Abstract
MicroRNAs (miRNA) are small non‐coding RNAs that target mRNAs and are consequently involved in the post‐transcriptional regulation of gene expression. Some miRNAs are ubiquitously expressed in tissue, while others are tissue‐specific or tissue‐enriched. miRNAs can be released by cells and are found in various biofluids, including serum and plasma. Thus, measuring miRNAs in the circulation may provide information on the originating tissue or cells. MyomiRs are described as striated muscle‐specific or muscle‐enriched miRNAs. Their circulating levels can be measured and have been proposed to be new biomarkers of physiological and pathological muscle processes. The aims of this review are to summarize the current knowledge of circulating myomiRs, to identify the types of information they can provide about skeletal muscle, and to determine how to apply that information in the fields of research and medicine.
Keywords: MicroRNA, Circulating microRNA, MyomiRs, Skeletal muscle, Biomarker
MicroRNA biogenesis and myomiRs
MicroRNAs (miRNAs) belong to the large family of non‐coding RNAs. They are transcribed as primary miRNAs (pri‐mRNA) that are very similar to mRNAs with a 5′ cap and a 3′ poly(A) tail. Pri‐miRNAs are processed by a protein complex that includes the endoribonuclease DROSHA, giving rise to a 60–70 bp hairpin precursor miRNA (pre‐miRNA). Once exported from the nucleus, the pre‐mRNA is cleaved by the endoribonuclease DICER1, releasing two short (18–22 nucleotides) mature miRNAs. The so‐called guide strand is loaded in the ribonucleoprotein complex miRNA‐induced silencing complex and binds to the 3′ untranslated region of target mRNAs, inducing degradation or blocking translation, whereas the other strand (‘passenger’ strand) is thought to be degraded.1 Therefore, miRNAs are important players in the post‐transcriptional control of gene expression. In terms of nomenclature, a prefix refers to the specie: ‘hsa’ for Homo sapiens, ‘mmu’ for Mus musculus, etc. (Table 1). Both precursor and mature miRNAs are given numbers; pre‐miRNAs are referred to as ‘mir‐number’, mature miRNA as ‘miR‐number’. Some miRNAs that only differ from one or two bases are given the same number, but a letter is added (for example, miR‐208a and miR‐208b). The two miRNAs released by pre‐miRNA cleavage are given the suffix ‘‐5p’ or ‘‐3p’ according to their 3′ or 5′ location on the pre‐miRNA. In many publications, both the prefix and the suffix are removed.
Table 1.
Genome context of muscle‐specific miRNAs (myomiRs) in humans
| MicroRNA | Pre‐miRNA | Genome context | Gene | Human chromosome | Clustered miRNA gene (distant from <10 kb) | Mature miRNA | Tissue specificity | |
|---|---|---|---|---|---|---|---|---|
| miR‐1 | hsa‐mir‐1‐1 | Intragenic | MIR1‐1 host gene | 20 | 20q13.33 | hsa‐mir‐133a‐2 | hsa‐miR‐1‐5p | Heart/skeletal muscle |
| hsa‐mir‐1‐2 | Intragenic | MIR133A1 host gene | 18 | 18q11.2 | hsa‐mir‐133a‐1 | hsa‐miR‐1‐3p | ||
| miR‐133a | hsa‐mir‐133a‐1 | Intragenic | MIR133A1 host gene | 18 | 18q11.2 | hsa‐mir‐1‐2 | hsa‐miR‐133a‐5p | Heart/skeletal muscle |
| hsa‐mir‐133a‐2 | Intragenic | MIR1‐1 host gene | 20 | 20q13.33 | hsa‐mir‐1‐1 | hsa‐miR‐133a‐3p | ||
| miR‐133b | hsa‐mir‐133b | Intergenic | 6 | 6p12.2 | hsa‐mir‐206 | hsa‐miR‐133b | Skeletal muscle | |
| miR‐206 | hsa‐mir‐206 | Intergenic | 6 | 6p12.2 | hsa‐mir‐133b | hsa‐miR‐206 | Skeletal muscle | |
| miR‐208a | hsa‐mir‐208a | Intragenic | MYH6 | 14 | 14q11.2 | hsa‐miR‐208a‐5p | Heart | |
| hsa‐miR‐208a‐3p | ||||||||
| miR‐208b | hsa‐mir‐208b | Intragenic | MYH7 | 14 | 14q11.2 | hsa‐miR‐208b‐5p | Heart/skeletal muscle | |
| hsa‐miR‐208b‐3p | ||||||||
| miR‐486 | hsa‐mir‐486‐1 | Intragenic | ANK1 | 8 | 8p11.21 | hsa‐mir‐486‐2 | hsa‐miR‐486‐5p | Heart/skeletal muscle |
| hsa‐mir‐486‐2 | Intergenic | hsa‐mir‐486‐1 | hsa‐miR‐486‐3p | |||||
| miR‐499a | hsa‐mir‐499a | Intragenic | MYH7B | 20 | 20q11.22 | hsa‐mir‐499b | hsa‐miR‐499a‐5p | Heart/skeletal muscle |
| hsa‐miR‐499a‐3p | ||||||||
| miR‐499b | hsa‐mir‐499b | Intergenic | 20 | 20q11.22 | hsa‐mir‐499a | hsa‐miR‐499b‐5p | Heart/skeletal muscle | |
Upon stimulation or injury, miRNAs can be released from the cell in an active (secretion) or passive (membrane leaking) manner.2 miRNAs have been found in most biofluids, including serum and plasma, and there is an increasing interest in studying circulating miRNAs (c‐miRNA) as biomarkers of physiological or pathological processes.3, 4 Furthermore, recent evidence shows that extracellular miRNAs can have direct biologic effects on other cells.5, 6 Extracellular vesicles, such as exosomes, microvesicles, high‐density lipoprotein, and apoptotic bodies, are secreted by most cells and contain various molecules, including miRNAs. The vesicles are released into the extracellular space or circulation and can be taken up by neighbouring cells. Their contents are released into the cytosol of recipient cells, where miRNAs can exert their effects. Thus, miRNAs may be involved in cell‐to‐cell communication in an autocrine, paracrine, or endocrine fashion.
