Abstract
We report the genomic characterization of a rare human G8P[14] rotavirus strain, identified in a stool sample from Guatemala (GTM) during routine rotavirus surveillance. This strain was designated as RV A/Human-wt/GTM/2009726790/2009/G8P[14], with a genomic constellation of G8-P[14]-I2-R2-C2-M2-A13-N2-T6-E2-H3. The VP4 gene occupied lineage VII within the P[14] genotype. Phylogenetic analysis of each genome segment revealed close relatedness to several zoonotic simian, guanaco and bovine strains. Our findings suggest that strain RVA/Human-wt/GTM/2009726790/2009/G8P[14] is an example of a direct zoonotic transmission event. The results of this study reinforce the potential role of interspecies transmission and reassortment in generating novel and rare rotavirus strains which infect humans.
Keywords: Rotavirus, G8P[14], Reassortant, Interspecies transmission
Group A rotavirus (RVA), a member of the Reoviridae family, is the major etiologic agent of severe, acute dehydrating diarrhea in young children and a wide variety of domestic animals (Estes and Kapikian, 2007). The RVA genome is composed of 11 double stranded RNA segments encoding six structural (VP1–VP4, VP6 and VP7) and five or six non-structural proteins (NSP1–NSP5/6). The segmented nature of the RVA genome permits reassortment events in which novel RVA strains are produced with new combinations of genome segments derived from parental virus strains (Estes and Kapikian, 2007). New strains regularly emerge as a result of genomic reassortment among co-circulating RVAs. Animal RVAs are regarded as a potential reservoir for genetic exchange with human RVAs and can infect humans, either by direct transmission of the virus or by contributing genes to reassortants (Muller and Johne, 2007). Reassortment and interspecies transmission events contribute significantly to RVA genetic diversity (Matthijnssens et al., 2011). A number of strains with unusual G and P types, regarded as animal-like strains, have been sporadically identified in humans in different parts of world (Gentsch et al., 2005; Santos and Hoshino, 2005).
Recently, more P[14] strains have been associated with gastroenteritis in humans and most of the reported P[14] strains have the Genogroup 2 gene constellation (Banyai et al., 2009; Banyai et al., 2010; Cowley et al., 2013; Donato et al., 2014; El Sherif et al., 2011; Ghosh et al., 2007; Matthijnssens et al., 2009; Medici et al., 2008; Mullick et al., 2013). Human P[14] strains have common origins with those of the even-toed ungulates belonging to the mammalian order Artiodactyla (Matthijnssens et al., 2009). Within P[14] strain genes, 7 lineages have been identified with human strains identified in all of them (Tam et al., 2014). Lineages I, III, IV and VII contain G6, G8 and G10 strains from bovines, guanacos, sheep and other ungulates. Animal RVA G8P[14] strains have been identified in Japan and India (bovine), Spain (ovine), and Argentina (guanaco) (Matthijnssens et al., 2009; Chitambar et al., 2011; Fukai et al., 2004; Ciarlet et al., 2008). RVA G8P[14] strains have been identified in human infections from Egypt, Italy, Belgium, Australia, Hungary, India, Denmark and Taiwan (Holmes et al., 1999; Medici et al., 2008; Matthijnssens et al., 2009; Chitambar et al., 2011; Swiatek et al., 2010; Banyai et al., 2010; Chitambar et al., 2011; Midgley et al., 2014; Midgley et al., 2012; Wu et al., 2012). The VP8* region of the P[14] VP4 protein has been shown to interact with type A histo blood group antigens of humans (Hu et al., 2012; Liu et al., 2012) as well as bovine and porcine mucins (Liu et al., 2012) and this is thought to play a role in cross-species transmission of P[14] RVAs.
RVA G8P[14] strains have not been reported in Latin America previously, thus it was of interest to us to characterize a G8P[14] rotavirus strain detected in Guatemala. Here we report the full genome sequence and phylogenetic analysis of RVA strain RVA/H uman-wt/GTM/2009726790/2009/G8P[14], detected in a stool sample from Guatemala.
