Abstract
Rationale: Successful transmission of tuberculosis depends on the interplay of human behavior, host immune responses, and Mycobacterium tuberculosis virulence factors. Previous studies have been focused on identifying host risk factors associated with increased transmission, but the contribution of specific genetic variations in mycobacterial strains themselves are still unknown.
Objectives: To identify mycobacterial genetic markers associated with increased transmissibility and to examine whether these markers lead to altered in vitro immune responses.
Methods: Using a comprehensive tuberculosis registry (n = 10,389) and strain collection in the Netherlands, we identified a set of 100 M. tuberculosis strains either least or most likely to be transmitted after controlling for host factors. We subjected these strains to whole-genome sequencing and evolutionary convergence analysis, and we repeated this analysis in an independent validation cohort. We then performed immunological experiments to measure in vitro cytokine production and neutrophil responses to a subset of the original strains with or without the identified mutations associated with increased transmissibility.
Measurements and Main Results: We identified the loci espE, PE-PGRS56, Rv0197, Rv2813–2814c, and Rv2815–2816c as targets of convergent evolution among transmissible strains. We validated four of these regions in an independent set of strains, and we demonstrated that mutations in these targets affected in vitro monocyte and T-cell cytokine production, neutrophil reactive oxygen species release, and apoptosis.
Conclusions: In this study, we identified genetic markers in convergent evolution of M. tuberculosis toward enhanced transmissibility in vivo that are associated with altered immune responses in vitro.
Keywords: tuberculosis, transmission, bacterial genomes, immunology
At a Glance Commentary
Scientific Knowledge on the Subject
Most studies on Mycobacterium tuberculosis transmission have been focused on host risk factors. However, variations in virulence and immunogenicity across M. tuberculosis lineages could also account for differences in transmissibility. To date, there has not been a systematic study of the genetic determinants of tuberculosis transmission at the level of individual strains.
What This Study Adds to the Field
We performed a genome-wide association study and identified five loci as targets of convergent evolution among transmissible strains, four of which were confirmed in an independent validation cohort. We subsequently demonstrated that mutations in these targets are associated with altered in vitro immune responses. To our knowledge, this is the first study to integrate molecular and conventional epidemiology with in vitro immunological assays to identify bacterial factors associated with tuberculosis transmission.
Transmission of pulmonary tuberculosis (TB) occurs through inhalation of small-droplet nuclei containing Mycobacterium tuberculosis bacilli that enter the lungs, evade killing by the innate immune system, and replicate intracellularly. If a series of transmission events occurs over a relatively short time, one can identify a group of patients with M. tuberculosis strains that are genotypically highly similar. Epidemiologists often use molecular fingerprinting to characterize the genetic similarity among a group of strains; strains that share a molecular fingerprint are described as “clustered” (1) and are inferred to be the result of recent transmission rather than the reactivation of a previous infection.
Host factors can affect TB transmission and disease progression (2), but recent molecular epidemiological studies have shown that M. tuberculosis strains also differ in their ability to cause pulmonary disease (3) and their proclivity to infect contacts (4) or cause secondary cases (5, 6). This variability may reflect the strains’ ability to subvert innate (7, 8) and/or adaptive (9, 10) immunity or their ability to exploit the host immune system by inducing a detrimental inflammatory response (11) leading to tissue damage (12, 13) and formation of cavities that enable disease spread (14). Cytokines play a pivotal role in these events; insufficient production of proinflammatory cytokines may lead to uncontrolled mycobacterial growth, whereas overproduction may lead to tissue damage (15).
Phylogenetic differences in cytokine response (16, 17) suggest that specific microbial genetic determinants may underlie transmission-related phenotypes. Several studies have used M. tuberculosis mutants in vitro and experimental models to identify the role of a few individual genes in transmission-associated phenotypes (18). However, further elucidation of the full spectrum of genes affecting transmission could improve our understanding of the host–pathogen relationship in TB.
We aimed to identify loci under positive selection for clustering by analyzing whole M. tuberculosis genomes from clustered and unclustered isolates for evidence of convergence. Following the hypothesis that clustered strains have consistent genetic differences compared with unclustered ones, and that the genes or intergenic regions implicated in these differences affect the host immune response, we performed a functional validation of the newly identified targets of independent mutation (TIMs) by measuring in vitro cytokine production and neutrophil responses.
