Abstract
The psychological effects of brain-expressed imprinted genes in humans are virtually unknown. Prader–Willi syndrome (PWS) is a neurogenetic condition mediated by genomic imprinting, which involves high rates of psychosis characterized by hallucinations and paranoia, as well as autism. Altered expression of two brain-expressed imprinted genes, MAGEL2 and NDN, mediates a suite of PWS-related phenotypes, including behaviour, in mice. We phenotyped a large population of typical individuals for schizophrenia-spectrum and autism-spectrum traits, and genotyped them for the single-nucleotide polymorphism rs850807, which is putatively functional and linked with MAGEL2 and NDN. Genetic variation in rs850807 was strongly and exclusively associated with the ideas of reference subscale of the schizophrenia spectrum, which is best typified as paranoia. These findings provide a single-locus genetic model for analysing the neurological and psychological bases of paranoid thinking, and implicate imprinted genes, and genomic conflicts, in human mentalistic thought.
Keywords: paranoia, Prader–Willi, magel2, necdin, genomic imprinting
1. Introduction
Genomic imprinting is the allele-specific expression of genes according to their parent of origin. By the kinship theory of imprinting, maternally and paternally expressed genes are in conflict over the expression of phenotypes that impose demands on asymmetrically related kin, especially the mother [1]. The kinship theory is well supported by empirical data for traits related to growth, but effects of imprinted genes remain virtually unstudied with regard to cognition and behaviour in typically developing humans [2].
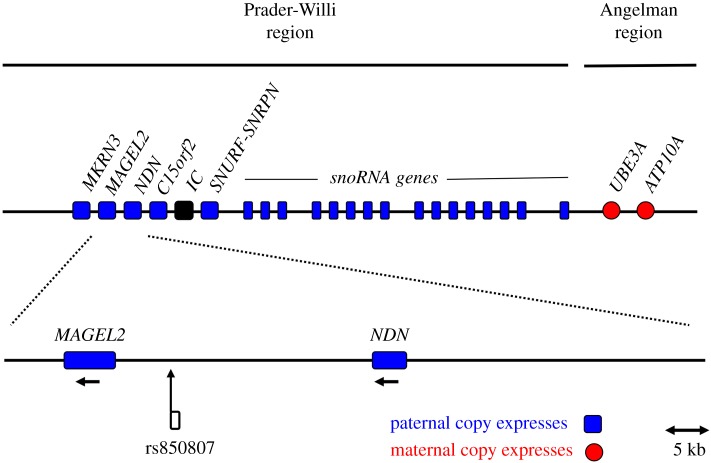
Most data on the effects of brain-expressed imprinted genes in humans come from study of the behavioural and psychological phenotypes expressed by individuals with rare neurogenetic conditions that involve losses or gains of imprinted gene expression. Prader–Willi syndrome (PWS), which is caused by some combination of absent paternal imprinted gene expression, or increased maternal imprinted gene expression, for an imprinted domain on chromosome 15 (figure 1), represents one of the best studied imprinted human neurogenetic conditions. This syndrome is characterized by poor feeding in infancy, excessive sleepiness and weak cry, followed in childhood by a high incidence of hyperphagia, and in adolescence and early adulthood by very high rates (over 20% in some studies) of psychosis and schizophrenia-associated traits, especially hallucinations and paranoia ([3,4]; electronic supplementary material, table S1), as well as autism [4]. The apparent absence of single-locus human or mouse mutations that recapitulate all major features of PWS indicates that its phenotypes are mediated by effects from multiple loci within the PWS region, though with clear behavioural effects from dosages of the paralogous genes MAGEL2 (MAGE Family Member L2) and NDN (Necdin) as indicated by knockouts in mice [5,6]. As such, specific loci within the PWS imprinted gene domain, especially in the MAGEL2–NDN region, are expected to mediate specific psychological aspects of this syndrome. Moreover, such psychological traits, related to the schizophrenia spectrum and autism spectrum, can be hypothesized to be underlain, in part, by segregating single-nucleotide polymorphism (SNP) variation within this region for typical populations, though in attenuated form.
Figure 1.
The Prader–Willi genomic region, in relation to the position of rs850807. (Online version in colour.)
In this study, we tested this hypothesis by phenotyping a large population of typical individuals for schizophrenia-spectrum and autism-spectrum traits, and genotyping them for the SNP rs850807, a polymorphism that is putatively functional and linked with MAGEL2 and NDN. We predicted in particular that genotypic variation in this SNP should be associated with one or more of the psychiatric phenotypes characteristic of PWS, though at non-clinical, personality-variation levels. Moreover, to the extent that rs850807 is associated with imprinted gene effects, the traits that it mediates should be relevant to the kinship theory of imprinting, for brain-expressed genes.
