Abstract
Preterm birth affects 1 out of every 10 infants in the United States, resulting in substantial neonatal morbidity and mortality. Currently, there are few predictive markers and few treatment options to prevent preterm birth. A healthy, functioning placenta is essential to positive pregnancy outcomes. Previous studies have suggested that placental pathology may play a role in preterm birth etiology. Therefore, we tested the hypothesis that preterm placentae may exhibit unique transcriptomic signatures compared to term samples reflective of their abnormal biology leading to this adverse outcome. We aggregated publicly available placental villous microarray data to generate a preterm and term sample dataset (n = 133, 55 preterm placentae and 78 normal term placentae). We identified differentially expressed genes using the linear regression for microarray (LIMMA) package and identified perturbations in known biological networks using Differential Rank Conservation (DIRAC). We identified 129 significantly differentially expressed genes between term and preterm placenta with 96 genes upregulated and 33 genes downregulated (P-value <0.05). Significant changes in gene expression in molecular networks related to Tumor Protein 53 and phosphatidylinositol signaling were identified using DIRAC. We have aggregated a uniformly normalized transcriptomic dataset and have identified novel and established genes and pathways associated with developmental regulation of the placenta and potential preterm birth pathology. These analyses provide a community resource to integrate with other high-dimensional datasets for additional insights in normal placental development and its disruption.
Keywords: placenta, transcriptomics, microarray, preterm birth, pregnancy
Summary Sentence
Distinct transcriptional profiles of spontaneous preterm birth placentae compared to term placentae identify gestational regulation or potential pathogenic mechanisms.
Introduction
Preterm birth is a devastating pregnancy outcome and the leading cause of death for children under 5 years of age. Every year 15 million children worldwide are born at less than 37 weeks’ gestation and 1 million of those die from complications related to their prematurity. In the United States, the incidence of prematurity is 9.6% [1]. In 2007, treating preterm birth cost about $26 billion dollars in the United States [2]. Globally, 60% of preterm births occur in Asia and Africa with rates as high as 15% [3]. The majority (70%) of preterm births are idiopathic and spontaneous, rather than being related directly to diagnosed medical causes (e.g. pre-eclampsia). While there are known risk factors, including smoking, stress, infection, and family history, there remains a lack of understanding of the key biological mechanisms and perturbed networks that underlie preterm birth [4].
Over the last decade, advances in the acquisition and analysis of high-dimensional “omics” data have allowed for the discovery of biomarkers and increased insight into various disease states, including placental pathologies [5–8]. The vast majority (76%) of transcriptomic placental studies to date have examined medically indicated preterm birth (i.e. from pre-eclampsia and chorioamnionitis), although these samples only represented 30% of the causes of preterm birth [9]. Only 18% of these studies focused on spontaneous preterm birth (sPTB), although this represents 45% of the causes of preterm delivery <37 weeks [9]. This discrepancy results in a gap in knowledge of the etiology of sPTB.
Analysis of omics data has been used to identify molecular signatures for pathologies in different organ systems [7]. Integrating disparate datasets from independent studies generally increases the robustness of the identified molecular signatures as the data represent a better approximation of population variation as well as a broader range of pathologies. It also amplifies the signal from the disease, while mitigating apparent signals that can be confounded with batch and lab effects [7,10]. Meta-analyses of placental gene expression have been used to identify validated biomarkers that were associated with different phenotypes of pre-eclampsia [11,12]. These types of systematic studies allow for qualitative exploration, and facilitate in silico modeling, hypothesis generation, and even the potential for prediction of disease.
Applying the same integrative systematic approaches utilizing placental villous tissue, we aimed to detect molecular signatures of preterm birth. During gestation, fetal growth and development are entirely dependent on the placenta, a transient organ that is a large part of the fetal/maternal interface. A fully developed and functional placenta is essential for a healthy and successful pregnancy. Understanding the biological differences between normal and pathological placentae is essential to ensuring positive pregnancy outcomes. To this end, we chose to initiate our study utilizing publicly available transcriptomes to identify potential molecular signatures through gene and network-level analyses of placental villous tissues. The differences we have observed provide insights into the etiology of sPTB and may point toward molecular signatures for therapeutic and clinical interventions.
Materials and methods
Sample selection
We performed a targeted search of Gene Expression Omnibus (GEO) and ArrayExpress for all publicly available microarray transcriptome studies that included term and preterm placental villous samples and identified 294 placental transcriptomes within six studies [11–16]. Four of these studies [12,14–16] examined the molecular basis of pre-eclampsia, utilizing preterm birth samples as gestational age (GA)-matched controls (<37 weeks’ GA) in addition to normal term controls (38–42 weeks’ GA). One study (GSE18809) [13] analyzed preterm birth specifically, using preterm samples (27.0–32.6. weeks’ GA) and term samples (38.4–40.0 weeks’ GA). The sixth study examined the inflammatory pathways between term and preterm (unpublished publicly available data, Genomic and Proteomic Network (GPN) for Preterm Birth Research (GSE73685)). These studies utilized both Affymetrix and Illumina microarray platforms, and analyses were performed with RNA extracted from placental villous tissues only. We removed all placental samples with known pregnancy complications, including chorioamnionitis (if status known), and pre-eclampsia from the raw data. We excluded samples between 36.0 and 37.6 weeks to mitigate inconsistencies in methods of dating GA. This final dataset included 55 preterm and 78 term placentae (total n = 133 placental samples). Preterm pregnancies were categorized as 25–36 weeks’ GA with term pregnancies categorized as 38–41 weeks’ GA. For studies lacking fetal sex data [13,15,16], fetal sex was imputed from expression of probes located within the Y chromosome using the Bioconductor package massiR [17].
Microarray data preprocessing, normalization, and aggregation
Microarray datasets were preprocessed using the methods described in Ramasamy et al. [18]. For each of the six selected studies, raw microarray data were reprocessed using only samples that met our inclusion criteria: sPTB <36 weeks’ GA, no other pathology (if known), and term birth <38 weeks’ GA with all other samples being discarded. Raw data from Affymetrix microarrays were normalized using Robust Multi-Array Average [19] and raw data from Illumina arrays were normalized using quantile normalization [20]. All data were log2 transformed and each dataset was assessed for within study batch effects through histograms and principle components analysis. For each specific dataset, individual probe identifiers were annotated with Ensembl Gene IDs using biomaRt [21]. Probes that did not map to any Ensembl Gene IDs or mapped to multiple Ensembl Gene IDs (cross-hybridizing probes) were discarded, as described in Ramasamy et al. [18] (Supplemental Table S1). For each individual array dataset, a gene level expression value was calculated by taking the mean expression value of all probes which mapped to an Ensemble Gene ID. While there are six datasets, two of the datasets were generated from the same array platform; thus, we only have five array platforms to consider for these analyses. An aggregated dataset was generated with only those genes that were present in all five array platforms used in the initial datasets (a total of 14,251 genes for analysis) (Figure 1).
Figure 1.