Methods
Clinical Isolates
We selected 100 mycobacterial strains with extreme phenotypes at both ends of a transmissibility spectrum. We considered strains to be highly transmissible if they came from clusters of active TB cases lacking known risk factors for being part of a cluster. Similarly, we considered strains to be minimally transmissible if they were unique (unclustered) and isolated from patients (e.g., homeless individuals with class 3 sputum smear–positive pulmonary TB) with increased risk for clustering. To classify strains as such, we used data on host risk factors for clustering to estimate the cluster propensity to propagate (CPP), a summary measure of risk for transmission of patients belonging to a particular TB cluster (see Table E1 in the online supplement) (19). This CPP was calculated for 10,389 patient isolates, with clusters defined by DNA fingerprinting (20, 21). CPP was significantly higher in clustered versus unclustered strains, although the CPP rapidly plateaus with increasing cluster size (Figure E1). Because there is no basis for sample size calculation in studies associating genomic variants with transmissibility (22, 23), we arbitrarily chose 100 strains for whole-genome sequencing (WGS): 66 unclustered strains and 34 clustered strains. Strains for the clustered phenotype were picked at random from 56 unique cluster fingerprints (5 pairs of strains came from within the same cluster). The 100 selected strains were all drug sensitive and belonged to patients originating from 44 different countries. At least one strain from both phenotypes (clustered and unclustered) and from each the four major M. tuberculosis lineages was represented. In an independent dataset (n = 143), we contrasted clustered and unclustered strains collected from patients of different geographical backgrounds. Most of these strains were drug resistant (Table E2).
Phylogeny Construction
Strains underwent WGS and variants were called (see online supplement). We then constructed a phylogeny on the basis of multiple-sequence alignment of the sequences, excluding single-nucleotide polymorphisms (SNPs) that occurred in repetitive elements, including PE/PPE and PGRS genes.
Phylogenetic Convergence Test for Selection
We used our previously developed method, the phylogenetic convergence test (PhyC) for selection, to identify genetic loci associated with clustering. For each nucleotide position in the genome, we counted the number of convergent SNPs and insertions/deletions (“Indels”) in clustered and unclustered branches, respectively, counting only one strain per cluster. We therefore assessed the significance of each convergent SNP or Indel compared with the empirical background distribution using a permutation test (22).
Protein Prediction
We used two protein prediction algorithms, I-Mutant v2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), to predict the functional impact of the significant SNPs on protein structure and function.
Immunological Experiments
Nineteen strains from the initial study were recultured, heat killed, and bead beated. Peripheral blood mononuclear cells from 12 healthy donors were stimulated with 3 μg/ml of lysate for production of tumor necrosis factor-α (TNF-α) (4 and 24 h); IL-1β, IL-1RA, IL-6, and IL-10 (24 h); and T-cell cytokines IL-17, IL-22, and IFN-γ (7 d) (17, 24). We also stimulated isolated polymorphonuclear cells (PMNs, largely consisting of neutrophils) for 1 hour and measured the production of reactive oxygen species (ROS) (six donors) after 6 hours using luminol-enhanced chemiluminescence, and neutrophil apoptosis and cell death with flow cytometry (eight donors). We constructed multivariate mixed models to exploit the covariance between assays and to control for lineage effect and interdonor variability and compared null models without assay-specific TIM indicators with full models with these indicators using likelihood ratio tests.
Results
TIMs
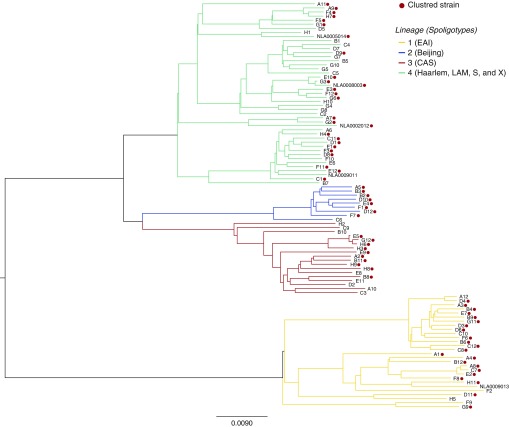
The primary set of 100 selected strains included 66 clustered isolates with a low predicted CPP (mean CPP, 0.75; SD, 0.01) and 34 unclustered isolates with a high predicted CPP (mean CPP, 1.02; SD, 0.30) (Figure E1). We conducted two parallel PhyCs to identify either individual nucleotide positions or genes and intergenic regions where cluster-associated mutations occur frequently along disparate locations in the phylogenetic tree (Figure 1). Region-level PhyC detected three genes and two intergenic regions as statistically significant TIMs (P < 0.05) (Table 1). A total of 12 SNPs, 2 insertions, and 31 deletions were found in these TIMs, including 1 SNP and 2 deletions that were also found to be significant by the site-level PhyC (Table E3). TIMs in the PE-PGRS56 gene occurred solely in clustered branches, whereas those in espE, Rv0197, Rv2813–2814c, and Rv2815–2816c were also found in unclustered branches, but at a lower rate than in clustered branches (depicted for espE in Figure E2).