2. Methods
We collected questionnaire and DNA data from 831 undergraduate students (553 females and 278 males). Levels and forms of schizotypal traits were quantified using the Schizotypal Personality Questionnaire-Brief Revised [7]. This questionnaire comprises 32 items using a 5-point Likert scale, with response choices that range from ‘strongly disagree’ to ‘strongly agree’. It includes seven subscales: (1) constricted affect; (2) social anxiety; (3) magical thinking; (4) unusual perceptions and (5) ideas of reference; (6) eccentric behaviour; and (7) odd speech, which sum to total schizotypy. Scores for one question in the odd speech subscale were missing due to technical error for 38 individuals, and were interpolated from the other questions in this subscale. The Autism Spectrum Quotient (AQ) [8] was used to quantify the extent to which participants endorsed autism spectrum phenotypes. This is a 50-item questionnaire that assesses autistic traits across five domains including: (1) sociality; (2) communication; (3) attention to detail; (4) attention switching; and (5) imagination, with total AQ score as the sum.
The SNP rs850807 was chosen for analysis based on two lines of evidence: (1) its patterns of linkage disequilibrium with SNPs in MAGEL2, in the CEU population (HapMap population with northern and western European ancestry) (D′ = 0.96, R2 = 0.83 with rs8920 at the 3′ end of the MAGEL2 exon) and NDN (D′ = 0.41, R2 = 0.16 with rs3743340 at the 5′ end of the NDN exon), and (2) the finding that this SNP was associated, nominally though not after statistical adjustment, with a psychotic–affective psychiatric phenotype, suicide attempts in bipolar disorder, in a previous study ([9], results from GWASdb, jjwanglab.org/gwasdb).
DNA was extracted from saliva using standard phenol–chloroform protocols.
Fluorophore-labelled primers for rs850807 fluorophore-labelled were used in TaqMan® genotyping using a Roche LightCycler® 96 Real-Time PCR machine. Fluorescence data were analysed under Endpoint Genotyping using LightCycler® 96 software, v. 1.1.0.1320, and genotyping success was greater than 98%.
Allele frequencies were identical to those in 1000 Genomes EUR Superpopulation (0.60 C and 0.40 T), and genotypes were in Hardy–Weinberg equilibrium (χ2 = 0.02, p > 0.50). Two-way ANOVAs of sex and genotype on each of the 12 AQ and schizotypal personality questionnaire (SPQ) subscales were used to test for sex by genotype interactions; only one of these interaction effects, for the SPQ magical thinking subscale, was nominally significant (p = 0.014); the sexes were therefore pooled for analyses. False discovery rate (FDR) adjustments were used for multiple testing, for the 12 AQ and SPQ subscales, using the 0.05 level of significance. Tests for phenotypic AQ and SPQ differences, using ANOVA and t-tests, among the rs850807 genotypes were conducted under three genetic models: (CC versus CT versus TT), (CC versus CT + TT) and (CC + CT versus TT). Statistics were conducted in R v. 3.3.2. Data have been deposited at http://dx.doi.org/10.5061/dryad.7g5s4 [10].
3. Results
The SPQ Ideas of Reference subscale showed highly significant differences (p < 0.01) between rs850807 genotypes, under two of the three genetic models (CC versus CT versus TT) and (CC versus CT + TT) (table 1). These differences remained significant (p < 0.05 and p < 0.01) after FDR adjustment that included all five of the AQ subscales and all seven SPQ subscales. After FDR adjustment, significance values for all AQ and SPQ subscales other than SPQ Ideas of Reference were above 0.35, indicating a high specificity of the effects for this subscale. A highly conservative threefold adjustment for the performance of the three (non-independent) genotypic model tests had no qualitative effect on the main results (p still less than 0.05 under two of the three models).
Table 1.