The stool sample was collected in 2008 from Guatemala, Santa Rosa as part of the facility based Rotavirus Strain Surveillance system in collaboration with the International Emerging Infections Program (IEIP), the Ministry of Public Health and Welfare and the Universidad del Valle de Guatemala. The sample was submitted to the Centers for Disease Control and Prevention (CDC) for rotavirus strain genotyping. The sample was from a 52 year old female with diarrhea for 2 days (5 episodes/day). RNA was extracted from the sample using the MagMax 96 Viral RNA Isolation kit (Applied Biosystems, Inc., Foster City, CA) on an automated KingFisher extraction system (Thermo Scientific, Waltham MA), or the MagNA pure compact RNA extraction kit on the MagNA pure compact instrument (Roche Applied Science, Indianapolis, IN) following manufacturer’s instructions. The extracted RNA was denatured at 95 °C for 5 min and RT-PCR was performed using the Qiagen OneStep RT-PCR kit (Qiagen, Inc., Valencia, CA) as described previously (Hull et al., 2011). Each rotavirus gene segment was amplified using previously published RVA specific consensus primers (Gouvea et al., 1990; Gentsch et al., 1992; Gomara et al., 2000). Additional primers were designed using published consensus sequences for genes which failed to generate a complete open reading frame (ORF). Oligonucleotides were designed in the consensus region of 5′ and 3′ ends with M13 and SP6 tails, respectively, for all eleven RVA genes to obtain the end sequences of the ORFs. (Mijatovic et al., manuscript in preparation). PCR reactions were analyzed by electrophoresis on 1% agarose gels as described previously (Hull et al., 2011). Specific PCR amplicons were excised from agarose gels and purified using the QIAquick Gel extraction kit (Qiagen, Inc., Valencia, CA) according to the manufacturer’s protocol. Cycle sequencing of each amplicon was performed with the same consensus primers used for RT-PCR, or M13 and SP6 tail sequences using Big Dye Terminator 3.1 Cycle Sequencing kit (Life technologies, Inc. Foster City, CA). Cycle sequencing products were purified using BioMag® carboxyl beads (Mijatovic-Rustempasic et al., 2012). Sequencing of cycle sequenced products was performed on an ABI Prism 3130XL Genetic Analyzer using 3130 POP-7 (Life technologies, Foster City, CA).
Sequence chromatogram files were edited and sequence contigs were assembled using Sequencher 5.0 software (Gene Codes Corporation, Inc., Ann Arbor, MI). Nucleotide similarity searches were performed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) to query the GenBank sequence database. The genotypes of each of the 11 genome segments were determined using the RotaC online classification tool (http://rotaCregatools.be/) (Maes et al., 2009). For phylogenetic analysis, nucleotide sequences of related strains for each gene were retrieved from GenBank and aligned using the MUSCLE program within MEGA version 5 software (http://www.megasoftware.net/). Once aligned, the JModelTest 2 program (Posada, 2008) was used to identify the optimal evolutionary model that best fitted the sequence datasets. Using optimal models identified by the corrected Akaike Information Criterion (AICc), maximum likelihood trees were constructed using PhyML 3.0 with aLRT statistics computed for branch support (Guindon et al., 2010). Nucleotide distance matrices were prepared using the p-distance algorithm of MEGA version 5 software.
The complete ORFs for all eleven genes of RVA/Human-wt/GT M/2009726790/2009/G8P[14] strain with the exception of 39 bases at the 5′ end of the VP7 ORF and 29 bases at the 3′ end of the VP4 ORF were sequenced and deposited into GenBank under accession numbers KP006506 to KP006516. Nucleotide identity of strain RVA/Human-wt/GTM/2009726790/2009/G8P[14] compared to strains available in the GenBank indicated that all eleven genes were closely related (95–98.1%) to animal strains (Table 1) including simian (NSP2), guanaco (NSP5 and VP7) and bovine (NSP1, NSP3 and NSP4, VP1, VP2, VP3, VP4 and VP6, genes).