Figure 1.
Consensus Bayesian phylogenetic tree. Clustered strains and Mycobacterium tuberculosis lineages are highlighted. CAS = Central Asian; EAI = East African-Indian; LAM = Latin American-Mediterranean.
Table 1.
Significant Genes or Intergenic Regions by Phylogenetic Convergence Test
| Gene/Region (Rv number) | Original Dataset (n = 100) |
Validation Dataset (n = 143) |
||||||
|---|---|---|---|---|---|---|---|---|
| Strains with Mutations, Deletions, and Insertions (n) |
P Value | Lineages with Cases | Strains with Mutations, Deletions, and Insertions (n) |
P Value | Lineages with Cases | |||
| Clustering | Nonclustering | Clustering | Nonclustering | |||||
| espE (Rv3864) | 10 | 1 | 0.0377 | 1, 3, 4 | 10 | 2 | 0.0232 | 1, 3, 4 |
| PE-PGRS56 (Rv3512) | 13 | 0 | 0.0052 | 1, 4 | 1 | 0 | 1 | 4 |
| Unnamed (Rv0197) | 20 | 12 | 0.0214 | 1, 2, 3, 4 | 26 | 12 | 0.0362 | 1, 2, 3, 4 |
| Unnamed (Rv2813–2814c) | 20 | 6 | 0.0458 | 1, 3, 4 | 22 | 3 | 0.0001 | 1, 3, 4 |
| Unnamed (Rv2815–2816c) | 18 | 4 | 0.0178 | 1, 4 | 22 | 5 | 0.0105 | 1, 4 |
Analysis of the validation set of 143 strains confirmed four of five genes or intergenic regions (Table 1), including Rv0197, in which PhyC detected the same nonsynonymous coding site (234,477TG). The TIMs occurring in the PE-PGRS56 gene could not be validated, because their occurrence in the original dataset was restricted mostly to lineage 1, which made up only 3.4% of the validation dataset.
Deleterious Effect of SNP TIMs on Proteins
All 12 SNPs in genes Rv0197 and espE are predicted to adversely affect the respective proteins (Table E4), including two TIMs in Rv0197 (234,265GT and 234,477TG) that result in a stop codon and truncation of the protein.
Association between TIMs and Induction of Cytokine Responses
Reasoning that genetic variation associated with transmissibility might be mediated through the host response, we next examined in vitro cytokine responses in strains with and without convergent changes. The distribution of TIMs across the strains is depicted in Figure E3 and Table E5.
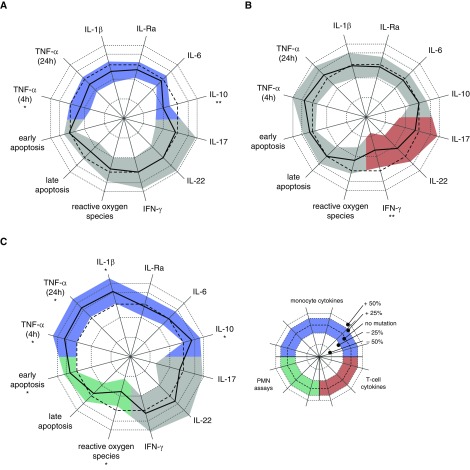
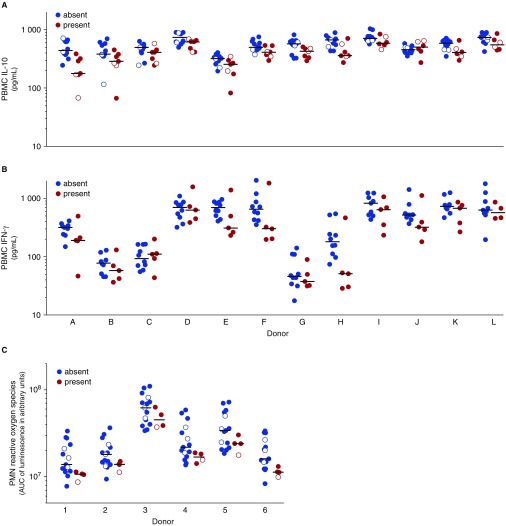
Mutations in two of the targets we identified, espE and Rv2813–2814, were associated with alterations in monocyte cytokine production (P < 10−4) (Table 2, Figure 2) in the multivariate mixed model. In the secondary analysis (see Table E6), mutations in espE were associated with decreased production of IL-10 (P = 1.7 × 10−8) (Figure 3A) and TNF-α (at 4 h; P = 8.0 × 10−3), and mutations in Rv2813–2814c were associated with increased production of TNF-α (P = 2.5 × 10−3), IL-1β (P = 7.7 × 10−3), and IL-10 (P = 1.9 × 10−3). Of the five genes or intergenic regions, only PE-PGRS56 affected T-cell cytokine responses (P = 5.4 × 10−3), and in our secondary analysis, this was associated with lower IFN-γ production (P = 1.6 × 10−3) (Figure 3B).
