Abstract
During pregnancy, fetal extravillous trophoblasts (EVT) play a key role in the regulation of maternal T cell and NK cell responses. EVT display a unique combination of human leukocyte antigens (HLA); EVT do not express HLA-A and HLA-B, but do express HLA-C, HLA-E, and HLA-G. The mechanisms establishing this unique HLA expression pattern have not been fully elucidated. The major histocompatibility complex (MHC) class I and class II transcriptional activators NLRC5 and CIITA are expressed neither by EVT nor by the EVT model cell line JEG3, which has an MHC expression pattern identical to that of EVT. Therefore, other MHC regulators must be present to control HLA-C, HLA-E, and HLA-G expression in these cells. CIITA and NLRC5 are both members of the nucleotide-binding domain, leucine-rich repeat (NLR) family of proteins. Another member of this family, NLRP2, is highly expressed by EVT and JEG3, but not in maternal decidual stromal cells. In this study, transcription activator-like effector nuclease technology was used to delete NLRP2 in JEG3. Furthermore, lentiviral delivery of shRNA was used to knockdown NLRP2 in JEG3 and primary EVT. Upon NLRP2 deletion, Tumor Necrosis Factor-α (TNFα)-induced phosphorylation of NF-KB p65 increased in JEG3 and EVT, and more surprisingly a significant increase in constitutive HLA-C expression was observed in JEG3. These data suggest a broader role for NLR family members in the regulation of MHC expression during inflammation, thus forming a bridge between innate and adaptive immune responses. As suppressor of proinflammatory responses, NLRP2 may contribute to preventing unwanted antifetal responses.
Keywords: pregnancy, MHC, HLA-G, inflammasome, NOD-like receptor
Summary Sentence
By modulating the NF-KB pathway and HLA-C expression on human trophoblasts, NLRP2, may contribute to preventing detrimental inflammatory responses at the maternal-fetal interface.
Introduction
Human extravillous trophoblasts (EVT) express a unique combination of human leukocyte antigens (HLA) and are key cells in the regulation of maternal T cell and NK cell responses at the maternal–fetal interface during pregnancy [1, 2]. Whereas nearly all cell types express HLA-A, HLA-B, HLA-C, and HLA-E, EVT lack expression of HLA-A and HLA-B, while expressing HLA-C and HLA-E. In addition, EVT uniquely express HLA-G. In fact, EVT are the only cells known to constitutively express HLA-G in healthy tissue [3, 4]. Despite the distinct HLA expression phenotype of EVT, the regulatory mechanisms that prevent HLA-A and HLA-B expression and establish HLA-C, HLA-E, and HLA-G expression on EVT have not been fully elucidated [5]. After the discovery of CIITA, the Class II Transactivator, as the transcriptional master regulator of major histocompatibility complex (MHC) class II genes in 1993 [6], NLRC5 (nucleotide-binding domain, leucine-rich repeat family, CARD domain-containing 5) has recently emerged as a transcriptional regulator of MHC class I genes [7–9]. Both CIITA and NLRC5 enter the nucleus despite lacking DNA-binding domains and activate MHC genes by forming a nucleoprotein complex, called the enhanseosome, together with transcription factors that include the RFX complex, CREB/ATF1, and the NF-Y factors [10–13]. Unlike CIITA, which affects constitutive as well as inducible MHC class II expression, NLRC5 mainly affects Interferon (IFNγ)-induced MHC class I expression and has a more limited effect on its constitutive expression [8, 9, 14, 15]. CIITA and NLRC5 are both members of the nucleotide-binding domain, leucine-rich repeat (NLR) family of proteins of which around 20 members have been identified in humans. Of these, NLRP12 (previously known as Monarch-1/Pan1/Pypaf2) was shown to enhance nonclassical and classical MHC class I expression at the level of the promoter, enhancing both mRNA and protein levels [16]. NLR family proteins also function as intracellular pattern recognition receptors and are key components of the various inflammasomes [17–19].
Microarray analysis demonstrated that NLRC5 and CIITA are expressed neither by EVT nor by the EVT model choriocarcinoma cell line JEG3 that has an identical MHC expression pattern to EVT [2]. Furthermore, RFX5 as well as other factors that bind the SXY-motif of the MHC class I promoter and cooperate with NLRC5 to induce MHC class I expression are expressed in low levels in trophoblasts [10, 20]. Therefore, other MHC regulators must be present in EVT and JEG3 to control expression of HLA-C, HLA-E, and HLA-G on these cells. Microarray data identified one particular member of the NLR family, NLRP2, as being highly expressed by villous trophoblast (VT), EVT, and JEG3 [2]. The activation of several members of the NLR gene family (e.g., NOD1, NOD2, NLRC4, NLRP1, and NLRP3) by pathogen-associated molecular patterns (PAMPs), (e.g., muramyl dipeptide), as well as danger-associated molecular patterns (e.g., extracellular ATP, uric acid and asbestos), elicits proinflammatory pathways, such as the phosphorylation of NF-ƙp p65, and can promote the assembly of macromolecular protein complexes termed inflammasomes. This process leads to activation of the cysteine protease, caspase-1, which in turn induces interleukin-1β (IL-1β) and IL-18 secretion [21–23], further enhancing inflammation. In contrast, overexpression of NLRP2, as well as NLRP4 (previously PAN2), NLRP10 (previously PYNOD), NLRP12, or NLRPX1, has been shown to inhibit NF-ƙB activation and reduce type I interferon responses [24, 25], possibly functioning as a negative feedback loop to control excessive inflammation [26–29]. Interestingly, NLRP2 inhibits NF-ƙ p65 signaling through interaction with proteins that control the activity of IƙB [24]. NLRP2 binds the IkB kinase (IKK) complex and thereby inhibits IƙB degradation induced by TNFα in macrophages and suppresses NF-ƙB p65 activation [28, 29]. Thus, the NLR family of proteins provides a complex system of inducers and inhibitors of key immune regulators such as NF-ƙB signaling. Furthermore, NLRP genes play important roles in mammalian oocytes and reproductive systems [30] and NLRP2 is expressed at much higher levels in placenta compared to other tissues [28]. In this study, we describe the immune suppressive function of NLRP2 in both JEG3 and primary human EVT. By suppressing NF-ƙw p65 phosphorylation and HLA-C expression, NLRP2 may contribute to the establishment of immune tolerance at the maternal–fetal interface and prevent detrimental antifetal responses by maternal NK and T cells. A broader role for NLR family members in the regulation of MHC expression during inflammation is suggested by these data.