Many miRNAs are ubiquitously expressed in most tissues and cell types, but some are tissue‐specific (mature miRNA expression is 20‐fold higher than the mean expression in other tissues) or tissue‐enriched (mature miRNA expression is higher than in other tissues but less than 20‐fold).7 Subsets of miRNAs can be described as striated muscle‐specific (miR‐1, miR‐133a, miR‐133b, miR‐206, miR‐208a, miR‐208b, and miR‐499) or muscle‐enriched (miR‐486). They are involved in myoblast proliferation/differentiation, muscle regeneration, or fibre‐type specification (for review, refer to Diniz and Wang8). The tissue specificity of myomiRs is due either to the genomic location of their coding DNA within introns of myosin heavy chain genes or to transcriptional control by muscle‐specific transcription factors such as MyoD, Mef2, or Srf.9, 10, 11, 12, 13 Therefore, several myomiRs are expressed in skeletal muscle as well as in cardiac muscle. However, miR‐206 and miR‐133b are specific to skeletal muscle, and miR‐208a is cardiac muscle‐specific. As with any miRNA, myomiRs can be released into the circulation and be measured in plasma or serum. In humans, baseline levels of circulating myomiRs are low,14, 15 but alterations in their levels have been reported in physiologic as well as in pathologic conditions. This review summarizes the current knowledge on circulating, cell‐free myomiRs and focuses on the physiological and medical significance of altered levels with regards to skeletal muscle.
Circulating myomiRs during exercise and recovery
Because miRNAs are involved in the regulation of gene expression, they can participate in the adaptation of muscles to exercise and training. In a mouse model of functional overload, an increase in the primary transcripts pri‐mir‐1‐2, pri‐mir‐133a‐2, and pri‐mir‐206 with a concomitant decrease in mature miR‐1 and miR‐133a was found within the muscle.16 This was the first demonstration of an alteration in miRNA expression in response to altered muscle load. Similar alterations are reported in humans, where resistance exercise reduced the levels of miR‐1, miR‐133a, or miR‐133b expression in skeletal muscle,17, 18 although no changes were found in another study.19 Conversely, an acute bout of endurance exercise (3 h of cycling) induced a modest but significant increase in muscle miR‐1, miR‐133a, or miR‐133b but not miR‐206 levels.20, 21 Therefore, it is now established that exercise modulates myomiR expression within muscle tissue, although the physiological significance of these alterations is still poorly understood. Whether modifications in muscle myomiRs could result in variations in their circulating levels is unknown. The first evidence that c‐miRNAs could be altered during exercise came from a study where the effect of both acute exhaustive exercise (short incremental cycling) and 9 weeks of endurance training (rowing) were examined.22 Several profiles of expression were described for inflammation or angiogenesis‐related miRNAs, but only one myomiR (miR‐133a) was measured, and no changes were found. Similarly, only miR‐486 slightly decreased in young men after an acute cycling exercise (60 min, 70% VO2max),14 and no changes in miR‐133 (now known as 133a) occurred in the serum of young athletes during and after a 4 h cycling test at 70% VO2max,23 showing that endurance exercise has limited effects on circulating myomiRs. However, several studies consistently reported increases in plasma levels of miR‐1, miR‐133a, miR‐133b, miR‐206, miR‐208b, or miR‐499 immediately after running a marathon or during the recovery period,23, 24, 25, 26 suggesting that the changes in circulating myomiRs may be dependent on the type of exercise. In a study that compared the effect of exercise modality on circulating miRNAs, we measured plasma myomiRs after eccentric (downhill treadmill walking) or concentric (uphill treadmill walking) exercise. We found an increase in plasma myomiRs in the eccentric group only, which was associated with a significant decrease in maximal voluntary force, a hallmark of muscle damage.15 Therefore, we proposed that circulating myomiRs could be biomarkers of exercise‐induced muscle damage (EIMD). After a marathon race, the levels of circulating myomiRs and serum creatine kinase (CK) activity are consistently increased, suggesting muscle damage,23, 26 whereas no increase in c‐miRNA and CK activity occurs after prolonged cycling with almost no eccentric component.23 It should be noted, although, that higher levels of cardiac damage or stress markers (CK‐MB, troponin, and pro‐BNP) were found after a marathon race,24, 26 raising the possibility that part of the myomiRs found in serum originated from cardiac muscle rather than skeletal muscle. Accordingly, an increase in the cardiac‐specific miR‐208a was reported.24 However, miR‐133b and miR‐206 are specifically expressed in skeletal muscle, and because muscle mass recruited for running is much higher than heart mass, the contribution of skeletal muscle to c‐miRNAs is possibly the most important.