Table 2.
Cytokine Profiles and Polymorphonuclear Neutrophil Responses to Mycobacterium tuberculosis according to the Presence or Absence of Mutations in the Five Targets of Independent Mutation
| Gene or Intergenic Region | Monocyte Cytokines (df = 6) | T-Cell Cytokines (df = 3) | PMNs (df = 3) |
|---|---|---|---|
| espE | 1.33 × 10−6 | 0.961 | 0.077 |
| PE-PGRS56 | 0.039 | 5.35 × 10−3 | 0.021 |
| Rv0197 | 0.017 | 0.224 | 0.343 |
| Rv2813–2814c | 7.47 × 10−6 | 0.309 | 5.79 × 10−8 |
| Rv2815–2816c | 0.025 | 0.027 | 0.151 |
Definition of abbreviations: df = degrees of freedom; PMNs = polymorphonuclear neutrophils.
Significance (in boldface type) is determined at α = 0.05/5 = 0.01, corrected for the five genes or intergenic regions tested.
Figure 2.
Response to Mycobacterium tuberculosis strain with or without mutations in the three targets of independent mutation that showed an effect in the primary analysis. Relative differences for individual assays in the secondary analysis are indicated by the difference between the thick black line (mutation present) and the thin reference line (no mutation) for each of the following targets of independent mutation that significantly influenced at least one assay group: (A) espE, (B) PE-PGRS56, or (C) Rv2813–2814c. Shaded area represents the 95% confidence interval, corrected for the fact that five genes or intergenic regions were tested for each assay. *P < 0.05/5 = 0.01. **Significant after further correcting for number of assays per group (i.e., 0.05/[5 × 6] for monocyte cytokines and P < 0.05/[5 × 3] for T-cell cytokines and polymorphonuclear neutrophil [PMN] assays). Significance in the primary analysis is indicated by a colored confidence interval for monocyte cytokines (blue), T-cell cytokines (red), and PMN assays (green). TNF = tumor necrosis factor.
Figure 3.
In vitro responses of selected assays for targets of independent mutation. Stimulation was performed with lysate of Mycobacterium tuberculosis strains from lineage 1 (filled circles) and lineage 4 (open circles) that did not harbor (blue) or harbored (red) a mutation in (A) espE, (B) PE-PGRS56, or (C) Rv2813–2814c. Peripheral blood mononuclear cells (PBMCs) of 12 healthy donors (A–L) were stimulated. (A) IL-10 was measured after 24 hours. (B) IFN-γ was measured after 7 days. (C) Polymorphonuclear cells (PMNs) of six healthy donors (1–6) and reactive oxygen species were measured by luminol-enhanced chemiluminescence and plotted in arbitrary units of the area under the curve (AUC) of the measurement over the first hour after stimulation. Circles in A show overlap because of limited variation; only lineage 1 strain results are shown in B, because no mutations occurred in PE-PGRS56 genes in strains from lineage 4.
Association between TIMs and Response of Neutrophils
We next examined the effects of TIMs on in vitro responses of neutrophils, given their putative role in transmission and clinical manifestation of TB in the same 19 strains. In the multivariate mixed effects model, we found that Rv2813–2814c affected PMN responses (P < 10−4) with lower ROS production (P = 4.8 × 10−4) (Figure 3C) and higher early apoptosis (P = 3.6 × 10−3) in secondary analysis.
Discussion
We identified five genes and intergenic regions (espE, PE-PGRS56, Rv0197, Rv2813–2814c, and Rv2815–2816c) as TIMs in clustered M. tuberculosis strains. We confirmed four of five genes and intergenic regions in a second dataset, despite differences in lineages and drug resistance profiles between the original and validation datasets. The TIMs we identified are predicted to alter the function of their respective proteins, and three of five identified genes or intergenic regions were associated with altered cytokine production or PMN responses, supporting the hypothesis that they confer a selective advantage for TB transmission.