Materials and methods
Generation of NLRP2 knockout cell lines
Transcription activator-like effector nucleases (TALENs) were designed to specifically target the first coding exon of the NLRP2 gene (Supplemental Figure S4A). TALEN genomic DNA-binding sites were chosen to be 15 bp in length such that the target sequence between the two binding sites was 14 bp in length; each binding site was anchored by a preceding T base in position “0” as has been shown to be optimal [31, 32]. Full-length TALENs were constructed as described previously [33]. JEG3 were cultured in RPMI containing glutamine, penicillin/streptomycin and 10% FBS. Transfection of the plasmids encoding for the TALEN pair into JEG3 was performed using Fugene 6 (Roche) in 10-cm tissue culture plates according to manufacturer's instructions. JEG3 were collected 48 h posttransfection by trypsin treatment, and GFP-expressing cells were sorted on a FACSAria III (BD). GFP+ JEG3 were plated on 10-cm culture dishes at 5000 cells/dish and allowed for recovery for 10 days. Single colonies were manually picked and dispersed and replated individually into 96-well plates. Colonies were allowed to grow to near confluence for 7 days, and split using trypsin and replica-plated to create two working stocks and one frozen stock. From one working stock, genomic DNA was extracted in 96-well format by adding lysis buffer (10 mM Tris pH 7.5, 10 mM EDTA, 10 mM NaCl, 0.5% Sarcosyl) containing proteinase K at 56°C overnight in a humidified chamber. Genomic DNA was precipitated by addition of 95% ethanol containing 75 mM NaCl for 1 h at 4°C. The DNA was washed twice in 70% ethanol, dried at RT and resuspended in Tris-EDTA (TE) buffer supplemented with RNAse A. Genotyping at the TALEN target site was then performed for each sample by PCR using FWD: TGATTGTCATCACAGCTCCCA; REV: TGCTTCCCATCAGCCTTGTC primers (Supplemental Figure 4A–C). Amplicons were loaded on 2.5% agarose gels to discriminate clones. Positive clones were identified by having a band or bands visibly shifted in size from the baseline (193 bp). A total of 39 positive clones out of 317 total clones were identified. The 39 clones were trypsinized and further expanded. Western blot lysates were made to determine the NLRP2 protein level in each of the 39 clones. One complete NLRP2 knockout clone was identified as well as multiple clones with reduced NLRP2 levels. To determine the mutations introduced into the NLRP2 KO clone and a subset of potentially heterozygous clones, PCR amplicons were sub cloned using the TOPO TA Cloning Kit (Invitrogen) and subjected to Sanger sequencing. In a similar fashion, four clones were confirmed to be wild type. One confirmed KO clone, four confirmed HZ, and four confirmed WT clones that were treated with TALENS but did not show the TALEN-mediated mutation were used in further experiments. In addition, one heterozygous clone with one mutant allele was expanded and subjected to a second round of TALEN targeting which yielded three more KO clones.
Tissue samples and trophoblast isolation
Discarded human placental material (gestational age 6–12 weeks) was obtained from women undergoing elective pregnancy termination in a local reproductive health clinic. All of the human tissue used for this research was deidentified, discarded clinical material. The Committee on the Use of Human Subjects (the Harvard IRB) determined that this use of placental and decidual material is Not Human Subjects Research. The procedure to isolate EVT has recently been described [2]. In short, villous tissue was gently scraped from the basal membrane. Thereafter, the tissue was digested for 8 min at 37°C with a trypsin (0.2%) EDTA (0.02%) solution. Trypsin was quenched with F12 medium containing 10% New Born Calf Serum (NCS) and pen/strep (all from Gibco). Cells were filtered over a gauze mesh and were washed once with complete F12 medium and layered on Ficoll (GE Healthcare) for density gradient centrifugation (20 min, 2000 rpm). Cells were collected, washed once, and incubated 20 min at 37°C in a tissue culture dish for removal of macrophages. Trophoblast isolates were stained for EGFR1 FITC (Serotec), HLA-G PE, (clone MEM-G/9, Abcam) CD14 PE-Cy7 (BD), and CD45 APC (BD) (Supplemental Table S1). CD45-EGFR1dimHLA-G+ EVT were sorted on FACS ARIA III. A purity of >95% HLA-G+ EVT was obtained and the median EVT yield was 1.5 × 105 (range 0.3–3.4 × 105). A minimum of 50.000 CD45-EGFR1dimHLA-G+ EVT were plated on 24-well cell culture plates (Costar) precoated with fibronectin (300 μl 20 ng/ml 45 min, BD), in DMEM/F12 medium (Gibco) supplemented with 10% NCS, pen, strep and glutamine, insulin, transferrin, selenium (Gibco), 5 ng/ml EGF (Peprotech), and 400 units human gonadotropic hormone (Sigma).
Lentivirus-mediated shRNA knockdown
Five NLRP2-shRNA constructs in pLKO.1 TRC cloning vector were obtained from Open Biosystems. A construct with a scrambled sequence was used as control. Packaging of lentivirus was achieved by transfecting 293T cells with one NLRP2-shRNA together with pLP1, pLP2, and pLP-VSVG plasmids according to the Viralpower Lentivirus expression system from Invitrogen. The supernatants from transfected 293T cells were harvested and used to infect JEG3 or EVT. Puromycin (5 μg/ml) was added 24 h after infection, and the efficiency of knocking down was verified by western blot.
Western blot analysis
Samples were lysed with RIPA lysis buffer (Sigma) for 30 min on ice, and spun 10 min 10,000 rpm. Supernatant was collected and boiled for 10 min with NuPage LDS sample buffer containing β-mercaptoethanol. Lysates were run on 4–12% Bis-Tris Polyacrylamide gel (Life Technologies), transferred to Nitrocellulose membranes on an iBlot transfer system (Life Technologies). Membranes were blocked for 1 h in 5% BSA in Tris-buffered saline–Tween (50 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween 20). The following antibodies were used for protein detection: NLRP2 (mAb 443625, R&D Systems), HLA-G (mAb MEM-G1, Abcam), Phospho-Ser563 NF-kB p65 (mAb 93H1), NF-kB p65 (mAb D14E12), and IkBα (mAb L35A5) all from Cell Signalling Technologies and HSP70 (mAb W27, Biolegend) (Supplemental Table S1). Antimouse IgG or anti-Rat IgG horseradish peroxidase (HRP)-conjugated secondary antibodies (GE Healthcare) were added for 1 h. Blots were developed using Immobilon Western HRP substrate Luminol reagent (Millipore Corporation). Quantification of western blots was done using ImageJ software.
