sequence, most notably the Chiroptera, some Eulipotyphla and some Rodentia (electronic supplementary material, table S2; figure 1). No CapA-like sequences were found in Marsupialia or Monotremata, which are the sister clades to Eutheria. These findings suggest that a single CapA-like sequence was present in the ancestor of eutherians and that it was likely the precursor sequence that gave rise to the CapA satDNA (figure 1).
Figure 1.
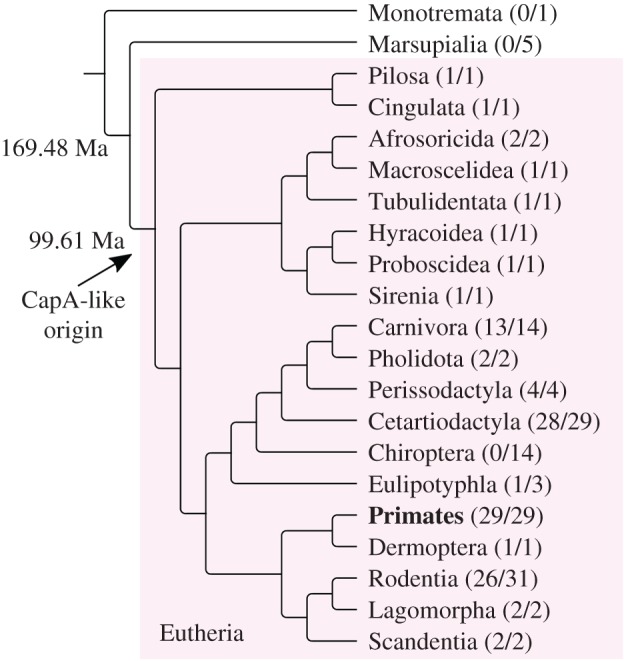
Mammalian phylogeny adapted from Foley et al. [6]. The CapA precursor probably predates the eutherian radiation (arrow). Numbers in parentheses indicate the genomes of species in which CapA was found and the number of species analysed. Within primates (bold) CapA was found as a satDNA only in Platyrrhini.
The only genomes in which we found multiple copies of CapA were those of NWMs from the family Cebidae, with some contigs revealing the presence of a few CapA TRs (electronic supplementary material, table S4). However, because assembly of TRs is usually incomplete, we also used raw Illumina data to estimate CapA abundance in available NWMs' genomes (electronic supplementary material, table S5). Strikingly, this approach revealed that CapA is an abundant TR in Cebus capucinus (genome proportion of 4.21%), Saimiri boliviensis (1.48%) and Aotus nancymaae (0.27%; electronic supplementary material, table S5). In fact, CapA divergence landscapes are very similar across the three Cebidae genera, despite the significant differences in abundance (electronic supplementary material, figure S1). A slightly higher overall divergence was detected in S. boliviensis (19.67%), compared with 17.8 and 18.46% in A. nancymaae and C. capucinus, respectively (electronic supplementary material, figure S1). In Callithrix jacchus, we found CapA in low copy numbers but also tandemly arranged (electronic supplementary material, table S4). The longest array we found in the Callithrix jacchus genome assembly is around 4.5 kb long, with nearly three monomers. Interestingly, even in the genomes where CapA achieved high abundance, the intronic ancestral locus persisted as an intact CapA-like monomer, with the exception of Callithrix jacchus. In this species, the intronic CapA on chromosome 1 suffered a rearrangement involving a small duplication and the insertion of an unrelated sequence of approximately 300 bp (electronic supplementary material, figure S2).