Descriptive statistics and significance tests, for differences among individuals by rs850807 genotype. Shown under p-value columns are FDR-adjusted significance values for the five AQ subscales, and the seven SPQ subscales; FDR adjustments that also include AQ and SPQ totals yield similar results, with FDR-adjusted significance for SPQ Ideas of Reference of 0.014 and 0.0028 for the first and third models, respectively, and a p-value of 0.28 for SPQ total under the CC versus (CT + TT) model.
| mean ± s.d. genotype |
p-values unadjusted/FDR-adjusted genotypic model |
|||||
|---|---|---|---|---|---|---|
| CC (N = 300) | CT (N = 397) | TT (N = 134) | CC versus CT versus TT (ANOVA) | (CC + CT) versus TT (t-test) | CC versus (CT + TT) (t-test) | |
| AQ social skills | 2.0±1.9 | 1.9±2.0 | 1.9±1.9 | 0.81/0.92 | 0.73/0.99 | 0.53/0.84 |
| AQ attention switching | 4.8±2.0 | 4.7±1.9 | 4.7±2.1 | 0.84/0.92 | 0.78/0.99 | 0.56/0/84 |
| AQ attention to detail | 5.2±2.2 | 5.4±2.2 | 5.5±2.2 | 0.27/0.65 | 0.40/0.91 | 0.12/0.36 |
| AQ communication | 2.3±1.9 | 2.2±1.8 | 2.3±1.9 | 0.44/0.88 | 0.46/0.91 | 0.45/0.84 |
| AQ imagination | 2.3±1.6 | 2.2±1.7 | 2.2±1.6 | 0.77/0.92 | 0.99/0.99 | 0.49/0.84 |
| AQ total | 16.6±5.6 | 16.4±5.6 | 16.7±5.7 | 0.87 | 0.73 | 0.79 |
| SPQ ideas of reference | 17.3±4.1 | 16.1±4.2 | 16.2±4.6 | 0.0010/0.012 | 0.35/0.91 | 0.00020/0.0024 |
| SPQ constricted affect | 14.5±4.6 | 14.5±5.0 | 14.4±5.3 | 0.99/0.99 | 0.91/0.99 | 0.98/0.98 |
| SPQ eccentric behaviour | 12.0±3.8 | 11.4±3.9 | 11.6±4.0 | 0.14/0.56 | 0.99/0.99 | 0.063/0.36 |
| SPQ social anxiety | 11.0±3.7 | 11.1±3.8 | 10.7±4.1 | 0.68/0.92 | 0.39/0.91 | 0.89/0.97 |
| SPQ magical thinking | 7.9±3.4 | 7.6±3.5 | 8.3±3.7 | 0.12/0.56 | 0.069/0.84 | 0.73/0.88 |
| SPQ odd speech | 13.1±2.7 | 12.8±3.1 | 12.7±3.2 | 0.26/0.65 | 0.30/0.91 | 0.12/0.36 |
| SPQ unusual perception | 9.8±2.8 | 9.7±3.1 | 9.6±3.3 | 0.80/0.92 | 0.53/0.91 | 0.67/0.88 |
| SPQ total | 85.5±14.8 | 83.1±16.2 | 83.6±18.2 | 0.15 | 0.69 | 0.055 |
As in Willour et al. [9], the ‘C’ allele was associated with elevated scores (on SPQ-Ideas of Reference, and suicidality risk in bipolar disorder). Homozygote CC individuals exhibited Ideas of Reference scores 6.6% higher than those of CT + TT individuals.
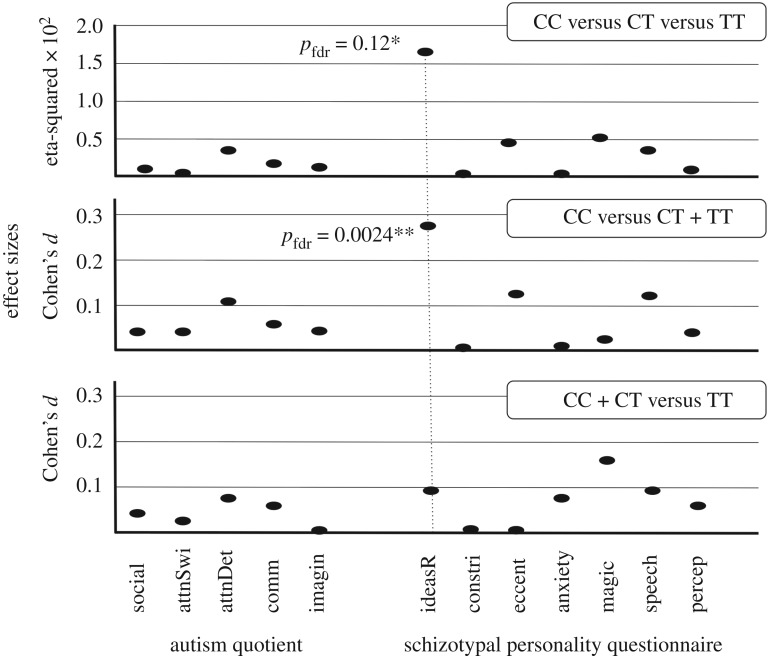
Effect sizes (eta-squared and Cohen's d) were small to moderate, under the two genetic models showing significant genotypic differences for SPQ Ideas of Reference; these effect sizes were at least two to three times greater than for any other AQ or SPQ subscale (figure 2).