Quantitative real-time PCR
Real-time PCR was performed on wild-type, heterozygous, and knockout JEG3 lines. RNA was directly isolated with a Stratagene Absolutely RNA Microprep Kit according to manufacturer's instructions. Purified RNA was stored at –80°C. RNA was reverse transcribed with Stratagene's AffinityScript QPCR cDNA Synthesis Kit and according to manufacturer's protocol. Amplification of specific PCR products for HLA-C, HLA-E, HLA-G, and GAPDH were detected using the SYBR green system (Applied Biosystems) in duplicates. The PCR program consisted of one cycle of 10 min at 95°C, 40 cycles of 15 s at 95°C, and 1 min at 60°C and was finalized with a melting curve analysis. Primer sets used for rtPCR: HLA-C-Fwd: GTGTCCACCGTGACCCCTGTC; HLA-C-Rev: ATTCAGGTTCTTAACTTCAT; HLA-E-Fwd: GCACAGATTTTCCGAGTGAAT; HLA-E-Rev: CAGCCATGCATCCACTGC; HLA-G-Fwd: GCTGCCCTGTGTGGGACTGAGTG; HLA-G-Rev: ACGGAGACATCCCAGCCCCTTT; GAPDH-Fwd: GAAGGTGAAGGTCGGAGT; GAPDH-Rev: GAAGATGGTGATGGGATTTC (all obtained from Eurofin MWG).
Confocal imaging
JEG3 or primary EVT were cultured on fibronectin-coated cover slips. Cells were fixed with 1% paraformaldehyde for 10 min and permeabilized using PBS/0.01% Triton X. Cells were stained with anti-NLRP2 (R&D systems) for 30 min and thereafter washed three times. Goat-antimouse Alexa-488 was added for 30 min and cells were further stained with DAPI (Biolegend) and Phalloidin-CF568 (Biotium). Extravillous trophoblasts were also stained with anti-HLA-G (MEM-G9 conjugated to CF647 using an antibody labeling kit (Biotium)). Anti-HLA-G was added to the culture 30 min before fixing and thereafter cells were stained with anti-NLRP2 and DAPI as described above. Acquisition was performed on the Zeiss LSM510 microscope.
Statistics
All data were analyzed using GraphPad prism software. To determine differences between two unpaired groups a nonparametric Mann–Whitney test was performed, whereas between two paired groups, a nonparametric Wilcoxon signed-rank test was performed. To determine differences between more than two groups a nonparametric Kruskal–Wallis one-way ANOVA with Dunn's multiple comparison post-test was performed. P-values <0.05 are considered to reflect significant differences.
Results
Regulation of major histocompatibility complex expression in trophoblasts
Expression of HLA proteins in somatic cells depends on the presence of the transcriptional regulators NLRC5 (MHC class I) and CIITA (MHC class II) and additional proteins that together form the MHC enhanceosome, allowing for HLA transcription (Supplemental Figure S1A and B; [34]). Analysis of a recently published microarray dataset (www.ebi.ac.uk/arrayexpress, accession number E-MTAB-3217([2]), that includes VT (MHC negative), EVT and JEG3 (HLA-C+, HLA-E+, HLA-G+), and decidual stromal cells (DSC) (HLA-A+, HLA-B+, HLA-C+, HLA-E+) demonstrated that the MHC regulator NLRC5 was expressed by DSC but not by VT, EVT, and JEG3. Thus, the pattern of NLRC5 parallels the expression of HLA-A and HLA-B, but not HLA-C and HLA-E (Figure 1A–C; Supplemental Figure S2). In addition, CIITA was absent in all cell types and NLRP12, previously implicated in the regulation of classical and nonclassical MHC genes, was not differentially expressed in VT, EVT, JEG3, and DSC (Figure 1A and D). Furthermore, RFX-5, RFX-ANK, and RFX-AP, key components of the MHC class I enhanceosome, are either absent or display lower expression levels in EVT compared to DSC (Supplemental Figure S1C and D). In JEG3, RFX5 expression is reduced, whereas RFX-ANK and RFX-AP have similar or increased expression compared to DSC. Other elements display equal (e.g., ATF1) or increased (e.g., NF-κB, IRF1) RNA expression levels in EVT and JEG3 compared to DSC (Supplemental Figure S1C and D). Besides transcriptional regulation, MHC protein stability and cell surface expression depend on the binding of peptides to the MHC molecules. Peptide loading to MHC class I molecules is a complex process that requires many proteins. The microarray data set revealed that key molecules required for peptide transport (TAP1, TAP2), peptide trimming (ERAP1 and ERAP2), and formation of the MHC/peptide loading complex (Tapasin, Calreticulin, Calnexin and ERP57), as well as beta-2 microglobulin (B2M), are expressed by EVT and are thus not limiting factors in MHC expression on the EVT cell surface (Supplemental Figure S3A and B). Thus, in the absence of NLRC5 and CIITA, other transcriptional regulators must be involved in controlling the expression of HLA-C, HLA-E, and HLA-G on trophoblasts. NLRC5 and CIITA are both members of the NLR family and another member, NLRP2, was found to be highly expressed by VT, EVT, and JEG3 but not by DSC (Figure 1E). NLRP2 is expressed at a much higher level in placenta compared to other tissues [28]. The expression of NLRP2 mRNA was about 2-fold higher in EVT and JEG3 compared to VT. NLRP2 expression was confirmed in JEG3 and primary EVT using confocal microscopy (Figure 2). Interestingly and in contrast to NLRC5 and CIITA, NLRP2 was not observed in the nucleus. Moreover, blocking nuclear export with Leptomycin B (11) did not result in nuclear accumulation of NLRP2, suggesting that NLRP2 does not enter the nucleus (data not shown).
Figure 1.
High expression of NLRP2, but not NLRC5 in EVT and JEG3. (A) Heat map depicts the global mRNA expression levels for all members of the NLR family in VT, EVT, JEG3, and DSC as found in (2). (B) Table shows MHC protein expression profile and the presence of MHC transcriptional regulators in the four cell types used. Graphs depict mRNA expression levels of (C) NLRC5, (D) NLRP12, and (E) NLRP2 in VT, EVT, JEG3, and DSC. Bars indicate median expression values.
Figure 2.
NLRP2 in JEG3 and EVT is not localized in the nucleus. (A) Confocal images of JEG3. Panels depict NLRP2 (top) and IgG2a (bottom), DAPI, F-actin, and composite images. (B) Confocal images of two isolates of primary EVT (EVT1 and EVT2). Panels depict NLRP2, HLA-G, DAPI, and composite image for EVT1 and NLRP2, DAPI and composite image for EVT2.