Previous experimental studies have established the importance of three of the identified genes or intergenic regions, including espE, which proved essential for M. tuberculosis virulence in a number of animal studies (25–27) (Table E7). Further support for our findings stems from other genomic epidemiological studies. Nonsynonymous SNPs (albeit different from the ones identified in this study) and a frameshift mutation in espE were found to be more common in Mycobacterium africanum strains relative to H37Rv (28) and to be implied in their reduced ability to induce a CD4-cell ESAT-6–induced IFN-γ host response (29, 30). Similarly, in a previous study, researchers identified a large sequence polymorphism associated with clustering in a gene (MT1801) encoding molybdopterin oxidoreductase, which is also encoded by Rv0197 (31). In another study, researchers reported that an M. tuberculosis strain responsible for a large outbreak in the United Kingdom harbored an insertion in position 3,121,877 of intergenic region Rv2815–2816c (32), adjacent to the 2-bp deletion in 3,121,879 observed in our own study.
Three of five genes or intergenic regions with TIMs associated with clustering of TB showed a clear and statistically significant effect on monocyte or T-cell cytokine production or PMN responses. M. tuberculosis strains with TIMs in PE-PGRS56 induced lower production of IFN-γ, which is unequivocally seen as a key factor in protection against TB (15, 33). Two other TIMs were associated with altered production of the monocyte cytokines IL-10 and TNF-α, which are involved in M. tuberculosis killing as well as damaging immunopathology, both of which may affect TB transmission (15). In line with our previous comparison of in vitro cytokine responses to different M. tuberculosis lineage strains (17), TNF-α and IL-6 induction in this study was higher in lineage 4 (ancient) than in lineage 1 (modern) strains. These lineage effects may depend on strain selection, as shown in a study by Reiling and colleagues (34), who found opposite results. The aim of our study was not to discern lineage effects; therefore, we corrected for these lineage effects in our statistical model.
With regard to neutrophils, strains harboring cluster-associated mutations in Rv2813–2814c induced significantly lower ROS production and early apoptosis. Neutrophils are considered protective during early infection, when they are recruited to the site of infection, phagocytose mycobacteria (35) or mycobacteria-infected macrophages (36), and resist mycobacterial growth using ROS (36). Children with chronic granulomatous disease have a reduced oxidative burst and are more susceptible to TB (37). Neutrophil ROS also correlates with apoptosis (7). Although speculative, lower induction of ROS production might as such contribute to disease progression and higher transmission of certain strains.
This study has several limitations. First, phenotype misclassification, most notably that of highly transmissible strains as unclustered, is a possibility among “imported” strains (those belonging to patients born outside the Netherlands: 53 vs. 88% in unclustered and clustered strains, respectively). Average follow-up time, however, as indicated by proxy data on “days resided within the Netherlands” at the time of diagnosis, among “imported” cases within the unclustered cohort was 3,607 days (ranging from 321 to 10,874 d). In other words, it is reassuring that, with the exception of one case (where the number of days resided within the Netherlands was 321 d), all the remaining “imported” cases classified as unclustered had been in the Netherlands for at least 4 years. Second, the difference in drug resistance profiles and related parameters, such as treatment efficacy between the original and validation cohorts of strains for the PhyC, could have introduced bias in measurement of the transmissibility phenotype (Table E2). The facts that transmission of drug-resistant strains has been widely documented (38, 39) and that mathematical models have estimated the transmission cost of drug resistance to be as low as 10% (40) suggest that the overall fitness for transmission of drug-resistant strains is comparable to that of sensitive strains. The possibility for epistasis indeed exists; in 2016, for example, multidrug-resistant strains in China with rpoC compensatory mutations were found to be more likely than their drug-sensitive counterparts to be clustered (41). Future PhyC tests stratified by drug resistance would identify such potential (positive or negative) epistatic mutations, as has already been attempted using epidemiological tools (42). In the present study, however, confirmation of four of the five genetic markers associated with transmission in the validation dataset reduces the risk of false-positive findings. Finally, the inclusion of additional key host factors that may influence disease transmissibility, such as the level of TB exposure (i.e., via prospective household contact data) and pulmonary cavitation, could improve the ability to isolate bacterial factors influencing transmissibility in the future.
Validating our findings for longitudinally collected strains from other low-burden settings and at a nationwide level could further increase the significance of our results. An adequately sized and publicly available WGS dataset as described was not available at the time of our analysis, however. In high-burden TB settings, we would expect crowding, treatment delays, and host risk factors (e.g., malnutrition, uncontrolled diabetes) to be more important for transmission. These factors could be controlled for using the CPP measure. Whether the sum of all these factors acting in synergy to facilitate transmission in high-burden settings translates to a reduced selective pressure for genomic adaptations in M. tuberculosis itself remains an interesting question.
Of note, we performed in vitro cellular stimulation, aiming to find biological support for the epidemiological associations identified through convergent evolutionary analysis and not to identify specific effects of individual TIMs on in vitro cellular responses. Such effects cannot be identified in this study, because multiple TIMs were present in single strains in this dataset (Table E5). For this purpose, additional studies using mutagenesis or recombination-mediated genetic engineering to isolate the mutational effects should be performed. It is no surprise that no single pattern of cytokine production or PMN response was found for the five genes or intergenic regions, because M. tuberculosis has different strategies to subvert or resist the host immune system or to use it to its advantage.