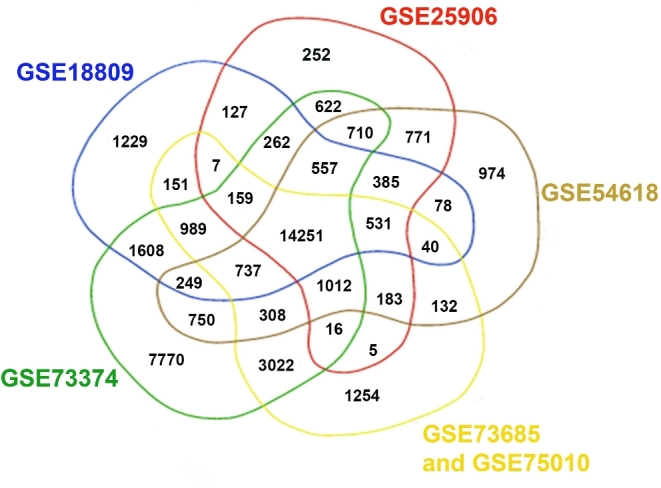
Representation of genes present in the datasets included in aggregated expression matrix: only genes present in the six datasets were included in the final aggregated dataset. GSE73685 and GSE75010 were conducted on the same array platform, and thus are represented by the same set of genes.
Principle components analysis was used to identify batch effects, both platform and study related, in the aggregated dataset. Batch effects were mitigated using parametrical empirical Bayesian adjustments implemented through utilization of the ComBat algorithm which selects effect parameter values related to batch and adjusted for them to mitigate expression related to study of origin [22]. Additional normalization to further mitigate batch effects was not conducted as it was unnecessary and would further diminish the ability to detect meaningful biological variation [22] (Supplemental Figure S1 and Supplemental Table S2).
Differential gene expression analysis
Differentially expressed genes were identified through a series of linear regression models fit for each gene within LIMMA (linear regression for microarray data) [23], which were adjusted for fetal sex. Statistically significant genes were identified based on the Q-value cutoff of <0.05, representing a false discovery rate after adjusting for multiple comparisons using the Benjamini–Hochberg approach [24]. We also examined differentially expressed genes using pairwise comparisons (sPTB versus term births) through t tests implemented within AltAnalyze [25]. Statistically significant genes were identified utilizing the Q-value cutoff of <0.05 after the Benjamini–Hochberg correction. The AltAnalyze results were concordant with the LIMMA results (data not shown).
Network level analysis
We used Differential Rank Conservation (DIRAC) [26] to identify gene networks where the relative expression of the genes in the networks was most significantly altered between placentae in term and preterm births. We calculated P-values based on permutation testing within DIRAC and reported on predictive ability of each tested network independently using DIRAC as a classifier and estimating accuracy using leave-one-out cross validation (LOO-CV). The networks used were defined in the Kyoto Encyclopedia of Genes and Genomes (KEGG), which contains 151 gene sets that are manually curated or computationally generated based on biological evidence [27]. For our analysis, only gene sets which contained between 5 and 100 genes and were curated in the Molecular Signatures Database [28]. Networks were considered statistically associated with preterm birth if they had adjusted P-value <0.05, after correction for multiple comparisons using the Benjamini–Hochberg approach, and a LOO-CV accuracy of 0.70 or higher. Here, LOO-CV uses rankings of genes within networks to predict expression of the remaining sample. Each network was assessed using the accuracy, with higher accuracy representing better predictive capacity. Differentially expressed genes within the significant networks were further characterized using unpaired t-tests with a P < 0.05.
All data were analyzed and visualized in R (version 3.3.1) using packages lumi [29], oligo [30], and bioMart [21] to process the microarray data, SVA [31] to adjust for batch effects, LIMMA [23] and AltAnalyze [25] to identify differentially expressed genes. Data were visualized within Cytoscape Version 3.2.1 [32] using the plugin Keggscape [27]. Specific codes used to aggregate data and perform DIRAC are available at https://github.com/alipaquette
Results
Dataset aggregation
We identified six GEO datasets with preterm and term data, which contained 133 samples (55 preterm and 78 term) after exclusion of data that did not meet our inclusion criteria. Three of the studies (GSE18809, GSE73374, and GSE54618) did not report fetal sex; therefore, we imputed fetal sex utilizing massiR, which uses Y chromosome gene expression to predict fetal sex [17] (Table 1). In the three studies that did report fetal sex, we also imputed sex using massiR to test the accuracy of this method, and found that massiR was able to predict sex in these samples with >95% accuracy. The average prediction accuracy for massiR is 96%–100% across various tissues depending on total sample number and skew of the sex ratio for each dataset [17]. The aggregated dataset contained a relatively even distribution of female and male samples (46% female and 54% male, see Table 1).
Table 1.
Publicly available studies selected for aggregated analyses.
| Original ref. | Array platform | No. of preterm (GA**) | No. of term (GA**) | No. of female | No. of male | Imputed* | Chorio-amnionitis status | Total | |
|---|---|---|---|---|---|---|---|---|---|
| GSE25906 | [14] | Illumina HT 6V2 | 5(24–36/33) | 18(38–40/38.7) | 9 | 14 | No | Unknown | 23 |
| GSE18809 | [13] | Affymetrix HG U133 | 5 (<34)1 | 5(38–39)1 | 7 | 3 | Yes | Unknown | 10 |
| GSE73374 | [16] | Affymetrix Human Gene 2.0 ST | 4(35–36/35.7) | 12(38–41/39.6) | 11 | 5 | Yes | Unknown | 16 |
| GSE54618 | [15] | Illumina HT 12V4 | 6(26–35.5/32.1 | 5(38–40.1/39.4) | 5 | 6 | Yes | Unknown | 11 |
| GSE75010 | [12] | Affymetrix Human Gene 1.0 ST | 29(26–36/31.5) | 27(38–41/38.7) | 24 | 32 | No | Status known | 56 |
| GSE73685 | Not yet published | Affymetrix Human Gene 1.0 ST | 6(25–34/30.1) | 11(38–40.1/38) | 5 | 11 | No | Unknown | 17 |
| TOTALS | 55 | 78 | 61 | 71 | 133 |
*Fetal sex was imputed from array data using massiR package.
**Gestational ages of samples in the dataset range/mean
1Study did not include exact gestational age for samples.
Initially within the aggregated dataset, transcriptome expression values were strongly associated with each study, with the majority of variation in expression (first principal component 81%) associated with study of origin (p < 0.0005, analysis of variance of first principal component based on study of origin (Supplemental Figure S1A and Supplemental Table S2)). As the studies were conducted on various array platforms, this variation is likely due to differences in experimental factors such as sample or array preparation; however, differences in study population cannot be ruled out. After utilizing the ComBat algorithm, such batch effects, whether due to platform or study population differences, were greatly reduced, as the first and second principal components were no longer associated with study of origin (Supplemental Figure S1B and Supplemental Table S2). Mean gene expression values for the top six differentially expressed genes before and after ComBat normalization are shown in Supplemental Table S3.