Generation of NLRP2-deficient JEG3 lines
To study the role of NLRP2 in JEG3 in more detail, NLRP2-deficient JEG3 lines were generated using TALEN technology [33]. Briefly, TALENs were designed to specifically target the first coding exon of the NLRP2 gene, as outlined in Methods and Supplemental Figure S4A and B. Next, JEG3 were transfected with the TALEN pair, FACS sorted and single colonies were isolated and expanded. PCR analysis identified 39 mutant clones out of a total of 319 clones, as assessed by the presence of a shifted PCR band compared to the parental JEG3 cell line. All 39 clones were analyzed by western blotting, and one clone out of these 39 clones showed no NLRP2 expression, whereas multiple clones had reduced NLRP2 expression as compared with parental JEG3. TOPO cloning and western blotting was used to confirm knockout, heterozygous, and wild-type clones (Supplemental Figure S4C–F). Four wild-type clones, four heterozygous clones, and the one knockout clone were selected for further experiments. In addition, one heterozygous clone was retargeted with the same TALEN pair and yielded three additional knockout clones that were confirmed by PCR and western blotting (Supplemental Figure S4G).
Deletion of NLRP2 increased HLA-C protein and mRNA expression
Knock-out, heterozygous, and wild-type clones, as well as parental JEG3, were analyzed in parallel for the expression of HLA-C (mAb clones TRA2G9 and DT9), HLA-E (mAb 3D12), and HLA-G (mAb MEM-G9) by flow cytometry. A significant increase in HLA-C expression on the NLRP2 KO clone compared to parental JEG3 and WT clones was observed using both TRA2G9 and DT9 mAbs (Figure 3A and B, E). In addition, a small, yet not statistically significant, increase in HLA-C expression was observed in the heterozygous versus the wild-type clones. Of note, no differences in expression of HLA-E and HLA-G between clones and parental JEG3 cells were observed (Figure 3C–E). The antibody used for detection of HLA-C (DT9) has been reported to cross-react with HLA-E [35]. However, HLA-E expression was not significantly affected by NLRP2 depletion, showing that the change in DT9 staining is due to changes in HLA-C expression. The three additional knockout clones generated by retargeting one heterozygous clone demonstrated a significant increase in HLA-C expression compared to retargeted but still heterozygous clones (Supplemental Figure S4H). To investigate whether loss of NLRP2 leads to increased transcription of HLA-C, total RNA was isolated from wild-type, heterozygous and knockout clones and analyzed for HLA-C, HLA-E, and HLA-G expression by qPCR. HLA-C mRNA was indeed found to be increased on all knockout clones compared to wild type, whereas HLA-E and HLA-G mRNA expression remained unaltered (Figure 3F–H). These data demonstrate that deletion of NLRP2 specifically increases HLA-C protein and mRNA expression in trophoblasts. Thus, NLRP2 functions as a suppressor of HLA-C expression, but does not influence expression of HLA-E and HLA-G molecules.
Figure 3.
HLA-C protein and mRNA expression is increased in NLRP2 knockout JEG3 clones. Graphs depict mean fluorescence intensity (MFI) for (A) HLA-C (mAb TRA2G9), (B) HLA-C (mAb DT9), (C) HLA-E (mAb 3D12), and (D) HLA-G (mAb MEM-G9) protein levels on parental JEG3 (JEG3) as well as on wild-type (WT) heterozygote (HZ), knockout (KO) clones. Five technical replicates for JEG3, four WT clones, and four HZ clones with a minimum of two replicates each, two replicates for four HZ clones and six replicates for KO clone 1 are depicted. (E) Representative FACS plots of HLA-C, HLA-E, and HLA-G on the parental and targeted cell lines. The lines indicate mean expression of HLA-C, HLA-E, and HLA-G in untreated JEG3 to visualize the shift in HLA-C but not HLA-E and HLA-G in HZ and KO cells. Relative mRNA expression of (F) HLA-C, (G) HLA-E, and (H) HLA-G normalized to GAPDH on wild-type (WT) heterozygote (HZ) and knockout (KO) JEG3 clones. Bars indicate median values and SEM. **P < 0.01.
Cytokine-induced major histocompatibility complex class I expression is affected NLRP2 knockout JEG3 clones
To investigate whether deletion of NLRP2 also affects TNFα- and IFNγ-induced MHC class I expression, NLRP2 knockout, heterozygous and wild-type clones, as well as the parental JEG3 cells, were stimulated with TNFα (20 ng/ml) or IFNγ (100 ng/ml) and analyzed by flow cytometry 48 h poststimulation. Stimulation with IFNγ significantly increased the expression of HLA-C on all JEG3 clones (Figure 4A and B). The parental JEG3 and wild-type clones had a significant greater fold change in HLA-C expression after IFNγ stimulation (∼4-fold and ∼3-fold, respectively) compared to the knockout clone (∼2-fold). However, despite this difference, the MFI for HLA-C remained higher on the knockout clone compared to the wild-type clones after IFNγ stimulation (Figure 4A and B). Stimulation with TNFα resulted in a significant increase of HLA-C on the parental JEG3 (∼3-fold increase) and wild-type clones (∼2.3-fold) but the knockout clones only increased HLA-C by ∼1.5-fold. IFNγ or TNFα stimulation did not increase the expression of HLA-E or HLA-G on any of the JEG3 clones (Figure 4C–E). Thus, NLRP2 negatively regulates constitutive HLA-C expression, but in its absence cytokine-induced HLA-C expression is impaired.
Figure 4.
Major histocompatibility complex class I expression on NLRP2 clones upon IFNγ or TNFα stimulation. (A) Graph depicts mean fluorescence intensity (MFI) of HLA-C (mAb clone DT9) protein expression on JEG3, wild-type (WT) heterozygote (HZ), and knockout (KO) clones in the absence of stimulation or stimulated with IFNγ (100 ng/ml) or TNFα (20 ng/ml). Relative protein expression for (B) HLA-C, (C) HLA-E, (D) HLA-G, and (E) W6/32. Relative protein expression levels are plotted as expression on stimulated cells relative to unstimulated cells. Graphs depict median and interquartile range of a least five independent experiments. *P < 0.05 **P < 0.01.
NLRP2 deletion in JEG3 increases NF-κB p65 Ser536 phosphorylation upon TNFα stimulation
NLRP2 was previously shown to inhibit activation of NF-ƙB p65 in macrophages through binding of the IKK complex [28, 29]. Wild-type and knockout JEG3 clones were stimulated with TNFα (20 ng/ml) and as a control with IFNγ (100 ng/ml) and analyzed by western blotting for presence of phosphorylated NF-ƙ (p65 Ser536 (NF-ƙB p65-P)) in a time-dependent manner (Figure 5A). NF-ƙB p65 Ser536 is targeted for phosphorylation by different kinases including IKKs upon TNFα stimulation [36, 37]. Stimulation with TNFα generated earlier and increased NF-ƙB p65-P in knockout versus wild-type clones (Figure 5A, C, and D). Furthermore, IƙBα protein decreased more rapidly in NLRP2 knockout compared to wild-type clones (Figure 5B–D), while NF-ƙB p65 levels did not change upon stimulation in both knockout and wild-type clones (Figure 5B–D). Basal protein levels of NF-ƙB p65-P, NF-ƙB p65, and IƙBα were not significantly different in unstimulated knockout and wild-type clones. This demonstrates the ability of NLRP2 to suppress phosphorylation of NF-ƙB p65 and IƙBα degradation in JEG3. IFNγ stimulation did not increase NF-ƙB p65-P in any of the JEG3 lines (data not shown).