Table 1.
Nucleotide sequence identities of RVA/Human-wt/GTM/2009726790/2009/G8P[14] strain to closely related strains.
| Gene | % identity to closest related strain | Closely related strain | Accession number | RotaC classification | Predominant host strain in phylogenetic lineage | % identity to RotaTeq vaccine strain |
|---|---|---|---|---|---|---|
| NSP1 | 95.0 | Bovine Dai-10 | AB573074 | A13 | * Mixed | 74.4 |
| NSP2 | 97.6 | Simian-RRV G3P3 | EU636931 | N2 | * Mixed | 95.7 |
| NSP3 | 98.1 | Bovine-BRV-106 | AB748599 | T6 | Bovine | 97.4 |
| NSP4 | 97.9 | Bovine-CBNU-1 | AF166353 | E2 | * Mixed | 92.9 |
| NSP5 | 96.1 | Guanaco/Arg/Chubut/1999 G8P[14] strain | FJ347110 | H3 | * Mixed | 94.8 |
| VP1 | 96.3 | Bovine-WC3 | GU565041 | R2 | * Mixed | 96.4 |
| VP2 | 96.9 | Bovine-Sun9 | AB374144 | C2 | * Bovine | 95.1 |
| VP3 | 95.0 | Bovine-CP-1 | FJ560906 | M2 | * Bovine | 92.2 |
| VP4 | 97.8 | Bovine-Tottori-SG-G15P[14] | AB853893 | P[14] | * Bovine | 69.0 |
| VP6 | 97.8 | Bovine-MVS-BRV4 | KC215501 | I2 | * Mixed | 97.1 |
| VP7 | 96.6 | Guanaco/Arg/Chubut/1999 G8P[14] strain | FJ347105 | G8 | * Mixed | 72.3 |
More than one predominant host strain represented in the lineage.
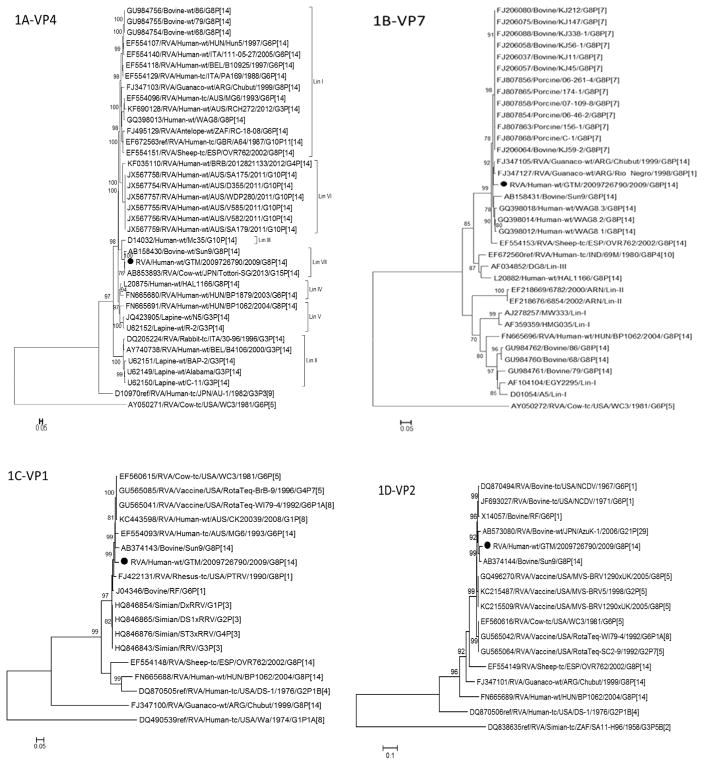
The VP4 gene of RVA/Human-wt/GTM/2009726790/2009/G8 P[14] strain was 97.8% identical to a bovine G15P[14] strain (Tottori) (Masuda et al., 2014), 86–96.1% identical to bovine G8P[14] strains and 83.7–87.5% identical to human G8P[14] strains. Strain RVA/Human-wt/GTM/2009726790/2009/G8P[14] VP4 gene occupies lineage VII shared with Tottori-G15P[14] and Sun9-G8P[14] strains from Japan (Fig. 1A). The VP7 gene of RVA/Human-wt/GTM/2009726790/2009/G8P[14] strain was closely related (93.6–96.5%) to bovine G8P[14] strain and 84.1–95.8% identical to VP7 genes of human G8P[14] strains (Fig. 1B).