Figure 2.
Effect sizes for genotypic differences, for the AQ and SPQ subscales. *p < 0.05; **p < 0.01.
4. Discussion
The main results of this study are twofold. First, the data support the hypothesis that the SNP rs850807 is associated strongly and specifically with ideas of reference, which indicate aspects of paranoia. As such, neurotypical individuals who differ in their rs850807 genotype exhibit psychological variation that reflects, in greatly reduced magnitude, this psychiatric characteristic that is commonly found among individuals with PWS who develop forms of psychosis. The specificity of these results, in that Ideas of Reference but not perceptual aberrations, magical thinking, other schizotypal or autism spectrum traits, or total schizotypy or autism, are associated with rs850807 genotype, implies that the neurogenetic circuitry mediated by this locus subserves paranoid ideation. In the SPQ, the Ideas of Reference subscale reflects endorsement of statements that others ‘are talking about me’, ‘have it in for me’, are ‘not trustworthy’, ‘take notice of me’, ‘are watching me’ and ‘want to take advantage of me’. As such, it includes multiple facets of paranoid thinking, including imagined and delusional thoughts, actions and plans.
The presence of specific neural systems subserving paranoid ideation is also consistent with the description of a paranoid subtype of schizophrenia (e.g. [11]), and with the existence of a personality disorder, Paranoid Personality Disorder, specific to this psychological domain [12]. Our findings may also help to explain the lack of significance in Fukuo et al.'s [13] test for association of three MAGEL2 SNPs with schizophrenia and mood disorders, given that in our study total schizotypy was not significantly associated with rs850807 (lowest p = 0.28 after FDR adjustment, model CC versus CT + TT), but the Ideas of Reference subscale was (p = 0.0028 after FDR under this model). Our data also indicate that autism spectrum traits are not associated with rs850807.
The discovery of genetic variation showing evidence of specific gene–phenotype correspondence for paranoia should be especially useful for neuroimaging genetics, to help discern the neurological basis of paranoid thinking. Such studies are important given the evidence that high levels of paranoia play a central role in motivating random acts of extreme violence (e.g. [14]), in addition to their deleterious effects on everyday functioning through enhanced fear of social threats.
Second, in the context of PWS, our findings implicate the rs850807 locus, which is linked strongly (D′ = 0.96, R2 = 0.83 in CEU) to SNPs in the gene MAGEL2 and weakly to moderately for NDN (D′ = 0.41, R2 = 0.16), in one component of psychosis. These results thus suggest that the psychiatric correlates of this syndrome exhibit a mosaic of genetic underpinnings that encompass at least two loci (since paranoia represents only one psychological aspect of PWS), and that presumably involve dosages of multiple brain-expressed imprinted genes. Future work should be directed towards replication in other populations, and elucidating the precise functional-genetic and epigenetic mechanisms of the psychological trait associations with rs850807 reported here, to determine what SNP, haplotype and gene are responsible for the patterns observed.
An apparent role for imprinted gene expression in paranoia is of particular interest given relevant evolutionary theory. Thus, by the kinship theory of imprinting applied to psychological variation [15], small maternal biases to brain-expressed imprinted gene expression may result in increased theory of mind-related cognition that, in childhood, is beneficial to maternally inherited genes and mothers. By contrast, however, large maternal biases are expected to lead to pathologically hyper-developed theory of mind (as in PWS), of which paranoia represents a paradigmatic case. This hypothesis predicts that individuals with high-paranoia genotypes of rs850807 exhibit lower expression or activity of the linked paternally expressed genes MAGEL2 or NDN, or both. More generally, variation in brain-expressed imprinted genes may mediate a range of human psychological traits related to mentalistic cognition.
Supplementary Material
Acknowledgements
We thank members of the Crawford Laboratory of Human Evolutionary Studies at Simon Fraser University for helpful discussions.
Ethics
The study was approved by the ethics boards at University of Alberta (Pro00015728) and Simon Fraser University (2010s0554), and participants provided prior written informed consent.