Figure 5.
Deletion of NLRP2 increases NF-ƙB p65-P upon TNFα stimulation. (A) Western blot images of NF-κB p65-P and HSP70 protein expression in NLRP2 wild-type (WT) and knockout (KO) JEG3 clones stimulated with 20 ng/ml TNFα for 0, 5, 15, 30, and 45 min. (B) Western blot images of HSP70, NF-κB p65, and IκBα expression in NLRP2 wild-type (WT, left) and knockout (KO, right) JEG3 clones stimulated with 20 ng/ml TNFα for 0, 5, 15, and 30 min. (C) Western blot quantification of NF-κB p65-P, NF-κB p65, and IκBα relative to HSP70 expression in NLRP2 wild-type (gray bars) and knockout (black bars) JEG3 clones stimulated with 20 ng/ml TNFα for 0, 5, 15, 30, and 45 min of one representative sample. (D) Graphs depict the fold change (FC) of NF-κB p65-P, NF-κB p65, and IκBα protein in KO clones relative to WT clones.
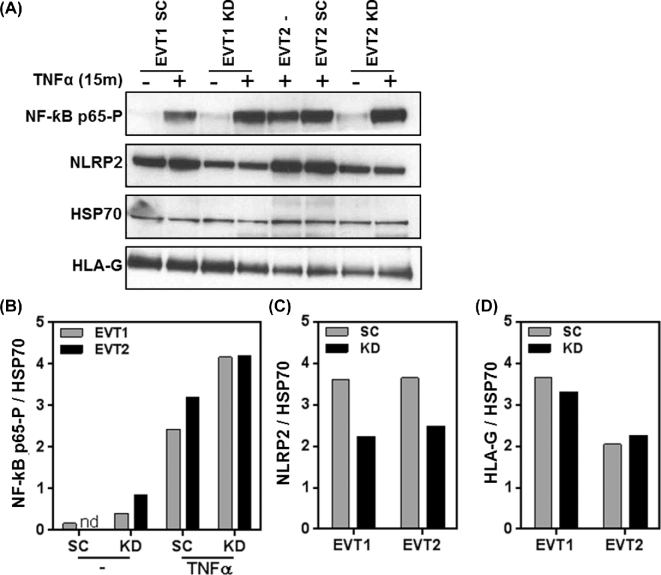
NLRP2 knockdown in primary extravillous trophoblasts leads to increased NF-κB p65 Ser536 phosphorylation upon TNFα stimulation
Although the MHC expression profile of JEG3 is similar to primary HLA-G+ EVT, many differences exist between JEG3 and EVT [2, 38]. Furthermore, and in contrast to JEG3, EVT are nonproliferating and short-lived cells in in vitro culture. The TALEN-mediated knockout system requires proliferating cells that can be cultured for multiple passages and is thus not suitable for use in primary EVT. Therefore, lentivirus containing NLRP2 shRNA as well as lentivirus with scrambled control shRNA were generated. To validate the lentivirus knockdown efficiency, JEG3 were infected with the five different lentiviral constructs and analyzed for NLRP2 expression. NLRP2 shRNA-4 containing lentivirus demonstrated the highest level of NLRP2 knockdown as examined by western blot analysis (Supplemental Figure S5A and B). HLA-G expression remained unchanged after knockdown of NLRP2 (Supplemental Figure S5C). In addition, NLRP2 knockdown by shRNA-4 also demonstrated increased NF-ƙH p65-P levels in JEG3 upon stimulation with TNFα (Supplemental Figure S5D and E). Primary HLA-G+ EVT were isolated as described previously [2] and infected with either NLRP2 shRNA-4 as well as scrambled control shRNA containing lentivirus. Three days after transduction with shRNA, EVT were stimulated with TNFα for 15 min and lysates were prepared and analyzed by western blotting. Transduction with NLRP2 shRNA-4 resulted in a ∼40% reduction of NLRP2 protein compared to transduction with scrambled control shRNA, while HLA-G levels remained unchanged (Figure 6A–C). Furthermore, TNFα stimulation of EVT resulted in an increase in NF-ƙB p65-P and most importantly NLRP2 shRNA-4 infected EVT had higher levels of NF-ƙB p65-P than EVT infected with scrambled control shRNA lentivirus (Figure 6A and D). As a control, uninfected EVT2 (Fig 6A, lane 5) and EVT2 infected with scrambled shRNA (Fig 6A, lane 6) were compared and no changes in protein expression was observed in these cell lysates. HLA-G levels did not change upon TNFα stimulation or NLRP2 knockdown. Thus, NLRP2 acts also as a suppressor of NF-ƙ, p65 phosphorylation in primary EVT.
Figure 6.
NLRP2 knockdown in primary EVT increases phosphorylation of NF-κB p65. (A) Western blots of NF-κB p65-P, NLRP2, HSP70, and HLA-G protein expression in primary EVT transduced with scrambled (SC) and NLRP2 (KD) shRNA stimulated without or with 20 ng/ml TNFα for 15 min. Graphs depict (B) NF-κB p65-P, (C) NLRP2, and (D) HLA-G relative to HSP70 in two primary EVT (EVT1 and EVT2) isolates transduced with scrambled (SC) and NLRP2 (KD) shRNA. NF-κB p65-P is depicted without or with 20 ng/ml TNFα for 15 min.
Discussion
Classical MHC class I molecules HLA-A, HLA-B, and HLA-C are highly polymorphic proteins of which the main function is peptide presentation to antigen-specific T cells. In contrast, the nonclassical HLA-E and HLA-G molecules have limited polymorphism. HLA-E has a limited peptide-binding repertoire and can present self and pathogen-derived peptides, whereas peptide-binding capacity for HLA-G remains to be confirmed [39–41]. HLA-C is the only classical MHC molecule expressed by EVT and has a unique role in pregnancy, as the only polymorphic MHC molecule that can elicit an allogeneic response by maternal T cells at the maternal–fetal interface [42]. In addition, HLA-C is also the only classical MHC molecule expressed by EVT that can present peptides to maternal T cells and can generate a protective immune response to intracellular pathogens [42]. This dual function of HLA-C may require tight (transcriptional) regulation to balance induction of immune tolerance and protective immunity to infections.
