Abstract
Intergeneric hybridization and introgression was reported from one of two populations of the recently discovered kipunji (Rungwecebus kipunji), a critically endangered African monkey species of southern Tanzania. Kipunjis of the introgressed population (from Mount Rungwe) carry a mitochondrial DNA (mtDNA) haplotype closely related to those of parapatric yellow baboons (Papio cynocephalus), whereas the second kipunji population, in the Udzungwa Mountains, carries the original kipunji mtDNA haplotypes, which diverged from the baboon lineage about 3 million years ago. Interestingly, in our study of yellow baboons in Tanzania, we found that baboons from the southeastern boundary of the Udzungwa Mountains carry mtDNA haplotypes closely related to the original kipunji haplotype, whereas baboons from the northern boundary, as expected, carry mtDNA haplotypes of the northern yellow baboon clade. These findings provide evidence for a case of inverted intergeneric admixture in primates: (i) a baboon mtDNA haplotype introgressed the Mount Rungwe kipunji population by mitochondrial capture and (ii) an Udzungwa Mountains kipunji mtDNA haplotype introgressed a small subpopulation of yellow baboons by either mitochondrial capture or nuclear swamping. The baboon–kipunji example therefore constitutes an interesting system for further studies of the effects of genetic admixture on fitness and speciation.
Keywords: primates, hybridization, mitochondrial capture, nuclear swamping, divergence-age
1. Introduction
Advances in molecular genetics have tremendously influenced our understanding of the impact of hybridization and introgression on the evolution of animal taxa [1,2]. Introgression is a long known phenomenon [3,4] and is defined as the infiltration of genetic material of one species into another through repeated backcrossing of hybrids to one or both parental species [5]. As a result of introgression certain genetic or phenotypical characters of species ‘A’ can be found in species ‘B’. Strong uni-parental backcrossing can lead to mitochondrial or Y-chromosomal capture so that species ‘A’ might carry mitochondria or the Y-chromosome of species ‘B’ [1,6]. First indications for introgression are incongruent phylogenies derived from different genetic markers or from genetic and phenotypic characters [1,7].
A remarkable case of intergeneric introgression in primates was reported from the kipunji (Rungwecebus kipunji), a critically endangered African monkey, discovered in 2003 in southern Tanzania [8]. Kipunjis occur in two isolated populations: in the forests of Mount Rungwe and the adjacent Livingstone Forest and about 350 km to the northeast in the Ndundulu Forest of the Udzungwa Mountains [9]. Analyses of mitochondrial DNA (mtDNA) of the Mount Rungwe population revealed that the kipunji mtDNA sequences are nested within the geographically proximate southern yellow baboon (Papio cynocephalus) clade [6,10,11]. In contrast, the population of the Udzungwa Mountains carry mitochondria concordant with a position as sister taxon to baboons [12]. This pattern was interpreted as a result of introgression with mitochondrial capture in the Mount Rungwe population. In contrast the Ndundulu haplotype from the Udzungwa Mountains was considered to represent the true, non-introgressed kipunji haplotype [12]. Here we report on the mtDNA phylogenetic relationships among the Udzungwa kipunjis and sym- and parapatric yellow baboons and a case of inverted intergeneric introgression.