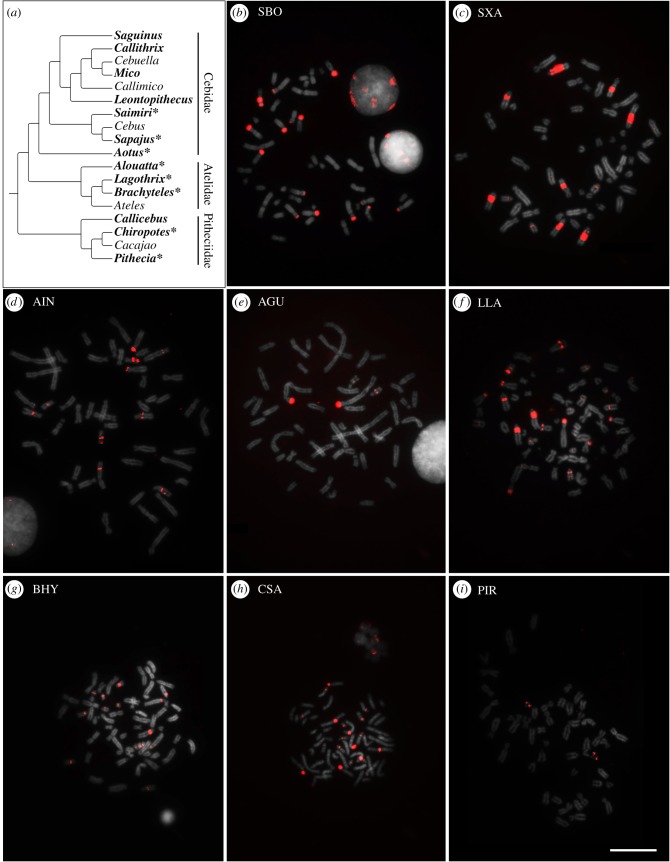
All currently available sequenced genomes of NWMs belong to species of Cebidae, preventing us from assessing the amplification status of CapA in the other two NWMs' families, Atelidae and Pitheciidae. To expand our analysis to species without sequence data, we assessed CapA abundance by performing fluorescent in situ hybridization (FISH) with the human intronic CapA-like sequence as a probe onto cells of several NWMs (table 1). We detected signs of CapA expansion in members of the three Platyrrhini families (figure 2). Within Cebidae, CapA was present in high abundance in Sapajus xanthosternos, S. boliviensis and Aotus infulatus and was not detected in representatives of Callithrichinae (a Cebidae subfamily). CapA also occurred as a high-copy-number sequence in Alouatta guariba, Lagothrix lagotricha and Brachyteles hypoxanthus, of the family Atelidae, in which CapA was less abundant in A. guariba than in L. lagotricha and B. hypoxanthus. In Pitheciidae, Chiropotes satanas and Pithecia irrorata also presented signs of CapA expansion, whereas Callicebus nigrifrons did not. CapA was found in only one chromosome pair of P. irrorata and was abundant in Chiropotes satanas. In the species in which CapA was abundant, it was mostly associated with heterochromatin revealed after CBG-banding (electronic supplementary material, table S6). In the species that did not display visible blocks of CapA it is possible that it occurs in low copy numbers or that the sequence has diverged considerably, preventing its detection by FISH.
Table 1.
Species in which FISH with the human intronic CapA-like was performed.
| species | family | procedence |
|---|---|---|
| Saguinus imperator (SIM) | Cebidae | Fundação Zoo-Botânica de Belo Horizonte |
| Callithrix penicillata (CPE) | Cebidae | Universidade Federal de Minas Gerais |
| Mico argentata (MAR) | Cebidae | Universidade de São Paulo |
| Leontopithecus rosalia (LRO) | Cebidae | Fundação Zoo-Botânica de Belo Horizonte |
| Sapajus xanthosternos (SXA) | Cebidae | Fundação Zoo-Botânica de Belo Horizonte |
| Saimiri boliviensis (SBO) | Cebidae | University of Florence |
| Aotus infulatus (AIN) | Cebidae | Fundação Zoo-Botânica de Belo Horizonte |
| Alouatta guariba (AGU) | Atelidae | Fundação Zoo-Botânica de Belo Horizonte |
| Lagothrix lagotricha (LLA) | Atelidae | Fundação Zoo-Botânica de Belo Horizonte |
| Brachyteles hypoxanthus (BHY) | Atelidae | Fundação Zoo-Botânica de Belo Horizonte |
| Callicebus nigrifrons (CNI) | Pitheciidae | Fundação Zoo-Botânica de Belo Horizonte |
| Chiropotes satanas (CSA) | Pitheciidae | Fundação Zoo-Botânica de Belo Horizonte |
| Pithecia irrorata (PIR) | Pitheciidae | Fundação Zoo-Botânica de Belo Horizonte |
Figure 2.
(a) Distribution of CapA among Platyrrhini families. Platyrrhini phylogeny adapted from Schneider & Sampaio [7]. Genera with representatives analysed are shown in bold and the asterisk indicates CapA amplification. Fluorescent in situ hybridization with CapA in metaphases of: (b) Saimiri boliviensis (SBO), (c) Sapajus xanthosternos (SXA), (d) Aotus infulatus (AIN), (e) Alouatta guariba (AGU), (f) Lagothrix lagotricha (LLA), (g) Brachyteles hypoxanthus (BHY), (h) Chiropotes satanas (CSA) and (i) Pithecia irrorata (PIR). Scale bar, 10 µm.
The data presented herein suggest that CapA suffered an expansion within Platyrrhini, less than approximately 43.5 million years ago (Ma), after the divergence between Platyrrhini and Catarrhini [8]. Assuming that CapA expansion predates the divergence of the NWMs' families, this satDNA would have been independently lost in some taxa, notably the Callithrichinae and C. nigrifrons. A less parsimonious hypothesis would be that CapA has become a satDNA independently multiple times, as only some species within each family showed CapA expansion (figure 2). To investigate the alternative possibilities more NWM species need to be analysed at the cytogenetic and genomic levels.