Gene level analyses
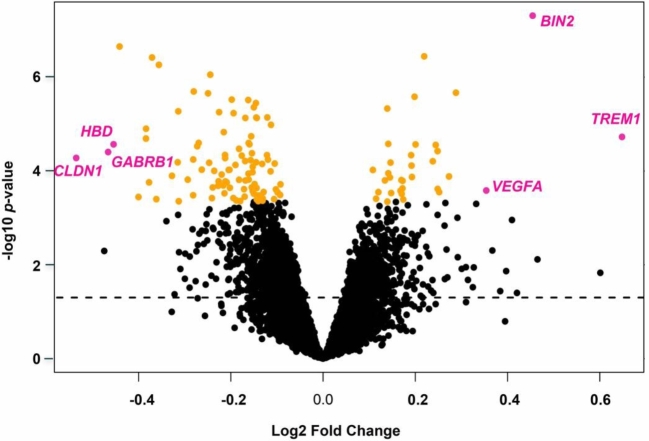
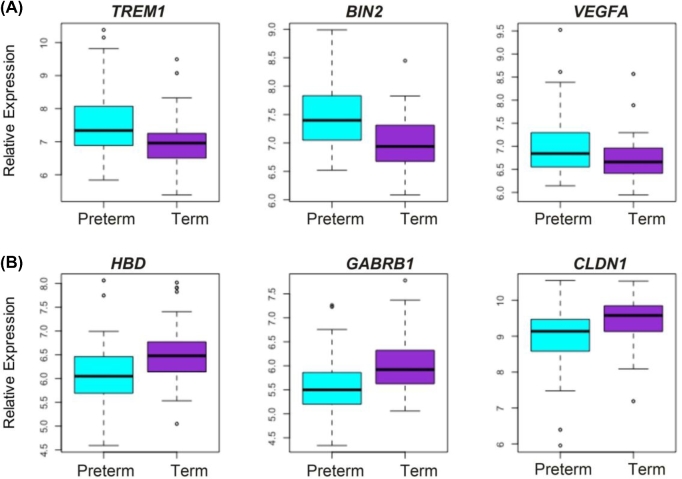
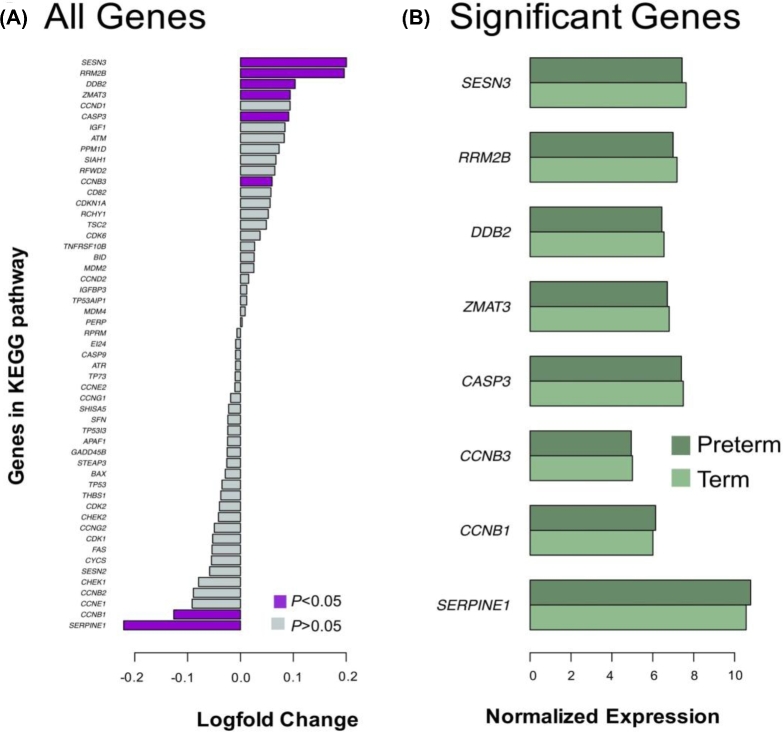
We performed a series of linear regressions using LIMMA to identify significant differences in gene expression based on prematurity status, adjusting for fetal sex as a confounding variable. Ninety-six genes were identified with significantly reduced expression in preterm samples and 33 genes with significantly higher expression in preterm samples after correcting for multiple comparisons using the Benjamini–Hochberg approach with a cutoff of Q < 0.05. The top genes with decreased expression in preterm placentae were HBD(hemoglobin subunit delta), GABRB1(gamma-aminobutyric acid type A receptor beta1 subunit), and CLDN1(claudin 1), and the top increased genes in preterm placentae were TREM1(triggering receptor expressed on myeloid cells 1), BIN2(bridging integrator 2), and VEGFA(vascular endothelial growth factor A) (Figures 2 and 3 and Table 2).
Figure 2.
Results of the LIMMA analysis on the aggregated data set adjusted for fetal sex as a confounding variable. The dotted line represents unadjusted P < 0.05. Significant samples adjusted for multiple comparisons by the Benjamini–Hochberg correction P < 0.05 are shaded in orange. The six most differentially expressed genes selected by fold-change are shaded in magenta.
Figure 3.
The six top differentially regulated genes in preterm placentae vs term placentae. Boxplots of the top three genes with the greatest positive and negative log/fold changes. Expression of preterm samples (n = 55) is shown in blue, and expression of term samples (n = 78) is shown in purple. (A) TREM1, BIN2, and VEGFA are all upregulated in preterm placentae compared to term. (B) HBD, GABRB1, and CLDN1 are all downregulated in preterm placentae compared to term.
Table 2.
Significantly differentially expressed genes with the largest fold-changes in relation to sPTB.
| Fetal sex adjusted model | ||
|---|---|---|
| Gene symbol | Log fold change | Adjusted P < value* |
| CLDN1 | –0.54 | 0.017 |
| GABRB1 | –0.47 | 0.014 |
| HBD | –0.45 | 0.011 |
| TREM1 | 0.65 | 9.9 × 10−4 |
| BIN2 | 0.45 | 7.0 × 10−4 |
| VEGFA | 0.35 | 0.036 |
Log-fold of preterm placentae vs term placentae, *Benjamini–Hochberg correction P < 0.05.
Identification and characterization of network level perturbations
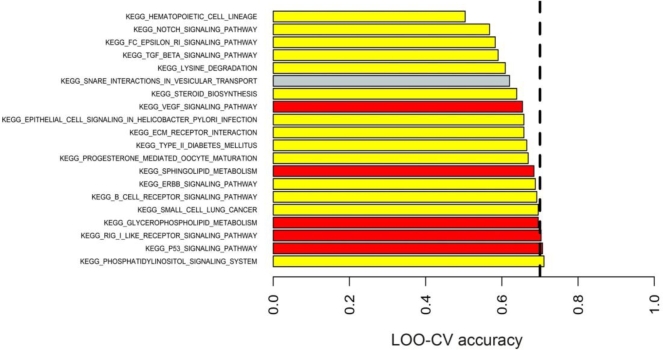
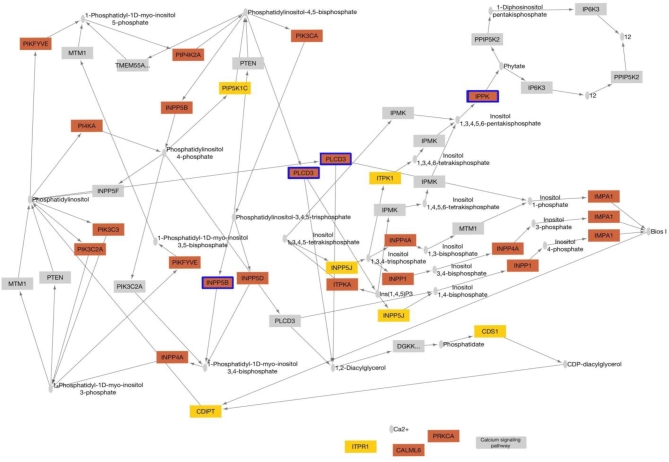
We identified two networks with significant differences between preterm and term placenate, phosphatidylinositol (PI3K) signaling and tumor protein 53 (TP53) signaling, which contained genes with the most significant and reliable reversals in relative gene expression based on rank (Figure 4 and Table 3). Networks were considered highly predictive if they had Benjamini–Hochberg adjusted P < 0.05 and a cross validation accuracy of >0.7 [33].