T cell activation strongly depends on the affinity of the TCR for its MHC/peptide complex. Low-affinity interactions predominantly result in T cell suppression or anergy, whereas high-affinity interactions result in effector T cell generation and cytolysis of target cells [43]. The affinity of TCR for MHC/peptide is dependent on the conformation of the MHC/peptide complex and the expression level of both TCR on the T cells and the MHC/peptide complexes on the target cells [44, 45]. IFNγ-mediated upregulation of HLA on host cells is a hallmark of the response to acute infection [46]. In this study, we demonstrate that NLRP2 is a suppressor of constitutive HLA-C expression on the cell surface. However, in the absence of NLRP2, the HLA-C induction by TNFα and IFNγ is reduced, suggesting that NLRP2 facilitates cytokine-induced HLA-C expression. Thus, NLRP2 may skew maternal T cell response to induce tolerance in the absence of these cytokines (and low HLA-C expression) while at the same time allowing maternal T cells to engage with high HLA-C levels when proinflammatory cytokines are present. High cell surface expression of HLA-C has been correlated with an increased control of HIV infection and associated with an increased cytotoxic T lymphocyte precursor frequency in hematopoietic stem cell transplant patients [47, 48]. In the presence of IFNγ (and to a lesser extent TNFα), HLA-C expression is increased on EVT and may result in higher affinity TCR engagement and effector T cell responses. Similarly, HLA-C is the ligand for killer cell immunoglobulin-like receptors (KIR) expressed by NK cells. HLA-C binding to KIR strongly depends on HLA-C expression levels and can result in activating (KIR2DS1) or inhibitory (KIR2DS1/2/3) responses by NK cells [49]. Recently, a skewing of KIR expression on decidual NK cells (dNK) at the maternal–fetal interface towards recognition of HLA-C was demonstrated [50]. In particular, the potential of dNK to develop an activating response through KIR2DS1 and its interaction with HLA-C2 was increased in comparison to peripheral blood NK cells [51]. Thus, the control of HLA-C by NLRP2 may facilitate immune tolerance in T cells and dNK at low levels but allow for dNK and T cell activation in the presence of proinflammatory signals (e.g., IFNγ) as found during infections.
Both NLRC5 and CIITA lack DNA-binding domains and activate MHC genes by forming a nucleoprotein complex together with transcription factors that include the RFX complex, CREB/ATF1, and the NF-Y factors [10–13]. NLRC5 and CIITA shuttle into the nucleus and point mutations in the nucleotide binding domain (NBD) prevented nuclear import and MHC transcription [15, 52, 53]. In this study, we present evidence that NLRP2 suppresses HLA-C mRNA transcription in JEG3. However, NLRP2 has not been shown to enter the nucleus, and does not have a nuclear localization signal nor a NBD. This suggests that NLRP2 is a regulator but not a transcriptional regulator.
Previous studies demonstrated that single nucleotide polymorphisms in NLRP2 were associated with Beckwith–Wiedemann syndrome, a fetal over growth and familial imprinting disorder [54], miscarriages [55], and rheumatoid arthritis [56]. In these studies, NLRP2 was suggested to regulate epigenetic modifications and gene methylation. More recently, NLRP5 was associated with reproductive wastage and multilocus imprinting disorders in humans [57]. Major histocompatibility complex expression is negatively correlated with promoter methylation [58, 59], although speculative NLRP2 may influence HLA-C expression through gene methylation. Another possibility is that the increased activation of NF-ƙa upon NLRP2 deletion is the proximal cause of activation of HLA-C transcription. Similar to HLA-A and HLA-B, the HLA-C promoter region contains a sequence (enhancer A) that allows NF-ƙf binding and enhances transcription [60, 61]. However, enhancer A in the HLA-C gene contains nucleotide variations that may alter the binding affinity of the various NF-ƙ-derived dimers and their ability to regulate HLA-C expression. The enhancer A motif is absent from HLA-G whose expression is not enhanced by knockout of NLRP2 [61, 62]. Activation of transcription of MHC class I genes by release of inhibitory factors from the MHC class I promoter region has been described [63]. Much remains to be learned about the mechanism of transcription of HLA-C that does not require NLRC5 in JEG3 or EVT.
NF-ƙB p65 is targeted for phosphorylation at Ser536 by different kinases including IKKs [36, 37]. IKK-mediated phosphorylation of Ser536 is induced by TNFα and LPS and enhances transcriptional activity of various proinflammatory genes. Phospho-Ser536 is an active mark for canonical NF-ƙ activation which is dependent on IƙBα. However, it can also be involved in noncanonical activation independent of IƙBα degradation [36]. NLRP2 has been shown to suppress NF-ƙ p65 activation in macrophages by binding the IKK complex and inhibiting IƙB degradation [28, 29]. Here, we demonstrate that NLRP2 suppresses NF-ƙB p65 Ser536 phosphorylation and inhibits IƙBα degradation in JEG3 and EVT. Interestingly, TNFα but not IFNγ induced phosphorylation of NF-kB p65 in JEG3 and EVT. However, both IFNγ and TNFα significantly increased HLA-C expression on JEG3. This suggests that NF-kB p65 activation and induction of HLA-C expression in JEG3 and EVT are not directly related. However, NLRP2 suppresses both pathways and thereby reduces the immunogenicity of EVT. NLRP2 may further suppress the immunogenicity of EVT by attenuation of the downstream NF-ƙp signaling pathways that include production of proinflammatory cytokines, chemokines, growth factors, and cell adhesion molecules. Suppression of these proinflammatory pathways within EVT may prevent maternal immune activation and detrimental immune responses that could lead to trophoblast rejection.
The ability of NLRP2 to suppress HLA-C expression and NF-ƙB activation confirms the anti-inflammatory nature of NLRP2. Moreover, this study supports the hypothesis that NLRP2 and possibly other proteins of the NLR family have additional functions beyond pattern recognition and induction of proinflammatory responses that includes the control of MHC expression [17]. Thus, NLR proteins may provide a bridge between innate and adaptive immunity whereas NLRP2 in particular helps to balance pro- and anti-inflammatory responses within EVT that may be crucial to establish maternal–fetal tolerance during pregnancy.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary data are available at BIOLRE online.
Acknowledgments
We wish to thank Anna Studwell and Killian Jansen for technical help with the experiments; Mary Carrington, NIH, Rockville, MD, for the HLA-C antibody clone DT9; and all past and current lab members for their helpful discussions.