Herein we found that the large CapA satDNA present in Platyrrhini has originated from an intronic single copy sequence still present in most eutherians. Nevertheless, it is difficult to reconstruct the steps of amplification. One possibility is that CapA arose through segmental duplications. In fact, segmental duplications have already been evoked to explain the hyperexpansion of sequences in primate genomes [9]. The single copy intronic precursor sequence is still present in the putative ancestral locus of CapA. Therefore, prior to amplification, this CapA sequence would have had to move to another genomic region. The TEs, such as L1 retrotransposons (long interspersed nucleotide element-1), may have also participated in the process through transduction events. L1 is associated with the indirect spread of other retrotranscripts, but it can also carry non-L1 DNA that flanks L1 3′ ends to new genomic locations in a process called transduction [10,11]. We analysed the original CapA locus in the species in which it became a satDNA and found no evidence of recurrent structural features that could foster its mobile potential.
Duplicative transposition followed by expansion of particular euchromatic segments has been described in the pericentromeric regions of human and the subterminal ends (generally heterochromatic) of great ape chromosomes [9,12]. CapA duplication and expansion in NWMs may be explained by a similar mechanism, in which transfer of the CapA intronic segment to heterochromatic regions in the ancestral Platyrrhini genome followed by hyperexpansion through unequal crossing over would have given rise to the CapA satDNA in some species. Although we favour this segmental duplication hypothesis, the incompleteness of current NWMs' genome assemblies prevents its scrutiny to exhaustion.
In conclusion, we characterized CapA, a satDNA formed less than 43.5 Ma, with approximately 1500 bp monomers present in species of the three Platyrrhini families. In Cebidae, with the exception of Callitrichines, CapA abundance ranges from 0.27 to 5%. The CapA-like ancestral sequence is present in most eutherians, embedded in the second intron of the NOS1AP gene, such as in H. sapiens. One hypothesis for the CapA expansion in NWMs is through duplicative transposition followed by expansion through unequal crossing over. To the best of our knowledge, this is the first report of a single copy intronic sequence giving origin to a satDNA, with as much as 100 000 copies in some genomes.
2. Material and methods
We searched for sequences similar to CapA in the non-redundant nucleotide collection of GenBank using the CapA monomer described in Fanning et al. [5] as query and the BLASTn tool [13]. This search returned hits from several mammals, particularly NWMs, and included a hit in the intron of the NOS1AP gene from H. sapiens (electronic supplementary material, table S1). After checking the human reference genome (hg38) at the University of California, Santa Cruz using BLAT, we confirmed that this CapA-like sequence exists at a single locus, inside the NOS1AP gene on chromosome 1. We then used this human intronic sequence as query in BLASTn searches against all vertebrate assembled genomes available at NCBI. A hit was included when query cover was greater than or equal to 30% and e-value ≤ e−5. Only Eutheria displayed CapA-like sequences. Number of hits with the CapA-like sequence, query cover, identity and e-value of each search are available in electronic supplementary material, table S2.
In the genomes where multiple hits of CapA were found, we used raw Illumina reads and RepeatMasker [14] to estimate CapA's abundance. We included data from an Old World monkey (Chlorocebus aethiops) and a great ape (H. sapiens) as negative controls. Sequence reads used here were obtained from the Short Read Archive at NCBI (available at http://www.ncbi.nlm.nih.gov/sra/), with accession numbers as follows: A. nancymaae SRR1692991, Callithrix jacchus SRR1746970, Cebus capucinus imitator SRR3144006, S. boliviensis SRR317821, Chlorocebus aethiops sabaeus SRR5251202 and H. sapiens ERR016352. Sequence reads were downloaded in fastq format using the software SRAToolkit (https://github.com/ncbi/sra-tools) and random samples of 2 million reads ranging from 101 to 125 bp were produced using the software seqtk (https://github.com/lh3/seqtk/). To determine the fraction of the reads similar to CapA, RepeatMasker was used in the following setup: sensitive mode, without search for low complexity or bacterial insertion sequences, wublast as search engine and a custom library containing the NOS1AP intronic CapA-like sequence. We used the alignment files generated by RepeatMasker to calculated Kimura distances of CapA fragments against the ancestral sequence (NOS1AP intronic CapA-like), using the utility script calcDivergenceFromAlign.pl from the RepeatMasker package. Results were imported to RStudio and plotted.