Figure 4.
The results of DIRAC analysis. The top 20 KEGG networks identified by DIRAC. The dashed line represents an LOO-CV accuracy equal to 0.70. The networks are shaded according to their adjusted P-value (Benjamini–Hochberg correction for multiple comparisons). Red P < 0.005, orange P < 0.01, yellow P < 0.05, gray bars are not significant.
Table 3.
KEGG networks identified in DIRAC analyses.
| Significantly different genes* | |||||
|---|---|---|---|---|---|
| KEGG network | Adjusted P-value | LOO-CV accuracy | Number of genes in network | Downregulated (PTB vs. term) | Upregulated (PTB vs. term) |
| Phosphatidylinositol signaling | 0.0214 | 0.71 | 66 | PIK3R1, PIK3CB | PLCB1, PLCB4, INPP5D, PIKFYVE, PIP5K1B, CDS1, CDS2, INPP4B, PI4KA |
| P53 signaling | <0.001 | 0.71 | 53 | SERPINE1, CCNB | ZMAT3, RRM2B, CASP3, SESN3, DDB2, CCNB3 |
*T-test, unadjusted P < 0.05.
Phosphatidylinositol signaling
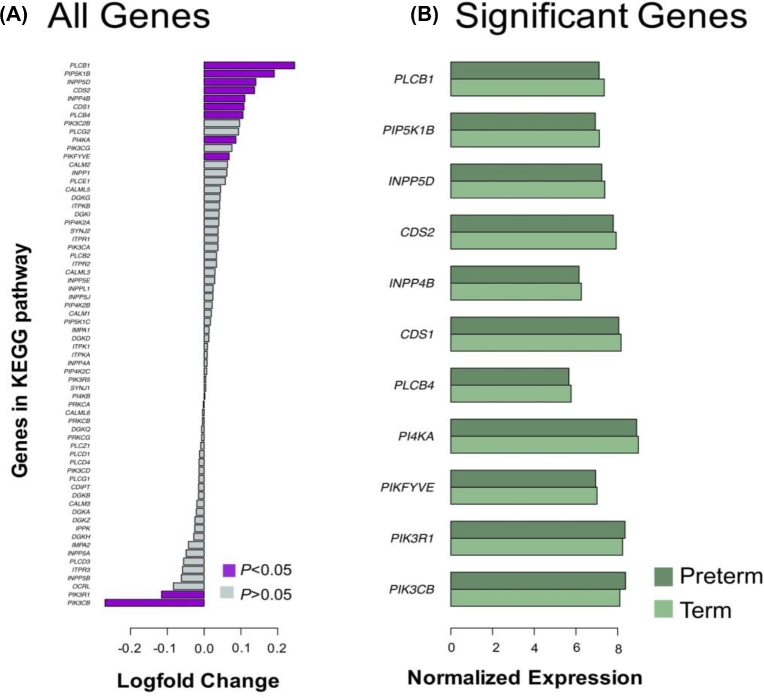
Relative changes of expression amongst the 66 genes within the PI3K signaling network were associated with preterm birth with a LOO-CV accuracy of 0.71. Expression of 11 of the 66 genes in this network was individually associated with preterm birth (unadjusted P < 0.05, t-test, Table 4, Figure 5A and B). Expression of PIK3R1 (phosphoinositide-3-kinase regulatory subunit 1) and PIK3CB (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta) was decreased in preterm placentae. PLCB1(phospholipase C beta 1), PLCB4(phospholipase C beta 4), INPP5D(inositol polyphosphate-5-phosphatase D), PIKFYVE(phosphoinositide kinase, FYVE-type zinc finger containing), PIP5K1B (phosphatidylinositol-4-phosphate 5-kinase type 1 beta), CDS1(CDP-diacylglycerol synthase 1), CDS2(CDP-diacylglycerol synthase 1), INPP4(inositol polyphosphate-4-phosphatase type II B ), and PI4KA(phosphatidylinositol 4-kinase alpha) expression was increased in preterm placentae compared to term placentae. Two of these genes (PIK3CB, CDS2) were considered statistically significant in our genome wide analysis (Benjamini–Hochberg adjusted P < 0.05), and three more genes showed trends toward genome wide significance (Benjamini–Hochberg adjusted P < 0.1). The potential impact of changes in gene expression on the network is shown in Figure 6.
Table 4.
Differentially expressed genes in the phosphatidylinositol signaling network.
| Gene symbol | Average expression of genes in PTB placentae | Average expression of genes in term placentae | Log fold change (PTB vs term) | Unadjusted P-values* | Adjusted P-values** |
|---|---|---|---|---|---|
| CDS1 | 8.16 | 8.06 | –0.11 | 0.011 | 0.12 |
| CDS2 | 7.93 | 7.79 | –0.14 | 1 × 10−4 | 0.0039 |
| INPP4B | 6.26 | 6.15 | –0.11 | 0.031 | 0.2 |
| INPP5D | 7.38 | 7.24 | –0.14 | 0.002 | 0.029 |
| PI4KA | 9.00 | 8.91 | –0.09 | 0.022 | 0.16 |
| PIK3CB | 8.11 | 8.38 | 0.27 | 5 × 10−4 | 0.017 |
| PIK3R1 | 8.24 | 8.36 | 0.12 | 0.020 | 0.16 |
| PIKFYVE | 7.01 | 6.95 | –0.07 | 0.015 | 0.14 |
| PIP5K1B | 7.12 | 6.93 | –0.19 | 9 × 10−4 | 0.019 |
| PLCB1 | 7.36 | 7.11 | –0.25 | 0.0012 | 0.02 |
| PLCB4 | 5.77 | 5.66 | –0.11 | 0.033 | 0.2 |
*T-test, unadjusted P < 0.05 ** Benjamini–Hochberg correction P < 0.05.
Figure 5.
Differentially expressed genes in phosphatidylinositol signaling network. (A) Changes in expression between preterm and term placentae for each of 66 genes in the network. Genes considered statistically different (P < 0.05, t-test) are purple. Nonsignificant genes are gray. (B) Average expression of statistically significant genes from panel A. Preterm placentae are shaded dark green and term placentae are shaded in light green.
Figure 6.
KEGG diagram of the phosphatidylinositol signaling network. Genes overexpressed in preterm birth placentae are orange, and downregulated genes are yellow. Genes with statistically significant changes in expression based on rank (P < 0.05, t-test, Small grey ovals represent molecules or chemical compounds. Arrows indicate molecular interactions.) are outlined in blue.