References
- 1. Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol 2002; 2:656–663. [DOI] [PubMed] [Google Scholar]
- 2. Tilburgs T, Crespo AC, van der Zwan A, Rybalov B, Raj T, Stranger B, Gardner L, Moffett A, Strominger JL. Human HLA-G+ extravillous trophoblasts: Immune-activating cells that interact with decidual leukocytes. Proc Natl Acad Sci USA 2015; 112:7219–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ellis SA, Sargent IL, Redman CW, McMichael AJ. Evidence for a novel HLA antigen found on human extravillous trophoblast and a choriocarcinoma cell line. Immunology 1986; 59:595–601. [PMC free article] [PubMed] [Google Scholar]
- 4. Kovats S, Main EK, Librach C, Stubblebine M, Fisher SJ, DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science 1990; 248:220–223. [DOI] [PubMed] [Google Scholar]
- 5. Holling TM, Bergevoet MW, Wierda RJ, van Eggermond MC, van den Elsen PJ. Genetic and epigenetic control of the major histocompatibility complex class Ib gene HLA-G in trophoblast cell lines. Ann NY Acad Sci 2009; 1173:538–544. [DOI] [PubMed] [Google Scholar]
- 6. Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell 1993; 75:135–146. [PubMed] [Google Scholar]
- 7. Neerincx A, Rodriguez GM, Steimle V, Kufer TA. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J Immunol 2012; 188:4940–4950. [DOI] [PubMed] [Google Scholar]
- 8. Meissner TB, Li A, Biswas A, Lee KH, Liu YJ, Bayir E, Iliopoulos D, van den Elsen PJ, Kobayashi KS. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proc Natl Acad Sci USA 2010; 107:13794–13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Staehli F, Ludigs K, Heinz LX, Seguin-Estevez Q, Ferrero I, Braun M, Schroder K, Rebsamen M, Tardivel A, Mattmann C, MacDonald HR, Romero P et al. . NLRC5 deficiency selectively impairs MHC class I- dependent lymphocyte killing by cytotoxic T cells. J Immunol 2012; 188:3820–3828. [DOI] [PubMed] [Google Scholar]
- 10. Meissner TB, Liu YJ, Lee KH, Li A, Biswas A, van Eggermond MC, van den Elsen PJ, Kobayashi KS. NLRC5 cooperates with the RFX transcription factor complex to induce MHC class I gene expression. J Immunol 2012; 188:4951–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev 2000; 14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 12. Gobin SJ, Peijnenburg A, van EM, van ZM, van den Berg R, van den Elsen PJ. The RFX complex is crucial for the constitutive and CIITA-mediated transactivation of MHC class I and beta2-microglobulin genes. Immunity 1998; 9:531–541. [DOI] [PubMed] [Google Scholar]
- 13. van den Elsen PJ, Peijnenburg A, van Eggermond MC, Gobin SJ. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today 1998; 19:308–312. [DOI] [PubMed] [Google Scholar]
- 14. Biswas A, Meissner TB, Kawai T, Kobayashi KS. Cutting edge: impaired MHC class I expression in mice deficient for Nlrc5/class I transactivator. J Immunol 2012; 189:516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meissner TB, Li A, Liu YJ, Gagnon E, Kobayashi KS. The nucleotide-binding domain of NLRC5 is critical for nuclear import and transactivation activity. Biochem Biophys Res Commun 2012; 418:786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Williams KL, Taxman DJ, Linhoff MW, Reed W, Ting JP. Cutting edge: Monarch-1: a pyrin/nucleotide-binding domain/leucine-rich repeat protein that controls classical and nonclassical MHC class I genes. J Immunol 2003; 170:5354–5358. [DOI] [PubMed] [Google Scholar]
- 17. Kufer TA, Sansonetti PJ. NLR functions beyond pathogen recognition. Nat Immunol 2011; 12:121–128. [DOI] [PubMed] [Google Scholar]
- 18. Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP et al. . The NLR gene family: a standard nomenclature. Immunity 2008; 28:285–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature 2012; 481:278–286. [DOI] [PubMed] [Google Scholar]
- 20. van den Elsen PJ, Gobin SJ, van der SN, Datema G, Vietor HE. Transcriptional control of MHC genes in fetal trophoblast cells. J Reprod Immunol 2001; 52:129–145. [DOI] [PubMed] [Google Scholar]
- 21. Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem 2001; 276:4812–4818. [DOI] [PubMed] [Google Scholar]
- 22. Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 2004; 20:319–325. [DOI] [PubMed] [Google Scholar]
- 23. Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature 2002; 416:194–199. [DOI] [PubMed] [Google Scholar]
- 24. Allen IC, Moore CB, Schneider M, Lei Y, Davis BK, Scull MA, Gris D, Roney KE, Zimmermann AG, Bowzard JB, Ranjan P, Monroe KM et al. . NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity 2011; 34:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams KL, Lich JD, Duncan JA, Reed W, Rallabhandi P, Moore C, Kurtz S, Coffield VM, Accavitti-Loper MA, Su L, Vogel SN, Braunstein M et al. . The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor alpha-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J Biol Chem 2005; 280:39914–39924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Hasegawa M, Imamura R, Kinoshita T, Kondo C, Konaka K, Suda T. PYNOD, a novel Apaf-1/CED4-like protein is an inhibitor of ASC and caspase-1. Int Immunol 2004; 16:777–786. [DOI] [PubMed] [Google Scholar]
- 27. Fiorentino L, Stehlik C, Oliveira V, Ariza ME, Godzik A, Reed JC. A novel PAAD-containing protein that modulates NF-kappa B induction by cytokines tumor necrosis factor-alpha and interleukin-1beta. J Biol Chem 2002; 277:35333–35340. [DOI] [PubMed] [Google Scholar]
- 28. Bruey JM, Bruey-Sedano N, Newman R, Chandler S, Stehlik C, Reed JC. PAN1/NALP2/PYPAF2, an inducible inflammatory mediator that regulates NF-kappaB and caspase-1 activation in macrophages. J Biol Chem 2004; 279:51897–51907. [DOI] [PubMed] [Google Scholar]
- 29. Fontalba A, Gutierrez O, Fernandez-Luna JL. NLRP2, an inhibitor of the NF-kappaB pathway, is transcriptionally activated by NF-kappaB and exhibits a nonfunctional allelic variant. J Immunol 2007; 179:8519–8524. [DOI] [PubMed] [Google Scholar]
- 30. Tian X, Pascal G, Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol Biol 2009; 9:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009; 326:1509–1512. [DOI] [PubMed] [Google Scholar]
- 32. Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science 2009; 326:1501. [DOI] [PubMed] [Google Scholar]