The release of myomiRs by damaged myofibres seems to be a key mechanism, although active secretion could also occur. Based on correlations between plasma myomiRs after a marathon and (i) an athlete's VO2max, (ii) individual speed at the lactate threshold, and (iii) cardiac dimensions, Mooren et al. proposed that c‐miRNAs could be markers of exercise adaptation.26 The hypothesis that myomiRs could be secreted to trigger paracrine or endocrine signalling has emerged, and a recent study supports this view.27 After separating extracellular vesicles from human plasma, Guescini et al. isolated a fraction of exosomes by immunoaffinity capture using an antibody raised against the muscle‐specific alpha‐sarcoglycan.27 This muscle‐specific fraction of exosomes contained miRNAs, including miR‐206. This strongly suggests that skeletal muscle secretes miRNA‐containing vesicles. Even though their biological role is not known, it raises the possibility that muscle fibres communicate with other cells/organs using miRNA‐containing vesicles. Whether exercise triggers the release of extracellular vesicles from muscle tissues is unclear in this study, because running exercise induced no changes in circulating myomiRs.27
In summary, there is evidence that exercise can modulate circulating myomiR levels, with two main physiological hypotheses. The first one suggests that myomiRs are actively released as mediators of muscle or cardiovascular adaptations, even though both the mechanisms of release and the biological functions are unknown. The second postulates that myomiRs are released in response to muscle damage in a passive manner (Figure 1). This hypothesis has been explored in the field of muscle pathology.
Figure 1.
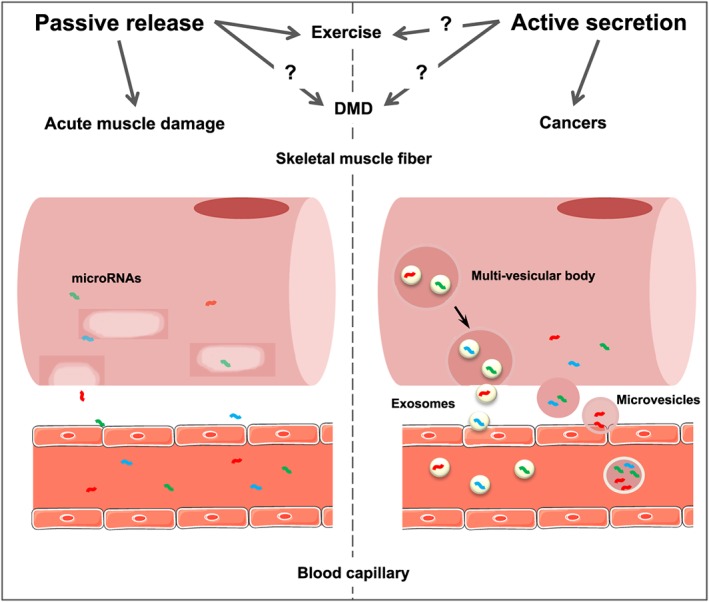
Possible mechanisms of myomiR release by skeletal muscle fibres. Skeletal muscle fibres can release myomiRs in their extracellular environment. The mechanisms of release are suspected to occur both in a passive and an active manner, depending on the stimulation. Myofibre necrosis induced by acute muscle damage is associated with a passive release of myomiRs in the blood through the plasma membrane. In the context of exercise or neuromuscular diseases, a passive release alone cannot explain the c‐miRNA profiles found in the blood; thus, it is probable that an active release of myomiRs occurs through secretion of extracellular vesicles (exosomes or microvesicles).
Circulating myomiRs and muscle damage
Neuromuscular diseases
Because Laterza et al. first proposed that tissue‐specific c‐miRNAs could be biomarkers of tissue injury,28 various applications have been proposed, and miRNAs have been shown to be reliable markers of liver or heart damage (for review, refer to Arrese et al. and Wronska et al. 29, 30). Concerning skeletal muscle, the first studies have focused on circulating myomiRs in animal models and human patients suffering from neuromuscular diseases. Duchenne muscular dystrophy (DMD), caused by mutations in the dystrophin gene, is one of the most severe neuromuscular diseases and is characterized by successive rounds of muscle degeneration and regeneration. Cacchiarelli et al. report that patients with DMD have elevated levels (up to 100‐fold) of serum miR‐1, miR‐133, and miR‐206.31 Interestingly, in patients with a milder form of dystrophin‐related disease, namely Becker muscular dystrophy, myomiR levels were in between those of patients with DMD and healthy controls. In addition, this study investigated and showed that, in a DMD animal model (mdx mice), treating the disease using an exon skipping strategy (intended to restore a truncated but functional dystrophin) allowed a histo‐morphological improvement of mouse muscle and a reduction of c‐miRNAs close to wild‐type levels. Therefore, miR‐1, miR‐133, and miR‐206 (also called dystromiRs) could be biomarkers of muscle dystrophy and treatment outcomes. Similarly, high dystromiR serum levels were found in other studies using mdx mice or dog models of DMD (CXMDJ, GRMD).32, 33, 34 When comparing different neuromuscular disease models, Vignier et al. found a common dystrophic profile (miR‐1, miR‐133a, miR‐133b, and miR‐378) in mdx mice and two mouse models of sarcoglycanopathy.35 Finally, miR‐208a, miR‐208b, and miR‐499 were also increased in the serum of GRMD dogs and patients with DMD, demonstrating that all muscle‐specific miRNAs are involved.36, 37 Therefore, it is now accepted that severe muscle dystrophies induce massive increases in plasma myomiRs, but limited or no alterations are described in moderate dystrophies such as Ullrich congenital muscle dystrophy, myotonic dystrophy type 1, or limb‐girdle muscle dystrophy.38, 39, 40 Whether c‐miRNAs are predictive of the severity of the dystrophy is unclear. Serum levels of miR‐1, miR‐133, and miR‐206 were inversely correlated with the clinical score in 10 young (3 to 6 year olds) ambulatory patients with DMD31 but not in a larger cohort of 26 patients (4 to 13 year olds).40 Both the age and the size of the groups may explain the discrepancy between the studies, but an additional explanation may be the fact that muscle mass decreases with the evolution of fibrosis and lipid deposition within dystrophic muscle, thereby reducing the number of muscle cells and myomiRs that are released. Consistent with this hypothesis, ambulant patients with DMD had higher circulating myomiRs than non‐ambulant patients, and because values decreased with age, c‐miRNAs may help monitor the remaining muscle mass in patients with DMD.40