Tumor protein 53 signaling
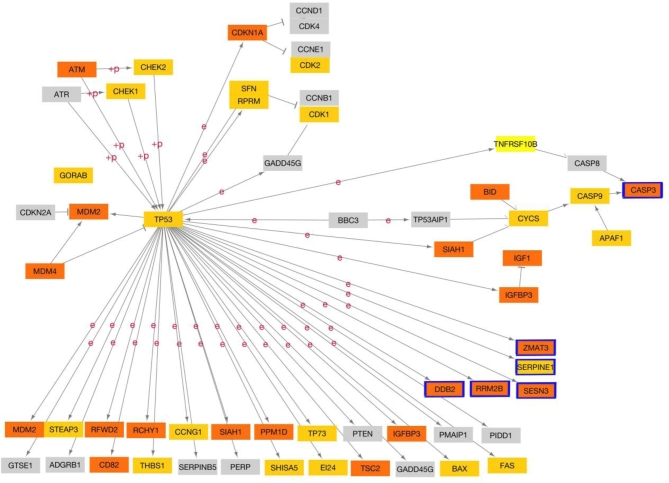
Relative changes of expression of the 53 genes within TP53 signaling network are potentially predictive of preterm birth with a LOO-CV accuracy of 0.71. Expression of 8 of the 53 genes in this network was individually associated with preterm birth (unadjusted P < 0.05, t-test, Table 5, Figure 7A and B). Expression of SERPINE1 (serpin family E member 1) and CCNB (cyclin B) was decreased and ZMAT3 (zinc finger matrin-type 3), RRM2B (ribonucleotide reductase regulatory TP53 inducible subunit M2B), CASP3(caspase 3), SESN3(sestrin 3), DDB2(damage specific DNA binding protein 2), and CCNB3(cyclin B3) was increased in preterm placentae compared to term placentae. The potential impact of changes in gene expression on the network is shown in Figure 8.
Table 5.
Differentially expressed genes in the TP53 signaling network.
| Gene symbol | Average expression of genes in PTB placentae | Average expression of genes in term placentae | Log fold change (PTB vs term) | Unadjusted P-values* | Adjusted P-values** |
|---|---|---|---|---|---|
| RRM2B | 6.99 | 7.19 | –0.20 | 5 × 10−6 | 0.00025 |
| DDB2 | 6.44 | 6.55 | –0.10 | 0.0029 | 0.076 |
| SESN3 | 7.43 | 7.63 | –0.20 | 0.011 | 0.19 |
| CCNB1 | 6.14 | 6.01 | 0.13 | 0.019 | 0.25 |
| CASP3 | 7.40 | 7.49 | –0.09 | 0.025 | 0.27 |
| SERPINE1 | 10.79 | 10.56 | 0.22 | 0.031 | 0.27 |
| CCNB3 | 4.95 | 5.01 | –0.06 | 0.042 | 0.32 |
| ZMAT3 | 6.71 | 6.80 | –0.09 | 0.049 | 0.32 |
*T-test, unadjusted P < 0.05 ** Benjamini–Hochberg correction P < 0.05.
Figure 7.
Differentially expressed genes in P53 signaling network. (A) Changes in expression between preterm and term placentae for each of 53 genes in network. Genes considered statistically different (P < 0.05, t-test) are purple. Nonsignificant genes are gray. (B) Average expression of statistically significant genes from panel A. Preterm placentae are shaded dark green and term placentae are shaded in light green.
Figure 8.
KEGG diagram of the P53 signaling network. Genes overexpressed in preterm birth placentae are orange, and downregulated genes are yellow. Genes with statistically significant changes in expression based on rank (P < 0.05, t-test, Arrows indicate molecular interaction. ``+P'' indicates phoshorylation events and ``e'' indicates expression.) are outlined in blue.
Discussion
The lack of molecular signatures for preterm birth etiology has made identification of biomarkers and potential therapeutic interventions difficult. Furthermore, there has been a shortage of studies specifically examining the role of the placenta in preterm birth [9]. To overcome this gap in knowledge, we have performed an aggregate analysis of six publicly available placental transcriptomic datasets and used known computational methodologies to examine gene expression and identify differences in molecular networks [23,26]. Using only the genes quantifiable in all six datasets, we identified genes and networks with significant changes in expression between the normal term and the preterm pathological placentae.
The placenta is a dynamic organ, changing over gestation via the interactions between mother and fetus within the in utero environment. The placenta matures, grows, and remodels throughout gestation to accommodate the growing fetus [34–36]. Essential placental developmental processes include cellular proliferation and differentiation as well as many others [37]. Placental insufficiency, which is defined as aberrant placental growth and function, has been implicated in adverse pregnancy outcomes including pre-eclampsia, intrauterine growth restriction (IUGR), and small for gestational age (SGA) [38]. Additionally, the placenta acts as a selective barrier, providing protection to the fetus from maternal factors including stress and sex hormones as well as xenobiotic factors [36]. If this barrier function is compromised, then the fetus is potentially exposed to adverse intrauterine conditions that could have an etiological role in preterm birth. Thus, by examining the transcriptional differences between term and preterm placentae, we may identify insights to the potential etiologies of preterm birth.
Gene level analyses
Several genes with the greatest differential expression (BIN2, GABRB1, and HBD) have not previously been associated with any adverse pregnancy outcomes or placental physiology. The remaining genes have either been implicated in adverse pregnancy outcomes or associated with specific placental physiological mechanisms.
Bridging integrator 2 (BIN2) is a member of the BAR (Bin/Amphiphysin/Rvs) family of proteins that possess a BAR domain, connecting the actin cytoskeleton and outer plasma membranes [39,40]. The BAR domain allows the plasma membrane to change its curvature promoting dynamic changes in membrane structure that accompanies cellular processes such as differentiation, cell interactions, and proliferation which are processes present in the developing and maturing placenta [39,40]. While BIN2 has not been previously studied in the context of placental physiology or adverse pregnancy outcomes, upregulation in preterm placentae could indicate any number of essential processes such as proliferation, differentiation, and syncytialization may be altered, thus leading to placental insufficiency.
Claudin 1 (CLDN1) is an integral membrane protein and is localized to the apical surface of the syncytiotrophoblast (STB) in placenta [41]. Upregulation is promoted by PPARG (peroxisome proliferator activated receptor gamma) and PKC (protein kinase C), which are central players in placental development [42,43], and it is repressed by TNF(tumor necrosis factor), NFKB1(nuclear factor kappa B subunit 1), IL1B (interleukin 1 beta), and TGFB1 (transforming growth factor beta 1), cytokines associated with initiation of parturition [42,44,45]. It is unclear if the reduction in CLDN1 expression is causative or the result of repression due to increased levels of cytokines at the initiation of parturition. A recent meta-analysis of pre-eclampsia [46] demonstrated that CLDN1 was also downregulated suggesting a potential underlying mechanism to both idiopathic spontaneous and medically induced pre-eclampsia (PE) preterm birth.
Hemoglobin subunit delta (HBD) codes for the hemoglobin delta subunit, which is expressed during the late third trimester and throughout adulthood, making up <3% of the adult hemoglobin [47,48]. The β-like hemoglobin locus consists of five paralogous genes—EPSILON, GAMMA-G, GAMMA-A, DELTA, and BETA—which code for the β-globin chains and are expressed in 5΄-3΄ order during development [48]. We cannot confirm the origin of the HBD mRNA. It could originate from maternal or fetal RBCs which are typically present in the villous samples or from the STB [49, 50]. Various hemoglobin subunits have been shown to be expressed outside erythroid lineages, but their function is unclear [47].
Vascular endothelial growth factor A (VEGFA) is a member of the VEGF growth factor family of proteins and shown to be essential in placental development and growth across gestation through its roles in vascularization and angiogenesis [51,52]. VEGFA expression peaks early and then declines over gestation in trophoblasts, while decidual and maternal serum levels rise over time [51]. Previous studies examining VEGFA expression levels in placental villous tissues have been inconclusive [13,53]. A study by Andraweera, et al. [53] demonstrated that VEGFA expression was reduced in sPTB placental villous tissues compared to normal villous tissues. However, Chim et al. [13] demonstrated that VEGFA expression increased in villous placental tissue from sPTB when compared to spontaneous term births. In our analysis, VEGFA is upregulated in sPTB (Table 2). The lack of age-matched controls for the placental tissues makes it difficult to distinguish if the changes are due to GA or are pathological.