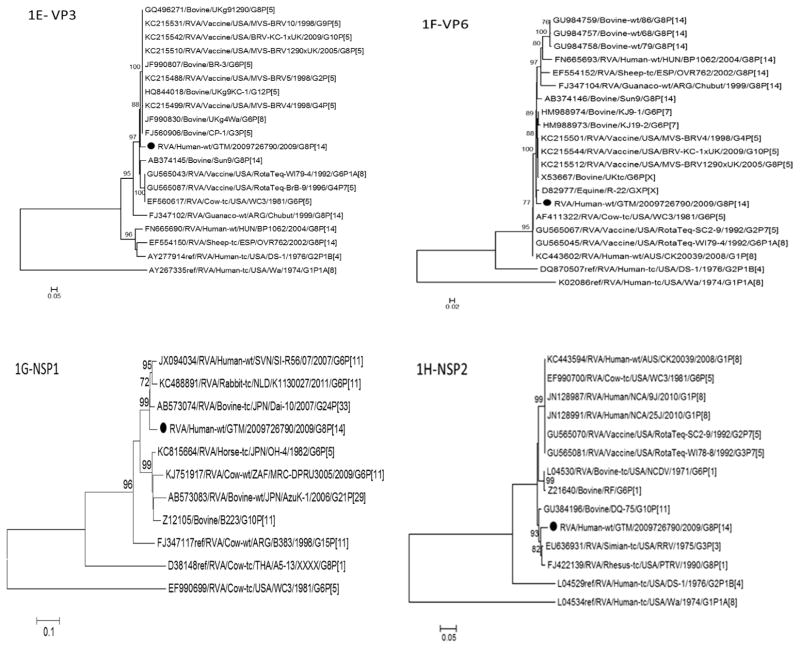
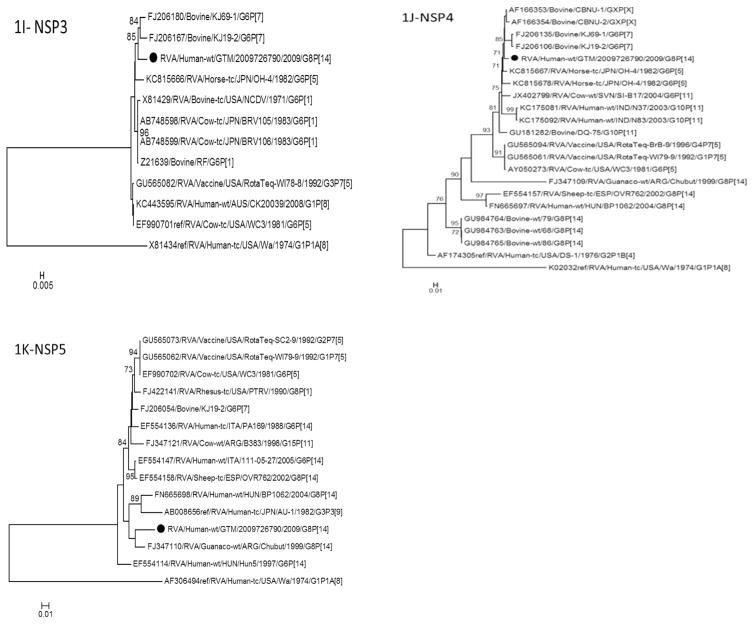
Fig. 1.