- 33. Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, Trevisan M, Gupta RM et al. . A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 2013; 12:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meissner TB, Li A, Kobayashi KS. NLRC5: a newly discovered MHC class I transactivator (CITA). Microbes Infect 2012; 14:477–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomas R, Apps R, Qi Y, Gao X, Male V, O’huigin C, O’Connor G, Ge D, Fellay J, Martin JN, Margolick J, Goedert JJ et al. . HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 2009; 41:1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004; 25:280–288. [DOI] [PubMed] [Google Scholar]
- 37. Huang B, Yang XD, Lamb A, Chen LF. Posttranslational modifications of NF-kappaB: another layer of regulation for NF-kappaB signaling pathway. Cell Signal 2010; 22:1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Apps R, Sharkey A, Gardner L, Male V, Trotter M, Miller N, North R, Founds S, Moffett A. Genome-wide expression profile of first trimester villous and extravillous human trophoblast cells. Placenta 2011; 32:33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pietra G, Romagnani C, Falco M, Vitale M, Castriconi R, Pende D, Millo E, Anfossi S, Biassoni R, Moretta L, Mingari MC. The analysis of the natural killer-like activity of human cytolytic T lymphocytes revealed HLA-E as a novel target for TCR alpha/beta-mediated recognition. Eur J Immunol 2001; 31:3687–3693. [DOI] [PubMed] [Google Scholar]
- 40. Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, Moretta L, Mingari MC. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci USA 2003; 100:10896–10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Diehl M, Munz C, Keilholz W, Stevanovic S, Holmes N, Loke YW, Rammensee HG. Nonclassical HLA-G molecules are classical peptide presenters. Curr Biol 1996; 6:305–314. [DOI] [PubMed] [Google Scholar]
- 42. Tilburgs T, Strominger JL. CD8+ effector T cells at the fetal-maternal interface, balancing fetal tolerance and antiviral immunity. Am J Reprod Immunol 2013; 69:395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faroudi M, Utzny C, Salio M, Cerundolo V, Guiraud M, Muller S, Valitutti S. Lytic versus stimulatory synapse in cytotoxic T lymphocyte/target cell interaction: manifestation of a dual activation threshold. Proc Natl Acad Sci USA 2003; 100:14145–14150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Heemskerk MB, Roelen DL, Dankers MK, van Rood JJ, Claas FH, Doxiadis II, Oudshoorn M. Allogeneic MHC class I molecules with numerous sequence differences do not elicit a CTL response. Hum Immunol 2005; 66:969–976. [DOI] [PubMed] [Google Scholar]
- 45. Rudolph MG, Wilson IA. The specificity of TCR/pMHC interaction. Curr Opin Immunol 2002; 14:52–65. [DOI] [PubMed] [Google Scholar]
- 46. Koeffler HP, Ranyard J, Yelton L, Billing R, Bohman R. Gamma-interferon induces expression of the HLA-D antigens on normal and leukemic human myeloid cells. Proc Natl Acad Sci USA 1984; 81:4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R, Yuki Y, Del Prete GQ, Goulder P, Brumme ZL, Brumme CJ, John M et al. . Influence of HLA-C expression level on HIV control. Science 2013; 340:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Israeli M, Roelen DL, Carrington M, Petersdorf EW, Claas FH, Haasnoot GW, Oudshoorn M. Association between CTL Precursor Frequency to HLA-C Mismatches and HLA-C Antigen Cell Surface Expression. Front Immunol 2014; 5:547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol 2005; 5:201–214. [DOI] [PubMed] [Google Scholar]
- 50. Male V, Sharkey A, Masters L, Kennedy PR, Farrell LE, Moffett A. The effect of pregnancy on the uterine NK cell KIR repertoire. Eur J Immunol 2011; 41:3017–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xiong S, Sharkey AM, Kennedy PR, Gardner L, Farrell LE, Chazara O, Bauer J, Hiby SE, Colucci F, Moffett A. Maternal uterine NK cell-activating receptor KIR2DS1 enhances placentation. J Clin Invest 2013; 123:4264–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harton JA, Cressman DE, Chin KC, Der CJ, Ting JP. GTP binding by class II transactivator: role in nuclear import. Science 1999; 285:1402–1405. [DOI] [PubMed] [Google Scholar]
- 53. Hake SB, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol 2000; 20:7716–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer E, Lim D, Pasha S, Tee LJ, Rahman F, Yates JR, Woods CG, Reik W, Maher ER. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet 2009; 5:e1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang JY, Su M, Lin SH, Kuo PL. A genetic association study of NLRP2 and NLRP7 genes in idiopathic recurrent miscarriage. Hum Reprod 2013; 28:1127–1134. [DOI] [PubMed] [Google Scholar]
- 56. Yang XL, Hu ZD, Wu Q, Liu X, Liu QJ, Zhang YC, Yang QR. Association of polymorphisms in SPARC and NLRP2 genes with rheumatoid arthritis in a Chinese Han population. Mod Rheumatol 2015; 25:67–71. [DOI] [PubMed] [Google Scholar]
- 57. Docherty LE, Rezwan FI, Poole RL, Turner CL, Kivuva E, Maher ER, Smithson SF, Hamilton-Shield JP, Patalan M, Gizewska M, Peregud-Pogorzelski J, Beygo J et al. . Mutations in NLRP5 are associated with reproductive wastage and multilocus imprinting disorders in humans. Nat Commun 2015; 6:8086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ramsuran V, Kulkarni S, O’huigin C, Yuki Y, Augusto DG, Gao X, Carrington M. Epigenetic regulation of differential HLA-A allelic expression levels. Hum Mol Genet 2015; 24:4268–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, Wang LD, Yang CS. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis 2001; 22:1615–1623. [DOI] [PubMed] [Google Scholar]
- 60. Gobin SJ, Keijsers V, van ZM, van den Elsen PJ. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor kappa B. J Immunol 1998; 161:2276–2283. [PubMed] [Google Scholar]
- 61. van den Elsen PJ. Expression regulation of major histocompatibility complex class I and class II encoding genes. Front Immunol 2011; 2:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gobin SJ, Biesta P, de Steenwinkel JE, Datema G, van den Elsen PJ. HLA-G transactivation by cAMP-response element-binding protein (CREB). An alternative transactivation pathway to the conserved major histocompatibility complex (MHC) class I regulatory routes. J Biol Chem 2002; 277:39525–39531. [DOI] [PubMed] [Google Scholar]
- 63. Israel A, Le BO, Hatat D, Piette J, Kieran M, Logeat F, Wallach D, Fellous M, Kourilsky P. TNF stimulates expression of mouse MHC class I genes by inducing an NF kappa B-like enhancer binding activity which displaces constitutive factors. EMBO J 1989; 8:3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at BIOLRE online.