It is known that a male bias exists with adverse pregnancy outcomes linked to placental insufficiency [54]. Buckberry et al. [55] performed a meta-analysis of publicly available data in one of the first large-scale studies of fetal sex differences and the potential impact on nonpathological placental physiology. These authors showed that sex differences are not limited to the X chromosomes, but genome wide and spread across the autosomes [55]. Our analyses identified 11 genes whose relationship in response to sPTB appears to be influenced by fetal sex. Of these 11 genes, LDLR (low density lipoprotein receptor), ENG (endoglin), MT1E (metallothionein 1), and KEAP1 (kelch like ECH associated protein 1) have either been implicated in preterm birth or as having essential roles in placental physiology [56–60].
Network level analyses
Network analyses can reveal mechanistic insights into gene functionality, which in turn allows for hypothesis generation and prioritization of potential functional experiments. We utilized DIRAC to perform network analyses, which is a combinatorial approach to examine networks in terms of relative changes in gene expression between cases and controls [26]. One strength of DIRAC is that it utilizes a rank-based approach that can detect statistically robust differences between phenotypes allowing for accurate detection of molecular signatures [26]. DIRAC uses LOO-CV to determine how accurately the statistically significant networks (as determined from the permutation testing) can predict the phenotype of interest in samples. The higher the LOO-CV accuracy, the more predictive a network's state is likely to be for the phenotype of interest [26]. In our study, none of the networks achieved a LOO-CV accuracy above 0.71; thus, their individual predictive capacity was limited. The accuracy of these networks to predict sPTB is likely limited by factors such as intrinsic differences in populations of individual studies including race and ethnicity, remaining undetected batch effects that were not removed through combat adjustment, lack of standardization of sample experimental protocols, and heterogeneity of the clinical subtypes of sPTB.
Phosphatidylinositol signaling
The phosphatidylinositol signaling network is composed of a family of membrane-bound phospholipid enzymes targeting multiple cell physiological processes. Through the activation of AKT1 (AKT serine/threonine kinase 1), additional subnetworks are activated and mediate physiological processes such as cell survival via BCL2 an apoptosis inhibitor, actin polymerization through AKT1, cell cycling through GSK3(glycogen synthase kinase 3), and protein synthesis and autophagy through MTOR(mechanistic target of rapamycin kinase) [61–63]. Furthermore, recent studies have indicated a role of PI3K signaling in modulating immune responses, with inhibition of the PI3K/AKT/MTOR pathway leading to a reduction in proinflammatory cytokines [64]. Each of these physiological processes is essential to placental physiology and birth timing, and thus alteration of the PI3K signaling pathway could directly alter preterm birth etiology through these molecular mechanisms [56,62,65–67].
Tumor protein 53 signaling network
TP53 is a master transcription factor regulating many physiological processes including tumor suppression, apoptosis, cell cycling arrest, and senescence among others [68–71]. Recent studies have illuminated the role of TP53 in placental physiology. TP53 localization, quantified through immunohistochemistry, was limited to the STB in normal placentae, while in the pathological placentae, TP53 was localized to both the cytotrophoblasts and STB. TP53 has been implicated in altered trophoblast physiology and adverse pregnancy outcome [69,70,72,73]. Expression of TP53 was increased in IUGR placentae with a corresponding increase in downstream apoptosis [73]. Studies of preeclamptic placenta showed an increase in TP53 protein levels but not mRNA suggesting that there may be a sequestration of the protein in response to stressors [72]. In addition, TP53 function in mouse decidua, demonstrated by conditional inactivation experiments, resulted in increased frequency of sPTB in mice, as well as a substantially augmented sensitivity to inflammation-induced preterm birth in this species [74]. Furthermore, a physiological role for TP53 in cellular senescence within human chorioamniotic membranes during parturition has been described [75]. While these findings reflect action on the maternal side component of the placenta, they serve to further highlight the importance of the TP53 pathway in preterm birth.
This study is limited by a lack of true normal placentae at earlier gestational time points. This is not unique to our study and is a known limitation in the fields of placental biology and preterm birth. The lack of entirely normal age-matched controls limits the distinction of gestational differences and preterm birth pathologies. Furthermore, because the villous tissue was collected at the time of delivery, regardless of status, it is unclear if the molecular signatures we have identified are due to etiology or a reflection of pathology. Additionally, these analyses are confounded by the mixed cell population of the placental villous samples which would be problematic even at earlier gestational ages. These tissue-specific limitations highlight the need to develop new methods to assess placental development and function through data acquired from noninvasive methodologies on mothers with normal term deliveries.
Like many multifactorial diseases, sPTB may have clinical subtypes that have yet to be identified. Several broad categories are proposed: spontaneous idiopathic preterm birth with and without premature rupture of membranes, sPTB leading to preterm premature rupture of membranes, and medically induced preterm birth for cases such as pre-eclampsia. While we selected studies based on the criteria of sPTB, we lacked essential covariate information that would help us further define the subtype of preterm birth and limited the types of analyses we could conduct. We were not able to categorize the samples by mode of delivery, indication of labor, or chorioamnionitis status, which was not available for all included samples. Ideally, cohorts of idiopathic spontaneous preterm with no additional underlying maternal pathologies should be utilized to identify molecular signatures of sPTB, which was beyond the scope of this study.
As the first aggregated transcriptomic study of PTB, our analysis acts as a resource in assessing the potential molecular differences between preterm and term placental villous tissues. While there are limitations to this study, we were able to detect gene and pathway level molecular signatures that may suggest a role for the placental villous tissue in preterm birth. Functional assessment of these signatures should provide insight into placental development and potentially preterm birth etiology. Future studies will include a genome-wide assessment of placental villous transcriptomes to assess essential placental genes. These genes, such as CGB (chorionic gonadotrophin beta) were beyond the scope of the current analysis. This was due to the limitation of microarray technologies and issues the aggregation of the data across platforms, both known issues with microarray meta-analyses. Additionally, these future studies will include expanded covariate information for further refinement of analyses and include a highly specific cohort of sPTB samples with better defined clinical characteristics.
Supplementary Material
Acknowledgments
We would like to thank the authors of the original manuscripts who provided data publicly through GEO and provided additional information about study design and sample collection for this manuscript. We would like to thank Helen Jones, members of the Muglia and Price laboratories, particularly Priyanka Baloni and Vineet Sangar for their review and editorial suggestions to this manuscript.
Grant Support: Bill and Melinda Gates Foundation: Systems Biology Approaches to Birth Timing and Preterm Birth Risk, Funding Period: 11/01/14-10/31/17 OPP1113966, March of Dimes Prematurity Research Center Ohio Collaborative, Funding Period 07/01/13–6/30/18, Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R01HD091527.
Conference presentation: Moderator/Invited Speaker: Nathan Price. Systems Analysis of OMICS Data in Pregnancy Conditions. Society of Gynecological Investigators Conference. Orlando Florida, March 2017
Alison Paquette. Aggregate transcriptional analysis of preterm infants identifies novel biomarkers and perturbed networks. Society of Gynecological Investigators Conference. Montreal, Canada, March, 2016
Supplementary data
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. Adjustment of aggregated dataset to eliminate batch effects. (A) Aggregated dataset without batch adjustment. (B) Aggregated dataset after adjustment with ComBat. Data points are colored based on the original study identifier. Preterm placentae are marked as circles and term placentae are marked with triangle.