Phylogenetic trees based on nucleotide sequences of complete open reading frames of 1A-VP4, 1B-VP7, 1C-VP1, 1D-VP2, 1E-VP3, 1F-VP6, 1G-NSP1, 1H-NSP2, 1I-NSP3, 1J-NSP4 and 1K-NSP5 rotavirus genes. The maximum likelihood trees were constructed using PhyML 3.0 with best model identified by J ModelTest2 program for each gene (NSP1-GTR + 1 + G; NSP2-TIM2 + I; NSP3-TIM2 + I; NSP4-TPM2uf + G; NSP5-HKY + G; VP1-TIM2 + I; VP2-GTR + I; VP3-TIM1 + I; VP4-GTR + G; VP6-TrN + G; VP7-TVM + G). The GenBank accession numbers, strain names and G and P-type associations are shown where available.
Full genome characterization of two animal G8P[14] strains, Arg/Chubut with genomic constellation G8-P[14]-I2-R5-C2-M2-A11-N2-T6-E12-H3 and OVR 762 with G8-P[14]-I2-R2-C2-M2-A11-N2-T6-E2-H3 have been reported (Matthijnssens et al., 2009). Eight genotypes (N2, T6, H3, C2, M2, P[14], I2 and G8) of the RVA/Human-wt/GTM/2009726790/2009/G8P[14] strain were the same as those in both the Arg/Chubut and OVR 762 strains. Eight out of eleven genes (NSP2, NSP3, NSP4, NSP5, VP1, VP2, VP3 and VP6) of RVA/Human-wt/GTM/2009726790/2009/G8P[14] strain also exhibited close identity (92.2–97.4%) with the WC3-derived genes of the RotaTeq® vaccine strain (Table 1).
Phylogenetic analysis of all eleven genes of RVA/Human-wt/G TM/2009726790/2009/G8P[14] showed that the VP7, VP1, VP6, NSP1, NSP2, NSP4 and NSP5 (Fig. 1B, C, F–H, J, and K, respectively) genes had more than one predominant host strain (human, bovine, simian, porcine, rhesus or guanaco) in their respective lineages. Bovines were the predominant hosts for strains in the phylogenetic lineages for the VP4, VP2, VP3 and NSP3 (Fig. 1A, D, E and I, respectively) genes.
Full genome classification of RVA/Human-wt/GT M/2009726790/2009/G8P[14] strain was G8-P[14]-I2-R2-C2-M2-A13-N2-T6-E2-H3 (Table 1). Full genome characterization of only one human G8P[14] strain has been reported in the literature to date (Banyai et al., 2010). The RVA/Human-wt/GT M/2009726790/2009/G8P[14] strain was 83.3–94.6% identical to all the genes except NSP1 gene with only 71.9% similarity to the published Hungarian G8P[14] strain which has a genomic constellation of G8-P[14]-I2-R2-C2-M2-A11-N2-T6-E2-H3. The NSP1 gene of RVA/Human-wt/GTM/2009726790/2009/G8P[14] was 94.9% identical to a Japanese bovine G24P[33] strain Dai-10 (Fig. 1G) exhibiting the A13 genotype for NSP1 gene (Abe et al., 2011). Predominantly, the A13 genotype for the NSP1 gene is found in bovine strains with an exception of a G6P[11] human strain (SI-R56) from a Slovenian patient (Steyer et al., 2013).
This report describes the characterization of RVA/Human-wt/G TM/2009726790/2009/G8P[14] strain isolated in Guatemala. With multiple hosts identified for G8P[14] strains in animals, including bovine, guanocos and sheep (Ciarlet et al., 2008; Fukai et al., 2004; Ghosh et al., 2007; Parreno et al., 2004), elucidating the original source of G8P[14] strains is complex and likely involves multiple animal-animal transmission and reassortment events. The genomic constellation of RVA/Human-wt/GT M/2009726790/2009/G8P[14] strain was closely related to two animal G8P[14] strains, suggesting one or more reassortment events followed by interspecies transmission of the G8P[14] strain from an animal to a human.