The hypothesis that myomiRs were passively released in response to myofibre degeneration31 has been debated because of inconsistencies between the myomiR profiles of muscle tissue and serum. The levels of miR‐1 and miR‐133 are lower, and the miR‐206 levels are higher in dystrophic muscle tissue than healthy muscle tissue, and the levels of all three are higher in serum than in muscle.33 Similarly, ubiquitous miRNAs, such as the let‐7 family members, are abundant in muscle tissue but are not elevated in the serum of patients with DMD or in the corresponding animal models.34, 36 Therefore, a passive leak alone cannot explain the c‐miRNA profiles reported, and an active secretion probably occurs,33, 36, 40 perhaps in response to local inflammatory signals.37 In mdx mice sera, dystromiRs were found primarily in the non‐vesicular fraction or bound to proteins.34 The authors of the study suggest that miRNAs may be released by degenerating fibre and secreted by regenerating fibre, sending signals to neighbouring cells. Although this is counterintuitive, because secretion often occurs via extracellular vesicles, the authors propose that such vesicles are gradually degraded in the plasma. Furthermore, another study found myomiRs in both the exosomal and non‐exosomal fractions from patients with DMD.38 These data demonstrate that more studies are needed to determine the contribution of either passive leaking or secretion to increased c‐miRNAs.
Duchenne muscular dystrophy is characterized by concomitant skeletal muscle and cardiac dystrophy, leading to life‐threatening cardiomyopathy and heart failure. As most myomiRs (except miR‐133b and miR‐206) are expressed in cardiac muscle, cardiac tissue may significantly contribute to high circulating levels of myomiRs. This is supported by the fact that high serum myomiR levels have been found in both acute (acute myocardial infarction) and chronic (heart failure) cardiovascular diseases (for review, refer to Shi and Yang and Vegter et al. 41, 42). In a dog model of DMD (GRMD), Jeanson‐Leh et al. found high levels of circulating miR‐208a (cardiac specific), although there was no correlation with cardiac function (left ventricular fractional shortening).36 Altogether, it is impossible to evaluate the contributions of skeletal and cardiac muscles to c‐miRNAs in patients with DMD, although because of its mass, the contribution of skeletal muscle may be critical.
Acute muscle damage
The literature on exercise suggests that c‐miRNAs could be markers of EIMD in healthy subjects. However, a metabolic or a mechanical effect of eccentric muscle contraction itself is still possible. To investigate the effect of an acute muscle injury in the absence of contraction on c‐miRNA levels, we studied plasma miRNA profiles in rats after an intramuscular injection of notexin, a myotoxin known to provoke massive myofibre necrosis. We found a substantial increase in circulating myomiRs that could efficiently discriminate between damaged and non‐damaged animals.43 Concomitant myofibre necrosis and a reduction in myomiRs within damaged muscle tissue suggest that a passive release by injured myofibres may account for high circulating levels. The increase in plasma levels started as early as 6 h after muscle injury, peaked at 12 h, and decreased back to control levels between 24 and 48 h. This kinetic differs from what is commonly found with classical markers and could therefore provide different information about acute muscle injury in humans. For example, myomiRs could help monitor the regression of an acute damaging process when CK activity remains elevated for a much longer period.
Another aspect of miRNA expression is that the abundance of myomiRs in muscle fibres can differ based on muscle phenotype. Higher levels of miR‐206, miR‐208b, and miR‐499 are reported in slow/oxidative muscles, whereas fast/glycolytic muscles express higher amounts of miR‐1.44, 45 We found that miR‐206 was significantly higher in plasma after soleus muscle (slow‐type) damage than after extensor digitorum longus (fast‐type) muscle damage. This demonstrates that, as opposed to using CK activity, circulating myomiRs could be informative of the phenotype of the damaged myofibres.43 In sum, the literature shows that circulating myomiRs are relevant biomarkers of muscle damage for neuromuscular diseases and exercise as well as other diseases.
It is unknown whether myomiRs could be markers of chronic toxic damage, and this deserves more attention because many drugs have myotoxic side effects that have to be monitored. In particular, statins are widely prescribed cholesterol‐lowering drugs that are sometimes associated with muscle complications ranging from mild muscle pain to rare but life‐threatening rhabdomyolysis.46 Serum CK activity is routinely used to detect statin intolerance but has some limitations, mainly due to high inter‐individual variability related to gender, ethnicity, or training status.47, 48 Therefore, alternative biomarkers would be of great interest. Circulating myomiRs are promising candidates, as suggested by both animal28 and human studies.49 In rats, plasma miR‐133a increased after 3 days of high‐dose statin administration.28 In humans, statins are reported to aggravate EIMD,50 and in marathon runners, plasma miR‐499, together with CK, was higher in statin users than in a control group during the recovery period.49 Further studies are needed to evaluate whether circulating myomiRs could be relevant markers of statin intolerance in large cohorts of non‐exercising subjects.