In summary, we present evidence based on an evolutionary convergence analysis that five M. tuberculosis genes or intergenic regions confer a selective advantage promoting the transmission of M. tuberculosis and/or TB disease progression, and that these genetic elements influence the response of the host to the mycobacteria. These findings serve as an important step forward in the quest for an improved understanding of the microbial genetic determinants of TB transmission.
Acknowledgments
Acknowledgment
The authors thank Jessica de Beer and Arnout Mulder for culturing of mycobacterial strains; Jeroen de Keijzer for processing them; Ekta Lachmandas, Bas Blok, Mark Gresnigt, and Cor Jacobs for assisting with immunological experiments; and Professor Jelle Goeman for statistical advice.
Footnotes
This study was funded by the Portuguese Foundation for Science and Technology (FCT) (SFRH/BD/33902/2009 [H.N.-G.]).
Author Contributions: H.N.-G., A.v.L., M.M., R.v.C., and D.v.S.: designed and conducted the study; H.N.-G., A.v.L., and M.R.F.: wrote the first draft of the manuscript; V.A.C.M.K., J.J.M., A.Z., S.A.F.T.v.H., M.G.N., M.M., R.v.C., and D.v.S.: contributed to the final version of the manuscript; V.A.C.M.K. and A.v.L.: conducted the immunological experiments; A.Z. and S.A.F.T.v.H.: provided support with bioinformatics; J.J.M.: performed the statistical analysis of the immunological assay results; and H.N.-G., A.v.L., M.R.F., V.A.C.M.K., J.J.M., M.G.N., M.M., R.v.C., and D.v.S.: contributed to analysis and interpretation.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201605-1042OC on December 20, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.World Health Organization (WHO) Global tuberculosis report 2012. Geneva, Switzerland: WHO; 2012. [Google Scholar]
- 2.Kik SV, Verver S, van Soolingen D, de Haas PEW, Cobelens FG, Kremer K, van Deutekom H, Borgdorff MW. Tuberculosis outbreaks predicted by characteristics of first patients in a DNA fingerprint cluster. Am J Respir Crit Care Med. 2008;178:96–104. doi: 10.1164/rccm.200708-1256OC. [DOI] [PubMed] [Google Scholar]
- 3.Kong Y, Cave MD, Zhang L, Foxman B, Marrs CF, Bates JH, Yang ZH. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. J Clin Microbiol. 2006;44:3940–3946. doi: 10.1128/JCM.01146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Albanna AS, Reed MB, Kotar KV, Fallow A, McIntosh FA, Behr MA, Menzies D. Reduced transmissibility of East African Indian strains of Mycobacterium tuberculosis. PLoS One. 2011;6:e25075. doi: 10.1371/journal.pone.0025075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kato-Maeda M, Kim EY, Flores L, Jarlsberg LG, Osmond D, Hopewell PC. Differences among sublineages of the East-Asian lineage of Mycobacterium tuberculosis in genotypic clustering. Int J Tuberc Lung Dis. 2010;14:538–544. [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, Jackson-Sillah DJ, Fox A, Deriemer K, Gagneux S, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romero MM, Balboa L, Basile JI, López B, Ritacco V, de la Barrera SS, Sasiain MC, Barrera L, Alemán M. Clinical isolates of Mycobacterium tuberculosis differ in their ability to induce respiratory burst and apoptosis in neutrophils as a possible mechanism of immune escape. Clin Dev Immunol. 2012;2012:152546. doi: 10.1155/2012/152546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, Peyron P, Mestre O, Kaplan G, van Soolingen D, Gao Q, Gicquel B, Neyrolles O. Innate immune response to Mycobacterium tuberculosis Beijing and other genotypes. PLoS One. 2010;5:e13594. doi: 10.1371/journal.pone.0013594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rakotosamimanana N, Raharimanga V, Andriamandimby SF, Soares JL, Doherty TM, Ratsitorahina M, Ramarokoto H, Zumla A, Huggett J, Rook G, et al. VACSEL/VACSIS Study Group. Variation in γ interferon responses to different infecting strains of Mycobacterium tuberculosis in acid-fast bacillus smear-positive patients and household contacts in Antananarivo, Madagascar. Clin Vaccine Immunol. 2010;17:1094–1103. doi: 10.1128/CVI.00049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manca C, Tsenova L, Bergtold A, Freeman S, Tovey M, Musser JM, Barry CE, III, Freedman VH, Kaplan G. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc Natl Acad Sci USA. 2001;98:5752–5757. doi: 10.1073/pnas.091096998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López B, Aguilar D, Orozco H, Burger M, Espitia C, Ritacco V, Barrera L, Kremer K, Hernandez-Pando R, Huygen K, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–37. doi: 10.1046/j.1365-2249.2003.02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aguilar D, Hanekom M, Mata D, Gey van Pittius NC, van Helden PD, Warren RM, Hernandez-Pando R. Mycobacterium tuberculosis strains with the Beijing genotype demonstrate variability in virulence associated with transmission. Tuberculosis (Edinb) 2010;90:319–325. doi: 10.1016/j.tube.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Kato-Maeda M, Shanley CA, Ackart D, Jarlsberg LG, Shang S, Obregon-Henao A, Harton M, Basaraba RJ, Henao-Tamayo M, Barrozo JC, et al. Beijing sublineages of Mycobacterium tuberculosis differ in pathogenicity in the guinea pig. Clin Vaccine Immunol. 2012;19:1227–1237. doi: 10.1128/CVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones-López EC, Kim S, Fregona G, Marques-Rodrigues P, Hadad DJ, Molina LPD, Vinhas S, Reilly N, Moine S, Chakravorty S, et al. Importance of cough and M. tuberculosis strain type as risks for increased transmission within households. PLoS One. 2014;9:e100984. doi: 10.1371/journal.pone.0100984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MPR. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 16.Portevin D, Gagneux S, Comas I, Young D. Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog. 2011;7:e1001307. doi: 10.1371/journal.ppat.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Laarhoven A, Mandemakers JJ, Kleinnijenhuis J, Enaimi M, Lachmandas E, Joosten LAB, Ottenhoff THM, Netea MG, van Soolingen D, van Crevel R. Low induction of proinflammatory cytokines parallels evolutionary success of modern strains within the Mycobacterium tuberculosis Beijing genotype. Infect Immun. 2013;81:3750–3756. doi: 10.1128/IAI.00282-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin DM, Jeon BY, Lee HM, Jin HS, Yuk JM, Song CH, Lee SH, Lee ZW, Cho SN, Kim JM, et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010;6:e1001230. doi: 10.1371/journal.ppat.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nebenzahl-Guimaraes H, Borgdorff MW, Murray MB, van Soolingen D. A novel approach - the propensity to propagate (PTP) method for controlling for host factors in studying the transmission of Mycobacterium tuberculosis. PLoS One. 2014;9:e97816. doi: 10.1371/journal.pone.0097816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Small PM, Hopewell PC, Singh SP, Paz A, Parsonnet J, Ruston DC, Schecter GF, Daley CL, Schoolnik GK. The epidemiology of tuberculosis in San Francisco: a population-based study using conventional and molecular methods. N Engl J Med. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- 21.Alland D, Kalkut GE, Moss AR, McAdam RA, Hahn JA, Bosworth W, Drucker E, Bloom BR. Transmission of tuberculosis in New York City: an analysis by DNA fingerprinting and conventional epidemiologic methods. N Engl J Med. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- 22.Farhat MR, Shapiro BJ, Sheppard SK, Colijn C, Murray M. A phylogeny-based sampling strategy and power calculator informs genome-wide associations study design for microbial pathogens. Genome Med. 2014;6:101. doi: 10.1186/s13073-014-0101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Read TD, Massey RC. Characterizing the genetic basis of bacterial phenotypes using genome-wide association studies: a new direction for bacteriology. Genome Med. 2014;6:109. doi: 10.1186/s13073-014-0109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YY, Chang JR, Huang WF, Hsu SC, Kuo SC, Sun JR, Dou HY. The pattern of cytokine production in vitro induced by ancient and modern Beijing Mycobacterium tuberculosis strains. PLoS One. 2014;9:e94296. doi: 10.1371/journal.pone.0094296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci USA. 2005;102:8327–8332. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 27.Fortune SM, Jaeger A, Sarracino DA, Chase MR, Sassetti CM, Sherman DR, Bloom BR, Rubin EJ. Mutually dependent secretion of proteins required for mycobacterial virulence. Proc Natl Acad Sci USA. 2005;102:10676–10681. doi: 10.1073/pnas.0504922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehre F, Otu J, DeRiemer K, de Sessions PF, Hibberd ML, Mulders W, Corrah T, de Jong BC, Antonio M.Deciphering the growth behaviour of Mycobacterium africanum PLoS Negl Trop Dis 20137e2220[Published erratum appears in PLoS Negl Trop Dis 2013;7(6).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong BC, Hill PC, Brookes RH, Gagneux S, Jeffries DJ, Otu JK, Donkor SA, Fox A, McAdam KPWJ, Small PM, et al. Mycobacterium africanum elicits an attenuated T cell response to early secreted antigenic target, 6 kDa, in patients with tuberculosis and their household contacts. J Infect Dis. 2006;193:1279–1286. doi: 10.1086/502977. [DOI] [PubMed] [Google Scholar]