Supplemental Table S1. Annotation indices for each individual dataset to be included in the final aggregated dataset.
Supplemental Table S2. Principle component analysis of aggregated dataset to identify batch effect variation.
Supplemental Table S3. Normalized expression values of highly significant genes before and after ComBat adjustments.
Author contributions
All authors contribute to the writing, editing of this manuscript and approval of the final version; AGP: conception of study design, performed all data aggregation, analysis, and assisted in interpretation; HMB: conception of study design, assisted with data analysis. Performed primary interpretation of results; NDP: conception of study design, provided guidance with data analysis and interpretation; LM: conception of Study Design, provided guidance with data analysis and interpretation.
REFERENCES
- 1. March of Dimes 2015 Premature Birth Report Card. 2015. https://www.marchofdimes.org/news/2015-premature-birth-report-card.aspx. [Google Scholar]
- 2. Institute of Medicine (US) Committee on Understanding Premature Birth and Assuring Healthy Outcomes Preterm Birth: Causes, Consequences, and Prevention, Behrman RE, Butler AS. 2007. https://www.nap.edu/catalog/11622/preterm-birth-causes-consequences-and-prevention. [PubMed] [Google Scholar]
- 3. Blencowe H, Cousens S, Chou D, Oestergaard M, Say L, Moller AB, Kinney M, Lawn J. Group BTSPBA Born too soon: the global epidemiology of 15 million preterm births. Reprod Health 2013; 10Suppl 1:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Monangi NK, Brockway HM, House M, Zhang G, Muglia LJ. The genetics of preterm birth: progress and promise. Semin Perinatol 2015; 39:574–83. [DOI] [PubMed] [Google Scholar]
- 5. Romero R, Espinoza J, Gotsch F, Kusanovic JP, Friel LA, Erez O, Mazaki-Tovi S, Than NG, Hassan S, Tromp G. The use of high-dimensional biology (genomics, transcriptomics, proteomics, and metabolomics) to understand the preterm parturition syndrome. BJOG 2006; 113Suppl 3:118–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gracie S, Pennell C, Ekman-Ordeberg G, Lye S, McManaman J, Williams S, Palmer L, Kelley M, Menon R, Gravett M, PREBIC "-Omics" Research Group. An integrated systems biology approach to the study of preterm birth using "-omic" technology - a guideline for research. BMC Pregnancy Childbirth 2011; 11: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sung J, Wang Y, Chandrasekaran S, Witten DM, Price ND. Molecular signatures from omics data: From chaos to consensus. Biotechnol J 2012; 7:946–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kedia K, Smith SF, Wright AH, Barnes JM, Tolley HD, Esplin MS, Graves SW. Global ‘Omics’ evaluation of human placental responses to preeclamptic conditions. Am J Obstet Gynecol 2016; 215:238.e1–238.e20. [DOI] [PubMed] [Google Scholar]
- 9. Eidem HR, Ackerman WE, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: a systematic review and meta-analysis. BMC Med Genomics 2015; 8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol 2017; 18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leavey K, Bainbridge SA, Cox BJ. Large scale aggregate microarray analysis reveals three distinct molecular subclasses of human preeclampsia. PLoS One 2015; 10:e0116508–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leavey K, Benton SJ, Grynspan D, Kingdom JC, Bainbridge SA, Cox BJ. Unsupervised placental gene expression profiling identifies clinically relevant subclasses of human preeclampsianovelty and significance. Hypertension 2016; 68:137–147. [DOI] [PubMed] [Google Scholar]
- 13. Chim SSC, Lee WS, Ting YH, Chan OK, Lee SWY, Leung TY. Systematic identification of spontaneous preterm birth-associated RNA transcripts in maternal plasma. PLoS One 2012; 7:e34328–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Thames BH, Piedrahita JA. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta 2011; 32:175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jebbink JM, Boot RG, Keijser R, Moerland PD, Aten J, Veenboer GJM, van Wely M, Buimer M, Ver Loren van Themaat E, Aerts JMFG, van der Post JAM, Afink GB et al. . Increased glucocerebrosidase expression and activity in preeclamptic placenta. Placenta 2015; 36:160–169. [DOI] [PubMed] [Google Scholar]
- 16. Martin E, Ray PD, Smeester L, Grace MR, Boggess K, Fry RC. Epigenetics and preeclampsia: defining functional epimutations in the preeclamptic placenta related to the tgf-β pathway. PLoS One 2015; 10:e0141294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buckberry S, Bent SJ, Bianco-Miotto T, Roberts CT. massiR: a method for predicting the sex of samples in gene expression microarray datasets. Bioinformatics 2014; 30:2084–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med 2008; 5:e184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003; 4:249–264. [DOI] [PubMed] [Google Scholar]
- 20. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
- 21. Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 2005; 21:3439–3440. [DOI] [PubMed] [Google Scholar]
- 22. Turnbull AK, Kitchen RR, Larionov AA, Renshaw L, Dixon JM, Sims AH. Direct integration of intensity-level data from Affymetrix and Illumina microarrays improves statistical power for robust reanalysis. BMC Med Genomics 2012; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benjamini Y. Discovering the false discovery rate. J Roy Stat Soc Series B 2010; 72:405–416. [Google Scholar]
- 25. Emig D, Salomonis N, Baumbach J, Lengauer T, Conklin BR, Albrecht M. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res 2010; 38:W755–W762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eddy JA, Hood L, Price ND, Geman D. Identifying tightly regulated and variably expressed networks by differential rank conservation (DIRAC). PLoS Comput Biol 2010; 6:e1000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014; 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008; 24:1547–1548. [DOI] [PubMed] [Google Scholar]
- 30. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010; 26:2363–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007; 3:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Series B 1995; 57:289–300. [Google Scholar]
- 34. Regnault TRH, Galan HL, Parker TA, Anthony RV. Placental development in normal and compromised pregnancies—a review. Placenta 2002; 23:S119–S129. [DOI] [PubMed] [Google Scholar]
- 35. Morgan T. Role of the placenta in preterm birth: a review. Am J Perinatol 2016; 33:258–266. [DOI] [PubMed] [Google Scholar]
- 36. Burton GJ, Fowden AL. The placenta: a multifaceted, transient organ. Philos Trans R Soc Lond B: Biol Sci 2015; 370:20140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maltepe E, Fisher SJ. Placenta: the forgotten organ. Annu Rev Cell Dev Biol 2015; 31:523–552. [DOI] [PubMed] [Google Scholar]
- 38. Mayhew TM. A stereological perspective on placental morphology in normal and complicated pregnancies. J Anat 2009; 215:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ge K, Prendergast GC. Bin2, a functionally nonredundant member of the BAR adaptor gene family. Genomics 2000; 67:210–220. [DOI] [PubMed] [Google Scholar]