Circulating myomiRs in cancer
As observed for proteins, there is a dysregulation of miRNAs in numerous cancers. Rhabdomyosarcomas are malignant tumours growing from skeletal muscle that overexpress myomiRs. They are released by the tumour, and patients with rhabdomyosarcoma have higher serum levels of miR‐1, miR‐133a, miR‐133b, and miR‐206 compared with healthy subjects or patients with non‐rhabdomyosarcoma tumours.51 However, alterations in circulating myomiRs are also reported in several non‐muscle cancers. Gastric cancer induced a 2‐fold increase in miR‐1,52 but in most malignant diseases studied, such as non‐small cell lung cancer, melanoma, astrocytoma, or osteosarcoma, circulating myomiRs were lower in patients than in the control population.53, 54, 55, 56, 57 Both miR‐206 and miR‐133 (a and b) have tumour suppressor properties, and they are down‐regulated within several tumours; a link between low expression and tumour development has been proposed (for review, refer to Nohata et al.58). However, it is unclear how low expression in tumours could result in decreased circulating levels in patients with cancer compared with healthy subjects. Thus, a more convincing hypothesis links cancer progression, muscle mass, and myomiRs. In many cancers, low circulating myomiRs are reported to predict unfavourable prognosis and survival.53, 54, 55, 57 Limited information on muscle damage biomarkers or muscle mass is given in those studies. Nevertheless, based on the fact that miR‐1 correlates with serum creatinine, Köberle et al. proposed that low miR‐1 levels may be associated with cachexia in advanced cancer disease.59 Thus, if skeletal muscle continuously releases muscle‐specific miRNAs, a substantial reduction in muscle mass could result in lower circulating myomiRs. Therefore, as previously shown in DMD, circulating myomiRs could be helpful in monitoring myofibre masses in cancer, where muscle quality correlates with overall survival.60 Loss of skeletal muscle is a common event in many other chronic diseases such as heart failure, pulmonary diseases, or rheumatologic diseases. Chronic muscle wasting can cause cachexia, increasing both morbidity and mortality in patients (review Drescher et al.61). Therefore, there is a constant effort to find new biomarkers of muscle wasting. The application of miRNAs as biomarkers of cachexia is interesting but needs further investigation specifically focused on the relationship between muscle mass/loss and circulating miRNAs in patients.
Considerations for clinical application
There are some problems to overcome before c‐miRNAs can be used routinely. First, many myomiRs are expressed in both cardiac and skeletal muscle, raising the possibility of confusion in situations where both cardiac and muscle injury occur. Measuring a judicious combination of myomiRs, such as miR‐208b or miR‐133a (good diagnostic accuracy), miR‐208a‐3p (cardiac‐specific, unaffected by muscle damage), and miR‐206‐3p or miR‐133b‐3p (skeletal muscle‐specific, unaffected by cardiac injury), could help overcome this problem. Second, technical considerations should be mentioned. Reverse transcriptase (RT) quantitative polymerase chain reaction (qPCR) is the gold standard technique. It is both reliable and reproducible but also time‐consuming and poorly adapted to clinical application, especially when a rapid result is required. Automated RNA isolation and RT solutions, as well as ultra‐fast PCR, could improve RT‐qPCR applicability.62, 63, 64 Moreover, novel probe‐based technologies are emerging and may allow quantitative measures that are much faster and easier to use in a clinical context.65, 66, 67 Third, we need to determine normal values in a wide population of healthy individuals in order to identify inter‐individual variability with regards to gender, age, ethnicity, or training status.
Conclusions
Monitoring the status of skeletal muscle has been a long‐time preoccupation, and there is constant, ongoing research to find new blood markers of muscle injury or muscle loss. Most studies have focused on proteins or peptides.61, 68 Circulating miRNAs have recently emerged in many clinical fields and are a new and promising class of biomarkers. Nucleic acids are structurally different from proteins or peptides; they have lower molecular weights and volumes, resulting in different properties in terms of release, diffusion, or stability in plasma. Therefore, circulating myomiRs can provide new and complementary information on skeletal muscle health status. The literature shows that they are robust biomarkers of both acute and chronic muscle damage and perhaps of adaptation to exercise. Many applications could emerge in both research and medicine. Exercise physiology and sports science could benefit from new biomarkers capable of evaluating muscle status in response to training. Whether c‐miRNAs could help monitor athletes' health and fitness throughout a season is still unclear but is a potential application. In the medical field, circulating myomiRs are reliable markers of muscle dystrophy and treatment outcome, although the significance of high circulating myomiRs in neuromuscular diseases is unclear. MyomiRs could be passively released through the compromised sarcolemma but also actively secreted, thereby playing a causal role in muscular dystrophy. Yet, for neuromuscular disorders as well as for exercise, it should be noted that strong experimental arguments supporting a signalling role of circulating myomiRs are currently lacking, and more studies are needed to elucidate that point. Acute muscle damage in sports medicine or intensive care medicine could also be monitored with c‐miRNAs as an interesting and complementary alternative to classical biomarkers. Finally, new data suggest that myomiRs could also reflect muscle mass or muscle loss. A deeper understanding of the mechanisms and regulation of miRNA release from healthy muscle tissue and muscle tissue in the early and late phases of cachexia is now necessary to fully understand the benefits and limitations of c‐miRNAs as biomarkers of muscle status. Muscle loss is a process and muscle mass a symptom; because both can often predict morbidity or mortality, circulating myomiRs are promising new diagnostic/prognostic tools in medicine.