2. Material and methods
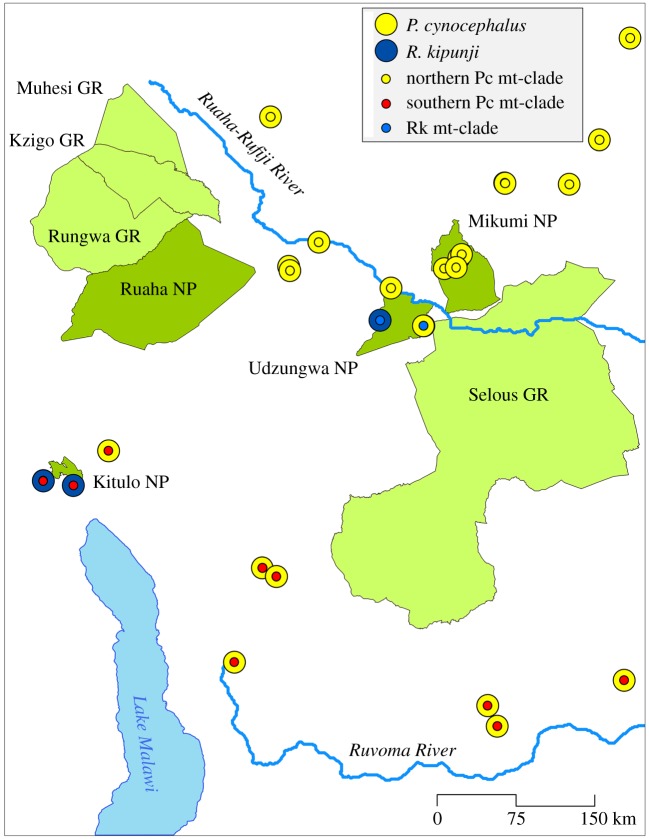
During a Tanzania-wide screening for infection of non-human primates with the pathogenic bacterium Treponema pallidum, we collected tissue samples from 17 yellow baboons in the Udzungwa Mountain National Park (see electronic supplementary material) (figure 1). After DNA extraction, we amplified and sequenced the Brown region [13] of the mtDNA genome using methods outlined in Newman et al. [14] and Zinner et al. [15]. Obtained haplotypes were aligned against orthologous sequences derived from GenBank of other baboons, kipunjis, and related taxa that were used as outgroups (electronic supplementary material, table S1). We reconstructed maximum-likelihood (ML) and Bayesian trees and estimated divergence ages. Full details of all analyses and an ethical note are provided in the electronic supplementary material.
Figure 1.
Map of southern Tanzania with yellow baboon (yellow circle) and kipunji (blue circle) sampling locations and geographical occurrence of mtDNA haplotypes of northern (small yellow circles) and southern (small red circles) yellow baboons and kipunjis (small blue circles). Protected areas: GR, game reserve; NP, national park, Pc, Papio cynocephalus; Rk, Rungwecebus kipunji. Details on samples can be found in the electronic supplementary material, table S1.
3. Results and discussion
As in earlier studies on baboon phylogeny using mtDNA markers [14–19], baboons segregate into various strongly supported clades (ML bootstrap support (BS): greater than 90%, Bayesian posterior probabilities (PP): greater than 0.90). However, the branching pattern among them is not well resolved. Moreover, these clades follow a geographical pattern and their distribution is not always concordant with the boundaries of the six species now generally recognized. As in previous studies, yellow, olive (Papio anubis), and chacma baboons (Papio ursinus) show para- or polyphylies, which were regarded as results of ancient introgressive hybridization events [15,20].
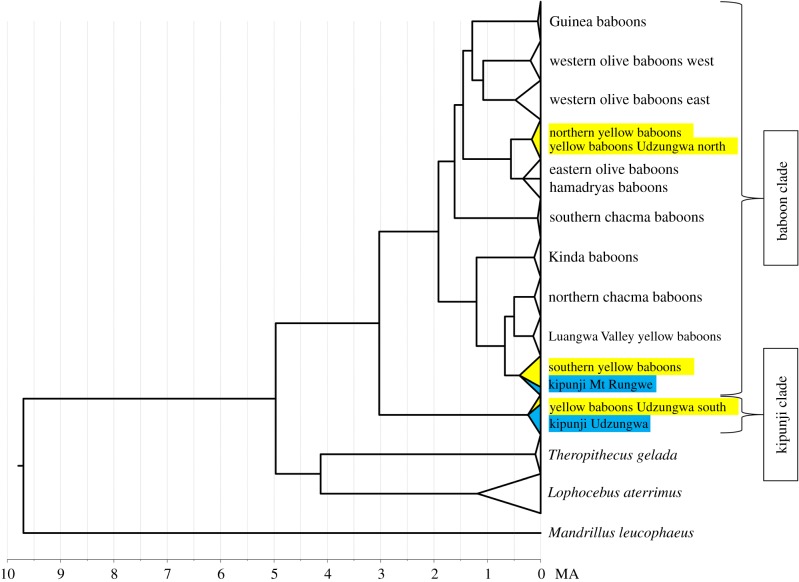
In our tree reconstruction, as previously found [10–12], the mtDNA lineage of Mount Rungwe kipunjis clusters with southern yellow baboons and the kipunji mtDNA lineage from Udzungwa constitutes a sister clade to the Papio clade, with a divergence time of 3.03 million years ago (Ma) (95% highest posterior density (HPD): 2.06–4.09 Ma) (figure 2; electronic supplementary material, table S2). Also expected is the grouping of yellow baboons from northern Udzungwa with conspecifics of the northern yellow baboon clade. However, yellow baboons from southern Udzungwa do not cluster either with southern or with northern yellow baboons. Instead, they represent a closely related sister clade to the mtDNA lineage of Udzungwa kipunjis. We estimated the split of the Mount Rungwe kipunji mtDNA lineage from its nearest southern yellow baboon lineage at 0.05 Ma (0.00–0.13 Ma), while the yellow baboon mtDNA clade from southern Udzungwa diverged from the Udzungwa kipunji lineage at 0.24 Ma (0.07–0.44 Ma) (electronic supplementary material, table S2).