Data accessibility
Data are in Dryad under: http://dx.doi.org/10.5061/dryad.7g5s4 [10].
Authors' contributions
B.C. and P.H. designed the study, S.R. and I.S. collected and analysed the data, and all authors contributed to writing and revision. All authors approved the final version of the manuscript and agree to be held accountable for its content.
Competing interests
The authors have no competing interests.
Funding
The project was funded by NSERC 2014-06505.
References
- 1.Haig D. 2014. Coadaptation and conflict, misconception and muddle, in the evolution of genomic imprinting. Heredity 113, 96–103. ( 10.1038/hdy.2013.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies W, Isles AR, Humby T, Wilkinson LS. 2007. What are imprinted genes doing in the brain? Epigenetics 2, 201–206. ( 10.4161/epi.2.4.5379) [DOI] [PubMed] [Google Scholar]
- 3.Haig D, Wharton R. 2003. Prader-Willi syndrome and the evolution of human childhood. Am. J. Hum. Biol. 15, 320–329. ( 10.1002/ajhb.10150) [DOI] [PubMed] [Google Scholar]
- 4.Cassidy SB, Driscoll DJ. 2009. Prader–Willi syndrome. Eur. J. Hum. Genet. 17, 3–13. ( 10.1038/ejhg.2008.165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muscatelli F, Abrous DN, Massacrier A, Boccaccio I, Le Moal M, Cau P, Cremer H. 2000. Disruption of the mouse Necdin gene results in hypothalamic and behavioral alterations reminiscent of the human Prader–Willi syndrome. Hum. Mol. Genet. 9, 3101–3110. ( 10.1093/hmg/9.20.3101) [DOI] [PubMed] [Google Scholar]
- 6.Schaaf CP, et al. 2013. Truncating mutations of MAGEL2 cause Prader–Willi phenotypes and autism. Nat. Genet. 45, 1405–1408. ( 10.1038/ng.2776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen AS, Matthews RA, Najolia GM, Brown LA. 2010. Toward a more psychometrically sound brief measure of schizotypal traits: introducing the SPQ-Brief Revised. J. Pers. Disord. 24, 516–537. ( 10.1521/pedi.2010.24.4.516) [DOI] [PubMed] [Google Scholar]
- 8.Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E. 2001. The autism-spectrum quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J. Autism Dev. Dis. 31, 5–17. ( 10.1023/A:1005653411471) [DOI] [PubMed] [Google Scholar]
- 9.Willour VL, et al. 2012. A genome-wide association study of attempted suicide. Mol. Psychiatry 17, 433–444. ( 10.1038/mp.2011.4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crespi B, Read S, Salminen I, Hurd P. 2017. Data from: A genetic locus for paranoia Dryad Digital Repository. ( 10.5061/dryad.7g5s4) [DOI] [PMC free article] [PubMed]
- 11.Montag C, Dziobek I, Richter IS, Neuhaus K, Lehmann A, Sylla R, Heekeren HR, Heinz A, Gallinat J. 2011. Different aspects of theory of mind in paranoid schizophrenia: evidence from a video-based assessment. Psychiatry Res. 186, 203–209. ( 10.1016/j.psychres.2010.09.006) [DOI] [PubMed] [Google Scholar]
- 12.Triebwasser J, Chemerinski E, Roussos P, Siever LJ. 2013. Paranoid personality disorder. J. Pers. Disord. 27, 795–805. ( 10.1521/pedi_2012_26_055) [DOI] [PubMed] [Google Scholar]
- 13.Fukuo Y, et al. 2010. Lack of association between MAGEL2 and schizophrenia and mood disorders in the Japanese population. Neuromolecular Med. 12, 285–291. ( 10.1007/s12017-010-8116-8) [DOI] [PubMed] [Google Scholar]
- 14.Coid JW, Ullrich S, Bebbington P, Fazel S, Keers R. 2016. Paranoid ideation and violence: meta-analysis of individual subject data of 7 population surveys. Schiz. Bull. 42, 907–915. ( 10.1093/schbul/sbw006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crespi B, Badcock C. 2008. Psychosis and autism as diametrical disorders of the social brain. Behav. Brain Sci. 31, 241–261. ( 10.1017/S0140525X08004214) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Crespi B, Read S, Salminen I, Hurd P. 2017. Data from: A genetic locus for paranoia Dryad Digital Repository. ( 10.5061/dryad.7g5s4) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data are in Dryad under: http://dx.doi.org/10.5061/dryad.7g5s4 [10].