Conflict of interest
None declared
Ethical guideline statement
The authors certify that they comply with the ethical guidelines for authorship and publishing of the Journal of Cachexia, Sarcopenia, and Muscle.69
Siracusa, J. , Koulmann, N. , and Banzet, S. (2018) Circulating myomiRs: a new class of biomarkers to monitor skeletal muscle in physiology and medicine. Journal of Cachexia, Sarcopenia and Muscle, 9: 20–27. doi: 10.1002/jcsm.12227.
References
- 1. Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- 2. Chen X, Liang H, Zhang J, Zen K, Zhang C‐Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol 2012;22:125–132. [DOI] [PubMed] [Google Scholar]
- 3. Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18:997–1006. [DOI] [PubMed] [Google Scholar]
- 4. De Guire V, Robitaille R, Tétreault N, Guérin R, Ménard C, Bambace N, et al. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: promises and challenges. Clin Biochem 2013;46:846–860. [DOI] [PubMed] [Google Scholar]
- 5. Mittelbrunn M, Gutiérrez‐Vázquez C, Villarroya‐Beltri C, González S, Sánchez‐Cabo F, González MÁ, et al. Unidirectional transfer of microRNA‐loaded exosomes from T cells to antigen‐presenting cells. Nat Commun 2011;2:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll‐like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci 2012;109:E2110–E2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 2007;14:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Diniz GP, Wang D‐Z. Regulation of skeletal muscle by microRNAs. Compr Physiol 2016;1279–1294. [DOI] [PubMed] [Google Scholar]
- 9. Kim VN, Nam J‐W. Genomics of microRNA. Trends Genet 2006;22:165–173. [DOI] [PubMed] [Google Scholar]
- 10. Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, et al. An intragenic MEF2‐dependent enhancer directs muscle‐specific expression of microRNAs 1 and 133. Proc Natl Acad Sci 2007;104:20844–20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle‐specific microRNAs. Proc Natl Acad Sci 2006;103:8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, et al. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR‐1, miR‐206 and miR‐133. Dev Biol 2008;321:491–499. [DOI] [PubMed] [Google Scholar]
- 13. Yeung F, Chung E, Guess MG, Bell ML, Leinwand LA. Myh7b/miR‐499 gene expression is transcriptionally regulated by MRFs and Eos. Nucleic Acids Res 2012;40:7303–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aoi W, Ichikawa H, Mune K, Tanimura Y, Mizushima K, Naito Y, et al. Muscle‐enriched microRNA miR‐486 decreases in circulation in response to exercise in young men. Front Physiol 2013;4:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Banzet S, Chennaoui M, Girard O, Racinais S, Drogou C, Chalabi H, et al. Changes in circulating microRNAs levels with exercise modality. J Appl Physiol 2013;115:1237–1244. [DOI] [PubMed] [Google Scholar]
- 16. McCarthy JJ, Esser KA. MicroRNA‐1 and microRNA‐133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol 2006;102:306–313. [DOI] [PubMed] [Google Scholar]
- 17. Drummond MJ, McCarthy JJ, Fry CS, Esser KA, Rasmussen BB. Aging differentially affects human skeletal muscle microRNA expression at rest and after an anabolic stimulus of resistance exercise and essential amino acids. Am J Physiol Endocrinol Metab 2008;295:E1333–E1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rivas DA, Lessard SJ, Rice NP, Lustgarten MS, So K, Goodyear LJ, et al. Diminished skeletal muscle microRNA expression with aging is associated with attenuated muscle plasticity and inhibition of IGF‐1 signaling. FASEB J 2014;28:4133–4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zacharewicz E, Della Gatta P, Reynolds J, Garnham A, Crowley T, Russell AP, et al. Identification of microRNAs linked to regulators of muscle protein synthesis and regeneration in young and old skeletal muscle. PLoS One 2014;9: e114009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nielsen S, Scheele C, Yfanti C, Åkerström T, Nielsen AR, Pedersen BK, et al. Muscle specific microRNAs are regulated by endurance exercise in human skeletal muscle: Muscle specific microRNAs and exercise. J Physiol 2010;588:4029–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Russell AP, Lamon S, Boon H, Wada S, Güller I, Brown EL, et al. Regulation of miRNAs in human skeletal muscle following acute endurance exercise and short‐term endurance training: miRNA in muscle after exercise. J Physiol 2013;591:4637–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Baggish AL, Hale A, Weiner RB, Lewis GD, Systrom D, Wang F, et al. Dynamic regulation of circulating microRNA during acute exhaustive exercise and sustained aerobic exercise training: circulating microRNA in exercise. J Physiol 2011;589:3983–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uhlemann M, Mobius‐Winkler S, Fikenzer S, Adam J, Redlich M, Mohlenkamp S, et al. Circulating microRNA‐126 increases after different forms of endurance exercise in healthy adults. Eur J Prev Cardiol 2014;21:484–491. [DOI] [PubMed] [Google Scholar]