Figure 2.
Chronogram showing phylogenetic relationships and divergence times among various baboon and kipunji mtDNA lineages. The mtDNA lineage of the Mount Rungwe kipunji clusters with southern yellow baboons, while yellow baboons from southern Udzungwa cluster with the Udzungwa kipunji. In contrast, yellow baboons from northern Udzungwa cluster with their conspecifics of the northern yellow baboon mtDNA clade (see also electronic supplementary material, figure S1 and table S2). (Online version in colour.)
The phylogenetic relationships of the two kipunji populations with respective para- or sympatric yellow baboons suggest three introgression scenarios. Alternative explanations for the observed pattern such as incomplete lineage sorting (ILS) are highly unlikely, because in small populations, as in kipunjis [21], lineage sorting should be relatively fast and the distribution pattern of haplotypes follows a geographical structure, while ILS should be random in respect to geography [15,22]. The first scenario assumes the mtDNA lineage found in Udzungwa kipunjis and baboons represents an ancient Papio mtDNA lineage, which would mean that this is the oldest Papio mtDNA lineage discovered so far and also that the Udzungwa kipunjis captured baboon mitochondria, similar to the scenario envisioned for the Mount Rungwe kipunji population [11]. The two other scenarios assume that the Udzungwa kipunji mtDNA lineage indeed is the original kipunji mtDNA lineage [12], which would mean that the southern Udzungwa yellow baboons either captured mitochondria from female kipunjis (scenario 2) where theoretically a single event of a female kipunji breeding successfully with a male baboon is sufficient or that baboon males introgressed a subpopulation (e.g. a small isolated group) of the Udzungwa kipunji population over generations leading to nuclear swamping (scenario 3). In scenario 3, male-mediated gene flow from baboons into kipunjis would alter the nuclear gene pool of the kipunjis, up to completely converting the kipunji phenotype into the baboon phenotype, while leaving the ancestral kipunji mtDNA sequences in place. We believe the first scenario is unlikely, because if the Udzungwa kipunji mtDNA lineage represented an old Papio lineage, we would expect to find it more frequently in the baboon population of southern Tanzania, which, however, was not the case [19]. It would be interesting to investigate whether this lineage can be also found in baboons of the adjacent Selous Game Reserve. To test the hypothetical scenarios, we would need to analyse whole nuclear genomes inferring amount and frequency of gene flow between kipunjis and baboons. It would be interesting to see which genes were exchanged, in which direction and whether similar mosaic genome patterns can be detected in both kipunji populations. Also, given that the baboon population carrying the kipunji mtDNA seems to be rather small, it might be interesting to examine whether these baboons show any indications of fitness loss related to genetic admixture, as compared with unadmixed conspecifics.
The kipunji–baboon case is, to our knowledge, the first reported case of inverted intergeneric introgression in primates, and maybe mammals overall. However, inverted introgression among species of the same genus has been reported e.g. in Lepus. Brown hares of northern Spain (L. europaeus) carry mtDNA lineages of mountain hares (L. timidus), whereas mountain hares in northern Russia carry mtDNA lineages of brown hares [23–26]. In the case of Spanish hares, ancient introgression is obvious, because the mountain hare went extinct in the region after the last glacial period, whereas introgressive hybridization is still ongoing in Sweden and Russia. Ongoing hybridization between Rungwecebus and Papio, however, is not reported.