- 30.Tientcheu LD, Sutherland JS, de Jong BC, Kampmann B, Jafali J, Adetifa IM, Antonio M, Dockrell HM, Ota MO. Differences in T-cell responses between Mycobacterium tuberculosis and Mycobacterium africanum-infected patients. Eur J Immunol. 2014;44:1387–1398. doi: 10.1002/eji.201343956. [DOI] [PubMed] [Google Scholar]
- 31.Talarico S, Ijaz K, Zhang X, Mukasa LN, Zhang L, Marrs CF, Cave MD, Bates JH, Yang Z. Identification of factors for tuberculosis transmission via an integrated multidisciplinary approach. Tuberculosis (Edinb) 2011;91:244–249. doi: 10.1016/j.tube.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yesilkaya H, Forbes KJ, Shafi J, Smith R, Dale JW, Rajakumar K, Barer MR, Andrew PW. The genetic portrait of an outbreak strain. Tuberculosis (Edinb) 2006;86:357–362. doi: 10.1016/j.tube.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 33.van Crevel R, Ottenhoff THM, van der Meer JWM. Innate immunity to Mycobacterium tuberculosis. Clin Microbiol Rev. 2002;15:294–309. doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiling N, Homolka S, Walter K, Brandenburg J, Niwinski L, Ernst M, Herzmann C, Lange C, Diel R, Ehlers S, et al. Clade-specific virulence patterns of Mycobacterium tuberculosis complex strains in human primary macrophages and aerogenically infected mice. MBio. 2013;4:e00250-13. doi: 10.1128/mBio.00250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe DM, Redford PS, Wilkinson RJ, O’Garra A, Martineau AR. Neutrophils in tuberculosis: friend or foe? Trends Immunol. 2012;33:14–25. doi: 10.1016/j.it.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Yang CT, Cambier CJ, Davis JM, Hall CJ, Crosier PS, Ramakrishnan L. Neutrophils exert protection in the early tuberculous granuloma by oxidative killing of mycobacteria phagocytosed from infected macrophages. Cell Host Microbe. 2012;12:301–312. doi: 10.1016/j.chom.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee PPW, Chan KW, Jiang L, Chen T, Li C, Lee TL, Mak PHS, Fok SFS, Yang X, Lau YL. Susceptibility to mycobacterial infections in children with X-linked chronic granulomatous disease: a review of 17 patients living in a region endemic for tuberculosis. Pediatr Infect Dis J. 2008;27:224–230. doi: 10.1097/INF.0b013e31815b494c. [DOI] [PubMed] [Google Scholar]
- 38.Arnold A, Witney AA, Vergnano S, Roche A, Cosgrove CA, Houston A, Gould KA, Hinds J, Riley P, Macallan D, et al. XDR-TB transmission in London: case management and contact tracing investigation assisted by early whole genome sequencing. J Infect. 2016;73:210–218. doi: 10.1016/j.jinf.2016.04.037. [DOI] [PubMed] [Google Scholar]
- 39.Xu P, Wu J, Yang C, Luo T, Shen X, Zhang Y, Nsofor CA, Zhu G, Gicquel B, Gao Q. Prevalence and transmission of pyrazinamide resistant Mycobacterium tuberculosis in China. Tuberculosis (Edinb) 2016;98:56–61. doi: 10.1016/j.tube.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luciani F, Sisson SA, Jiang H, Francis AR, Tanaka MM. The epidemiological fitness cost of drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2009;106:14711–14715. doi: 10.1073/pnas.0902437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li QJ, Jiao WW, Yin QQ, Xu F, Li JQ, Sun L, Xiao J, Li YJ, Mokrousov I, Huang HR, et al. Compensatory mutations of rifampin resistance are associated with transmission of multidrug-resistant Mycobacterium tuberculosis Beijing genotype strains in China. Antimicrob Agents Chemother. 2016;60:2807–2812. doi: 10.1128/AAC.02358-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvatore PP, Becerra MC, Abel zur Wiesch P, Hinkley T, Kaur D, Sloutsky A, Cohen T. Fitness costs of drug resistance mutations in multidrug-resistant Mycobacterium tuberculosis: a household-based case-control study. J Infect Dis. 2016;213:149–155. doi: 10.1093/infdis/jiv347. [DOI] [PMC free article] [PubMed] [Google Scholar]