- 24. Baggish AL, Park J, Min P‐K, Isaacs S, Parker BA, Thompson PD, et al. Rapid upregulation and clearance of distinct circulating microRNAs after prolonged aerobic exercise. J Appl Physiol 2014;116:522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gomes CPC, Oliveira‐Jr GP, Madrid B, Almeida JA, Franco OL, Pereira RW. Circulating miR‐1, miR‐133a, and miR‐206 levels are increased after a half‐marathon run. Biomarkers 2014;19:585–589. [DOI] [PubMed] [Google Scholar]
- 26. Mooren FC, Viereck J, Kruger K, Thum T. Circulating micrornas as potential biomarkers of aerobic exercise capacity. Am J Physiol Heart Circ Physiol 2013;306:H557–H563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guescini M, Canonico B, Lucertini F, Maggio S, Annibalini G, Barbieri E, et al. Muscle releases alpha‐sarcoglycan positive extracellular vesicles carrying miRNAs in the bloodstream. PLoS One 2015;10: e0125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laterza OF, Lim L, Garrett‐Engele PW, Vlasakova K, Muniappa N, Tanaka WK, et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin Chem 2009;55:1977–1983. [DOI] [PubMed] [Google Scholar]
- 29. Arrese M, Eguchi A, Feldstein A. Circulating microRNAs: emerging biomarkers of liver disease. Semin Liver Dis 2015;35:043–054. [DOI] [PubMed] [Google Scholar]
- 30. Wronska A, Kurkowska‐Jastrzebska I, Santulli G. Application of microRNAs in diagnosis and treatment of cardiovascular disease. Acta Physiol 2015;213:60–83. [DOI] [PubMed] [Google Scholar]
- 31. Cacchiarelli D, Legnini I, Martone J, Cazzella V, D'Amico A, Bertini E, et al. miRNAs as serum biomarkers for Duchenne muscular dystrophy. EMBO Mol Med 2011;3:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mizuno H, Nakamura A, Aoki Y, Ito N, Kishi S, Yamamoto K, et al. Identification of muscle‐specific microRNAs in serum of muscular dystrophy animal models: promising novel blood‐based markers for muscular dystrophy. PLoS One 2011;6: e18388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roberts TC, Blomberg KEM, McClorey G, Andaloussi SE, Godfrey C, Betts C, et al. Expression analysis in multiple muscle groups and serum reveals complexity in the microRNA transcriptome of the mdx mouse with implications for therapy. Mol Ther Nucleic Acids 2012;1: e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roberts TC, Godfrey C, McClorey G, Vader P, Briggs D, Gardiner C, et al. Extracellular microRNAs are dynamic non‐vesicular biomarkers of muscle turnover. Nucleic Acids Res 2013;41:9500–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vignier N, Amor F, Fogel P, Duvallet A, Poupiot J, Charrier S, et al. Distinctive serum miRNA profile in mouse models of striated muscular pathologies. PLoS One 2013;8: e55281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jeanson‐Leh L, Lameth J, Krimi S, Buisset J, Amor F, Le Guiner C, et al. Serum profiling identifies novel muscle miRNA and cardiomyopathy‐related miRNA biomarkers in golden retriever muscular dystrophy dogs and Duchenne muscular dystrophy patients. Am J Pathol 2014;184:2885–2898. [DOI] [PubMed] [Google Scholar]
- 37. Li X, Li Y, Zhao L, Zhang D, Yao X, Zhang H, et al. Circulating muscle‐specific miRNAs in Duchenne muscular dystrophy patients. Mol Ther Acids 2014;3: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Matsuzaka Y, Kishi S, Aoki Y, Komaki H, Oya Y, Takeda S, et al. Three novel serum biomarkers, miR‐1, miR‐133a, and miR‐206 for limb‐girdle muscular dystrophy, facioscapulohumeral muscular dystrophy, and Becker muscular dystrophy. Environ Health Prev Med 2014;19:452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perfetti A, Greco S, Bugiardini E, Cardani R, Gaia P, Gaetano C, et al. Plasma microRNAs as biomarkers for myotonic dystrophy type 1. Neuromuscul Disord 2014;24:509–515. [DOI] [PubMed] [Google Scholar]
- 40. Zaharieva IT, Calissano M, Scoto M, Preston M, Cirak S, Feng L, et al. Dystromirs as serum biomarkers for monitoring the disease deverity in Duchenne muscular dystrophy. PLoS One 2013;8: e80263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi Q, Yang X. Circulating microRNA and long noncoding RNA as biomarkers of cardiovascular diseases. J Cell Physiol 2016;231:751–755. [DOI] [PubMed] [Google Scholar]
- 42. Vegter EL, van der Meer P, de Windt LJ, Pinto YM, Voors AA. MicroRNAs in heart failure: from biomarker to target for therapy. Eur J Heart Fail 2016;18:457–468. [DOI] [PubMed] [Google Scholar]
- 43. Siracusa J, Koulmann N, Bourdon S, Goriot M‐E, Banzet S. Circulating miRNAs as biomarkers of acute muscle damage in rats. Am J Pathol 2016;186:1313–1327. [DOI] [PubMed] [Google Scholar]
- 44. Liu Y, Li M, Ma J, Zhang J, Zhou C, Wang T, et al. Identification of differences in microRNA transcriptomes between porcine oxidative and glycolytic skeletal muscles. BMC Mol Biol 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Muroya S, Taniguchi M, Shibata M, Oe M, Ojima K, Nakajima I, et al. Profiling of differentially expressed microRNA and the bioinformatic target gene analyses in bovine fast‐ and slow‐type muscles by massively parallel sequencing. J Anim Sci 2013;91:90–103. [DOI] [PubMed] [Google Scholar]
- 46. Taha DA, De Moor CH, Barrett DA, Gershkovich P. Translational insight into statin‐induced muscle toxicity: from cell culture to clinical studies. Transl Res 2014;164:85–109. [DOI] [PubMed] [Google Scholar]