Supplementary Material
Acknowledgements
We are grateful to Julius D. Keyyu (Tanzania Wildlife Research Institute, TAWIRI) and Inyasi Y. Lejora (Tanzania National Parks, TANAPA) for their support and help with the logistics and advice. We thank TAWIRI, TANAPA and the Commission for Science and Technology, Tanzania for necessary permissions. Hassan Mohamed Nguluma, Christina Kibwe and the other Udzungwa National Park staff are thanked for supporting sampling. We thank Simone Lüert and Christiane Schwarz for help with the laboratory work.
Ethics
No animals were captured specifically for this study. The study was in line with the Veterinary Act of 2003 and Tanzania Wildlife Research Institute's (TAWIRI) Guideline for Conducting Wildlife Research (2001). Further information is available in the electronic supplementary material.
Data accessibility
Detailed methods are available as electronic supplementary material. Newly generated haplotypes have been deposited in GenBank (accession numbers MG569923–MG569944).
Authors' contributions
D.Z. and C.R. conceived the study. I.S.C. collected samples; S.K. did laboratory work; C.R. did the phylogenetic analyses. All authors discussed the results, collaborated in writing the paper, approved the final version of the manuscript and agreed to be held accountable for the content therein.
Competing interests
The authors declare no competing interests.
Funding
This study was supported by grants of the German Research Foundation (DFG): KN1097/3-1 (to S.K.) and RO3055/2-1 (to C.R.).
References
- 1.Abbott RJ, Barton NH, Good JM. 2016. Genomics of hybridization and its evolutionary consequences. Mol. Ecol. 25, 2325–2332. ( 10.1111/mec.13685) [DOI] [PubMed] [Google Scholar]
- 2.Tung J, Barreiro LB. 2017. The contribution of admixture to primate evolution. Curr. Opin. Genet. Dev. 47, 61–68. ( 10.1016/j.gde.2017.08.010) [DOI] [PubMed] [Google Scholar]
- 3.Anderson E. 1949. Introgressive hybridization. New York, NY: Wiley. [Google Scholar]
- 4.Heiser CB. 1973. Introgression re-examined. Bot. Rev. 39, 347–366. ( 10.1007/BF02859160) [DOI] [Google Scholar]
- 5.Anderson E, Hubricht L. 1938. Hybridization in Tradescantia. III. The evidence for introgressive hybridization. Am. J. Bot. 25, 396–402. ( 10.2307/2436413) [DOI] [Google Scholar]
- 6.Zinner D, Arnold ML, Roos C. 2011. The strange blood: natural hybridization in primates. Evol. Anthropol. 20, 96–103. ( 10.1002/evan.20301) [DOI] [PubMed] [Google Scholar]
- 7.Funk DJ, Omland KE. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 34, 397–423. ( 10.1146/annurev.ecolsys.34.011802.132421) [DOI] [Google Scholar]
- 8.Davenport TRB, Jones T. 2008. Rungwecebus kipunji. The IUCN red list of threatened species 2008 , e.T136791A4340286. 10.2305/IUCN.UK.2008.RLTS.T136791A4340286.en (accessed 18 December 2017). [DOI]
- 9.Jones T, Ehardt CL, Butynski TM, Davenport TRB, Mpunga NE, Machaga SJ, De Luca DW. 2005. The highland mangabey Lophocebus kipunji: a new species of African monkey. Science 308, 1161–1164. ( 10.1126/science.1109191) [DOI] [PubMed] [Google Scholar]
- 10.Burrell AS, Jolly CJ, Tosi AJ, Disotell TR. 2009. Mitochondrial evidence for the hybrid origin of the kipunji, Rungwecebus kipunji (Primates: Papionini). Mol. Phylogenet. Evol. 51, 340–348. ( 10.1016/j.ympev.2009.02.004) [DOI] [PubMed] [Google Scholar]
- 11.Zinner D, Arnold ML, Roos C. 2009. Is the new primate genus Rungwecebus a baboon? PLoS ONE 4, e4859 ( 10.1371/journal.pone.0004859) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts TE, Davenport TRB, Hildebrandt KBP, Jones T, Stanley WT, Sargis EJ, Olson LE. 2010. The biogeography of introgression in the critically endangered African monkey Rungwecebus kipunji. Biol. Lett. 6, 233–237. ( 10.1098/rsbl.2009.0741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown WM, Prager EM, Wang A, Wilson AC. 1982. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J. Mol. Evol. 18, 225–239. ( 10.1007/BF01734101) [DOI] [PubMed] [Google Scholar]