- 40. Sánchez-Barrena MJ, Vallis Y, Clatworthy MR, Doherty GJ, Veprintsev DB, Evans PR, McMahon HT. Bin2 is a membrane sculpting N-BAR protein that influences leucocyte podosomes, motility and phagocytosis. PLoS One 2012; 7:e52401–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liévano S, Alarcón L, Chávez Munguía B, González Mariscal L. Endothelia of term human placentae display diminished expression of tight junction proteins during preeclampsia. Cell Tissue Res 2006; 324:433–448. [DOI] [PubMed] [Google Scholar]
- 42. Günzel D, Yu ASL. Claudins and the modulation of tight junction permeability. Physiol Rev 2013; 93:525–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huppertz B, Ghosh D, Sengupta J. An integrative view on the physiology of human early placental villi. Prog Biophys Mol Biol 2014; 114:33–48. [DOI] [PubMed] [Google Scholar]
- 44. Tossetta G, Paolinelli F, Avellini C, Salvolini E, Ciarmela P, Lorenzi T, Emanuelli M, Toti P, Giuliante R, Gesuita R, Crescimanno C, Voltolini C et al. . IL-1β and TGF-β weaken the placental barrier through destruction of tight junctions: An in vivo and in vitro study. Placenta 2014; 35:509–516. [DOI] [PubMed] [Google Scholar]
- 45. Stockert J, Adhikary T, Kaddatz K, Finkernagel F, Meissner W, Muller-Brusselbach S, Muller R. Reverse crosstalk of TGF and PPAR / signaling identified by transcriptional profiling. Nucleic Acids Res 2011; 39:119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vaiman D, Miralles F. An integrative analysis of preeclampsia based on the construction of an extended composite network featuring protein-protein physical interactions and transcriptional relationships. PLoS One 2016; 11:e0165849–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Saha D, Koli S, Patgaonkar M, Reddy KVR. Expression of hemoglobin-α and β subunits in human vaginal epithelial cells and their functional significance. PLoS One 2017; 12:e0171084–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ohls RK. Core concepts: The biology of hemoglobin. Neoreviews 2011; 12:e29–e38. [Google Scholar]
- 49. Uhlen M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson I, Edlund K et al. . Tissue-based map of the human proteome. Science 2015; 347:1260419. [DOI] [PubMed] [Google Scholar]
- 50. Kabanova S, Kleinbongard P, Volkmer J, Andree B, Kelm M, Jax TW. Gene expression analysis of human red blood cells. Int J Med Sci 2009; 6:156–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Burton GJ, Charnock-Jones DS, Jauniaux E. Regulation of vascular growth and function in the human placenta. Reproduction 2009; 138:895–902. [DOI] [PubMed] [Google Scholar]
- 52. Chen D-B, Zheng J. Regulation of placental angiogenesis. Microcirculation 2014; 21:15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Andraweera PH, Dekker GA, Thompson SD, North RA, McCowan LME, Roberts CT. The interaction between the maternal BMI and angiogenic gene polymorphisms associates with the risk of spontaneous preterm birth. Mol Hum Reprod 2012; 18:459–465. [DOI] [PubMed] [Google Scholar]
- 54. Murji A, Proctor LK, Paterson AD, Chitayat D, Weksberg R, Kingdom J. Male sex bias in placental dysfunction. Am J Med Genet 2012; 158A:779–783. [DOI] [PubMed] [Google Scholar]
- 55. Buckberry S, Bianco-Miotto T, Bent SJ. Integrative transcriptome meta-analysis reveals widespread sex-biased gene expression at the human fetal-maternal interface. Mol Hum Reprod 2014; 20:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang R, Dong S, Ma W-W, Cai X-P, Le Z-Y Xiao R, Zhou Q, Yu H-L. Modulation of cholesterol transport by maternal hypercholesterolemia in human full-term placenta. PLoS One 2017; 12:810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Steffen KM, Cooper ME, Shi M, Caprau D, Simhan HN, Dagle JM, Marazita ML, Murray JC. Maternal and fetal variation in genes of cholesterol metabolism is associated with preterm delivery. J Perinatol 2007; 27:672–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gregory AL, Xu G, Sotov V, Letarte M. Review: The enigmatic role of endoglin in the placenta. Placenta 2014; 35:S93–S99. [DOI] [PubMed] [Google Scholar]
- 59. Gundacker C, Hengstschläger M. The role of the placenta in fetal exposure to heavy metals. Wien Med Wochenschr 2012; 162:201–206. [DOI] [PubMed] [Google Scholar]
- 60. Sussan TE, Sudini K, Talbot CC, Wang X, Wills-Karp M, Burd I, Biswal S. Nrf2 regulates gene-environment interactions in an animal model of intrauterine inflammation: Implications for preterm birth and prematurity. Sci Rep 2017; 7:40194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cantley LC. The Phosphoinositide 3-Kinase Pathway. Science 2002; 296:1655–1657. [DOI] [PubMed] [Google Scholar]
- 62. Martini M, De Santis MC, Braccini L, Gulluni F, Hirsch E. PI3K/AKT signaling pathway and cancer: an updated review. Ann Med 2014; 46:372–383. [DOI] [PubMed] [Google Scholar]
- 63. Muralimanoharan S, Gao X, Weintraub S, Myatt L, Maloyan A. Sexual dimorphism in activation of placental autophagy in obese women with evidence for fetal programming from a placenta-specific mouse model. Autophagy 2016; 12:752–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xie S, Chen M, Yan B, He X, Chen X, Li D. Identification of a role for the PI3K/AKT/mTOR signaling pathway in innate immune cells. PLoS One 2014; 9:e94496–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Forbes K, Westwood M. Maternal growth factor regulation of human placental development and fetal growth. J Endocrinol 2010; 207:1–16. [DOI] [PubMed] [Google Scholar]
- 66. Howell KR, Powell TL. Effects of maternal obesity on placental function and fetal development. Reproduction 2017; 153:R97–R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Díaz P, Powell TL, Jansson T. The role of placental nutrient sensing in maternal-fetal resource allocation. Biol Reprod 2014; 91:82–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hu W, Feng Z, Atwal GS, Levine AJ. p53: A new player in reproduction. Cell Cycle 2008; 7:848–852. [DOI] [PubMed] [Google Scholar]
- 69. Toro AR, Maymó JL, Ibarbalz FM, Pérez-Pérez A, Maskin B, Faletti AG, Sánchez-Margalet V, Varone CL. Leptin is an anti-apoptotic effector in placental cells involving p53 downregulation. PLoS One 2014; 9:848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Toro AR, Pérez-Pérez A, Corrales Gutiérrez I, Sánchez-Margalet V, Varone CL. Mechanisms involved in p53 downregulation by leptin in trophoblastic cells. Placenta 2015; 36:1266–1275. [DOI] [PubMed] [Google Scholar]
- 71. Luo Q, Beaver JM, Liu Y, Zhang Z. Dynamics of p53: A master decider of cell fate. Genes 2017; 8:66. [Google Scholar]
- 72. Sharp AN, Heazell AEP, Crocker IP, Mor G. Placental apoptosis in health and disease. Am J Reprod Immunol 2010; 64:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Heazell AEP, Bernatavicius G, Warrander L, Brown MC, Dunn WB. A metabolomic approach identifies differences in maternal serum in third trimester pregnancies that end in poor perinatal outcome. Reprod Sci 2012; 19:863–875. [DOI] [PubMed] [Google Scholar]
- 74. Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Invest 2010; 120:803–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gomez-Lopez N, Romero R, Plazyo O, Schwenkel G, Garcia-Flores V, Unkel R, Xu Y, Leng Y, Hassan SS, Panaitescu B, Cha J, Dey SK. Preterm labor in the absence of acute histologic chorioamnionitis is characterized by cellular senescence of the chorioamniotic membranes. Am J Obstet Gynecol 2017; 217:592.e1–592.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.