- 47. Kenney K, Landau ME, Gonzalez RS, Hundertmark J, O'Brien K, Campbell WW. Serum creatine kinase after exercise: drawing the line between physiological response and exertional rhabdomyolysis. Muscle Nerve 2012;45:356–362. [DOI] [PubMed] [Google Scholar]
- 48. Nicholson G, Morgan G, Meerkin M, Strauss E, McLeod J. The creatine kinase reference interval: an assessment of intra‐and inter‐individual Variation. J Neurol Sci 1985;71:225–231. [DOI] [PubMed] [Google Scholar]
- 49. Min P‐K, Park J, Isaacs S, Taylor BA, Thompson PD, Troyanos C, et al. Influence of statins on distinct circulating microRNAs during prolonged aerobic exercise. J Appl Physiol 2016;120:711–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thompson PD, Zmuda JM, Domalik LJ, Zimet RJ, Staggers J, Guyton JR. Lovastatin increases exercise‐induced skeletal muscle injury. Metabolism 1997;46:1206–1210. [DOI] [PubMed] [Google Scholar]
- 51. Miyachi M, Tsuchiya K, Yoshida H, Yagyu S, Kikuchi K, Misawa A, et al. Circulating muscle‐specific microRNA, miR‐206, as a potential diagnostic marker for rhabdomyosarcoma. Biochem Biophys Res Commun 2010;400:89–93. [DOI] [PubMed] [Google Scholar]
- 52. Liu R, Zhang C, Hu Z, Li G, Wang C, Yang C, et al. A five‐microRNA signature identified from genome‐wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 2011;47:784–791. [DOI] [PubMed] [Google Scholar]
- 53. Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum microRNA signatures identified in a genome‐wide serum microRNA expression profiling predict survival of non‐small‐cell lung cancer. J Clin Oncol 2010;28:1721–1726. [DOI] [PubMed] [Google Scholar]
- 54. Li M, Zhang Q, Wu L, Jia C, Shi F, Li S, et al. Serum miR‐499 as a novel diagnostic and prognostic biomarker in non‐small cell lung cancer. Oncol Rep 2014;31:1961–1967. [DOI] [PubMed] [Google Scholar]
- 55. Tian R, Liu T, Qiao L, Gao M, Li J. Decreased serum microRNA‐206 level predicts unfavorable prognosis in patients with melanoma. Int J Clin Exp Pathol 2015;8:3097. [PMC free article] [PubMed] [Google Scholar]
- 56. Yang C, Wang C, Chen X, Chen S, Zhang Y, Zhi F, et al. Identification of seven serum microRNAs from a genome‐wide serum microRNA expression profile as potential noninvasive biomarkers for malignant astrocytomas. Int J Cancer 2013;132:116–127. [DOI] [PubMed] [Google Scholar]
- 57. Zhang C, Yao C, Li H, Wang G, He X. Serum levels of microRNA‐133b and microRNA‐206 expression predict prognosis in patients with osteosarcoma. Int J Clin Exp Pathol 2014;7:4194–4203. [PMC free article] [PubMed] [Google Scholar]
- 58. Nohata N, Hanazawa T, Enokida H, Seki N. microRNA‐1/133a and microRNA‐206/133b clusters: dysregulation and functional roles in human cancers. Oncotarget 2012;3:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Köberle V, Kronenberger B, Pleli T, Trojan J, Imelmann E, Peveling‐Oberhag J, et al. Serum microRNA‐1 and microRNA‐122 are prognostic markers in patients with hepatocellular carcinoma. Eur J Cancer 2013;49:3442–3449. [DOI] [PubMed] [Google Scholar]
- 60. Shachar SS, Williams GR, Muss HB, Nishijima TF. Prognostic value of sarcopenia in adults with solid tumours: a meta‐analysis and systematic review. Eur J Cancer 2016;57:58–67. [DOI] [PubMed] [Google Scholar]
- 61. Drescher C, Konishi M, Ebner N, Springer J. Loss of muscle mass: current developments in cachexia and sarcopenia focused on biomarkers and treatment: loss of muscle mass: current developments. J Cachexia Sarcopenia Muscle 2015;6:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mackay JF. Taking it to the extreme: PCR at wittwerspeed. Clin Chem 2015;61:4–5. [DOI] [PubMed] [Google Scholar]
- 63. Neuzil P. Ultra fast miniaturized real‐time PCR: 40 cycles in less than six minutes. Nucleic Acids Res 2006;34:e77–e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Son JH, Cho B, Hong S, Lee SH, Hoxha O, Haack AJ, et al. Ultrafast photonic PCR. Light Sci Appl 2015;4: e280. [Google Scholar]
- 65. Lee JH, Kim JA, Kwon MH, Kang JY, Rhee WJ. In situ single step detection of exosome microRNA using molecular beacon. Biomaterials 2015;54:116–125. [DOI] [PubMed] [Google Scholar]
- 66. Liu Q, Shin Y, Kee JS, Kim KW, Mohamed Rafei SR, Perera AP, et al. Mach–Zehnder interferometer (MZI) point‐of‐care system for rapid multiplexed detection of microRNAs in human urine specimens. Biosens Bioelectron 2015;71:365–372. [DOI] [PubMed] [Google Scholar]
- 67. Shin Y, Lim SY, Lee TY, Park MK. Dimethyl adipimidate/thin film sample processing (DTS): a simple, low‐cost, and versatile nucleic acid extraction assay for downstream analysis. Sci Rep 2015;5:14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nedergaard A, Sun S, Karsdal MA, Henriksen K, Kjaer M, Lou Y, et al. Type VI collagen turnover‐related peptides‐novel serological biomarkers of muscle mass and anabolic response to loading in young men. J Cachexia Sarcopenia Muscle 2013;4:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: update 2015. J Cachexia Sarcopenia Muscle 2015;6:315–316. [DOI] [PMC free article] [PubMed] [Google Scholar]