- 14.Newman TK, Jolly CJ, Rogers J. 2004. Mitochondrial phylogeny and systematics of baboons (Papio). Am. J. Phys. Anthropol. 124, 17–27. ( 10.1002/ajpa.10340) [DOI] [PubMed] [Google Scholar]
- 15.Zinner D, Groeneveld LF, Keller C, Roos C. 2009. Mitochondrial phylogeography of baboons (Papio spp.)—indication for introgressive hybridization? BMC Evol. Biol. 9, 83 ( 10.1186/1471-2148-9-83) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wildman DE, Bergman TJ, al-Aghbari A, Sterner KN, Newman TK, Phillips-Conroy JE, Jolly CJ, Disotell TR. 2004. Mitochondrial evidence for the origin of hamadryas baboons. Mol. Phylogenet. Evol. 32, 287–296. ( 10.1016/j.ympev.2003.12.014) [DOI] [PubMed] [Google Scholar]
- 17.Zinner D, Buba U, Nash S, Roos C. 2011. Pan-African voyagers. The phylogeography of baboons. In Primates of Gashaka (eds Sommer V, Ross C), pp. 267–306. New York, NY: Springer. [Google Scholar]
- 18.Zinner D, Wertheimer J, Liedigk R, Groeneveld LF, Roos C. 2013. Baboon phylogeny as inferred from complete mitochondrial genomes. Am. J. Phys. Anthropol. 150, 133–140. ( 10.1002/ajpa.22185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinner D, Keller C, Nyahongo JW, Butynski TM, de Jong YA, Pozzi L, Knauf S, Liedigk R, Roos C. 2015. Distribution of mitochondrial clades and morphotypes of baboons Papio spp. (Primates: Cercopithecidae) in eastern Africa. J. East Afr. Nat. Hist. 104, 143–168. ( 10.2982/028.104.0111) [DOI] [Google Scholar]
- 20.Keller C, Roos C, Groeneveld LF, Fischer J, Zinner D. 2010. Introgressive hybridization in southern African baboons shapes patterns of mtDNA variation. Am. J. Phys. Anthropol. 142, 125–136. ( 10.1002/ajpa.21209) [DOI] [PubMed] [Google Scholar]
- 21.Davenport TRB, De Luca DW, Jones T, Mpunga NE, Machaga SJ, Kitegile A, Phillipps GP. 2008. The critically endangered kipunji Rungwecebus kipunji of southern Tanzania: first census and conservation status assessment. Oryx 42, 352–359. ( 10.1017/S0030605308000422) [DOI] [Google Scholar]
- 22.Avise JC. 2004. Molecular markers, natural history, and evolution. Sunderland, MA: Sinauer Associates. [Google Scholar]
- 23.Alves PC, Melo-Ferreira J, Freitas H, Boursot P. 2008. The ubiquitous mountain hare mitochondria: multiple introgressive hybridization in hares, genus Lepus. Phil. Trans. R. Soc. B 363, 2831–2839. ( 10.1098/rstb.2008.0053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marques JP, Farelo L, Vilela J, Vanderpool D, Alves PC, Good JM, Boursot P, Melo-Ferreira J. 2017. Range expansion underlies historical introgressive hybridization in the Iberian hare. Sci. Rep. 7, 40788 ( 10.1038/srep40788) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melo-Ferreira J, Farelo L, Freitas H, Suchentrunk F, Boursot P, Alves PC. 2014. Home-loving boreal hare mitochondria survived several invasions in Iberia: the relative roles of recurrent hybridisation and allele surfing. Heredity 112, 265–273. ( 10.1038/hdy.2013.102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thulin CG, Fang M, Averianov AO. 2006. Introgression from Lepus europaeus to L. timidus in Russia revealed by mitochondrial single nucleotide polymorphisms and nuclear microsatellites. Hereditas 143, 68–76. ( 10.1111/j.2006.0018-0661.01952.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Detailed methods are available as electronic supplementary material. Newly generated haplotypes have been deposited in GenBank (accession numbers MG569923–MG569944).