To investigate CapA abundance in species for which no genome data were available, we performed FISH with the intronic CapA probe. This probe was obtained after PCR amplification of human DNA with primers flanking the CapA-like intronic sequence from NOS1AP (CapA-F: ACTTCCTCACTGACCTGTCTT; CapA-R: GGGCTGATGCTTAATGTAGCA). The PCR products were purified, cloned and sequenced to ensure specificity (accession number: MG264524). Chromosome spreads of several NWMs, mostly with unknown geographic origin, were obtained from fibroblast or lymphocyte cultures (table 1). FISH was performed with 200 ng of biotin-labelled probes, following Araújo et al. [15].
Supplementary Material
Acknowledgements
The authors thank the following people for their assistance in obtaining samples for this study: Yatiyo Yonenaga-Yassuda and Camila do Nascimento Moreira (Universidade de São Paulo); Roscoe Stanyon (University of Florence); Alan Lane de Melo (Universidade Federal de Minas Gerais).
Ethics
The work did not involve the direct use of animals, so ethical permission was not required.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material.
Authors' contributions
M.P.V. and G.B.D. carried out bioinformatics, cytogenetic and molecular analyses, participated in data analysis, designed the study and drafted the manuscript; V.S.P. collected samples; G.B.D., G.C.S.K. and M.S. conceived and coordinated the study, and helped drafting the manuscript. All authors gave final approval for publication and agree to be accountable for all content therein.
Competing interests
The authors declare they have no competing interests.
Funding
This work was supported by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq process 407262/2013-0) to M.S. M.P.V. and G.B.D. received fellowships from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
References
- 1.Plohl M, Meštrović N, Mravinac B. 2012. Satellite DNA evolution. Genome Dyn. Basel 7, 126–152. ( 10.1159/000337122) [DOI] [PubMed] [Google Scholar]
- 2.Biscotti MA, Olmo E, Heslop-Harrison JS. 2015. Repetitive DNA in eukaryotic genomes. Chromosome Res. 23, 415–420. ( 10.1007/s10577-015-9499-z) [DOI] [PubMed] [Google Scholar]
- 3.Meštrović N, Mravinac B, Pavlek M, Vojvoda-Zeljko T, Šatović E, Plohl M. 2015. Structural and functional liaisons between transposable elements and satellite DNAs. Chromosome Res. 23, 583–596. ( 10.1007/s10577-015-9483-7) [DOI] [PubMed] [Google Scholar]
- 4.Malfoy B, Rousseau N, Vogt N, Viegas-Pequignot E. 1986. Nucleotide sequence of an heterochromatic segment recognized by the antibodies to Z-DNA in fixed metaphase chromosomes. Nucleic Acids Res. 14, 3197–3214. ( 10.1093/nar/14.8.3197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fanning TG, Seuánez HN, Forman L. 1993. Satellite DNA sequences in the new World primate Cebus apella (Platyrrhini, Primates). Chromosoma 102, 306–311. ( 10.1007/BF00661273) [DOI] [PubMed] [Google Scholar]
- 6.Foley NM, Springer MS, Teeling EC. 2016. Mammal madness: is the mammal tree of life not yet resolved? Phil. Trans. R. Soc. B 371, 20150140 ( 10.1098/rstb.2015.0140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schneider H, Sampaio I. 2015. The systematics and evolution of New World primates—a review. Mol. Phylogenet. Evol. 82, 348–357. ( 10.1016/j.ympev.2013.10.017) [DOI] [PubMed] [Google Scholar]
- 8.Perelman P, et al. 2011. A molecular phylogeny of living primates. PLoS Genetics 7, e1001342 ( 10.1371/journal.pgen.1001342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, et al. 2005. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93. ( 10.1038/nature04000) [DOI] [PubMed] [Google Scholar]
- 10.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 10, 411–415. ( 10.1101/gr.10.4.411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostertag EM, Kazazian HH. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538. ( 10.1146/annurev.genet.35.102401.091032) [DOI] [PubMed] [Google Scholar]
- 12.Eichler EE, Budarf ML, Rocchi M, Deaven LL, Doggett NA, Baldini A, Nelson DL, Mohrenweiser WH. 1997. Interchromosomal duplications of the adrenoleukodystrophy locus: a phenomenon of pericentromeric plasticity. Hum. Mol. Genet. 6, 991–1002. ( 10.1093/hmg/6.7.991) [DOI] [PubMed] [Google Scholar]
- 13.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 14.Smit AFA, Hubley R, Green P.2015. RepeatMasker Open-4.0. 2013–2015. Institute for Systems Biology. See http://repeatmasker.org .
- 15.Araújo NP, de Lima LG, Dias GB, Kuhn GCS, de Melo AL, Yonenaga-Yassuda Y, Stanyon R, Svartman M. 2017. Identification and characterization of a subtelomeric satellite DNA in Callitrichini monkeys. DNA Res. 24, 377–385. ( 10.1093/dnares/dsx010) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material.