In conclusion, this is the first report of a G8P[14] RVA strain detected in Guatemala, Latin America. Also this is the first report of a G8P[14] strain with a genomic constellation of G8-P[14]-I2-R2-C2-M2-A13-N2-T6-E2-H3 and this strain occupied lineage VII within the P[14] genotype. Close similarity of eight genes of RVA/ Human-wt/GTM/2009726790/2009/G8P[14] strain with RotaTeq® vaccine strain genes suggests that RotaTeq® vaccine would be highly effective against this genotype. Future RVA surveillance is required to monitor for unusual animal-like RVA strains and to determine whether vaccine-induced immunity would provide protection against interspecies transmission of strains.
Acknowledgments
We would like to thank Ms. Leanne Ward for her critical review of the manuscript.
Abbreviations
- RVA
Group A rotaviruses
- WHO
World Health Organization
- ORF
open reading frame
- GTM
Guatemala
Footnotes
Institution where work was completed: National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. Names of specific vendors, manufacturers, or products are included for public health and informational purposes; inclusion does not imply endorsement of the vendors, manufacturers, or products by the Centers for Disease Control and Prevention or the US Department of Health and Human Services.
References
- Abe M, Ito N, Masatani T, Nakagawa K, Yamaoka S, Kanamaru Y, Suzuki H, Shibano K, Arashi Y, Sugiyama M. Whole genome characterization of new bovine rotavirus G21P[29] and G24P[33] strains provides evidence for interspecies transmission. J Gen Virol. 2011;92:952–960. doi: 10.1099/vir.0.028175-0. [DOI] [PubMed] [Google Scholar]
- Banyai K, Martella V, Molnar P, Mihaly I, Van Ranst M, Matthijnssens J. Genetic heterogeneity in human G6P[14] rotavirus strains detected in Hungary suggests independent zoonotic origin. J Infect. 2009;59:213–215. doi: 10.1016/j.jinf.2009.06.009. [DOI] [PubMed] [Google Scholar]
- Banyai K, Papp H, Dandar E, Molnar P, Mihaly I, Van Ranst M, Martella V, Matthijnssens J. Whole genome sequencing and phylogenetic analysis of a zoonotic human G8P[14] rotavirus strain. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2010;10:1140–1144. doi: 10.1016/j.meegid.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Chitambar SD, Arora R, Kolpe AB, Yadav MM, Raut CG. Molecular characterization of unusual bovine group A rotavirus G8P[14] strains identified in western India: emergence of P[14] genotype. Vet Microbiol. 2011;148:384–388. doi: 10.1016/j.vetmic.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Ciarlet M, Hoffmann C, Lorusso E, Baselga R, Cafiero MA, Banyai K, Matthijnssens J, Parreno V, de Grazia S, Buonavoglia C, Martella V. Genomic characterization of a novel group A lamb rotavirus isolated in Zaragoza, Spain. Virus Genes. 2008;37:250–265. doi: 10.1007/s11262-008-0257-6. [DOI] [PubMed] [Google Scholar]
- Cowley D, Donato CM, Roczo-Farkas S, Kirkwood CD. Novel G10P[14] rotavirus strain, northern territory, Australia. Emerg Infect Dis. 2013;19:1324–1327. doi: 10.3201/eid.1908.121653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato CM, Manuelpillai NM, Cowley D, Roczo-Farkas S, Buttery JP, Crawford NW, Kirkwood CD. Genetic characterization of a novel G3P[14] rotavirus strain causing gastroenteritis in 12 year old Australian child. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2014;25:97–109. doi: 10.1016/j.meegid.2014.04.009. [DOI] [PubMed] [Google Scholar]
- El Sherif M, Esona MD, Wang Y, Gentsch JR, Jiang B, Glass RI, Abou Baker S, Klena JD. Detection of the first G6P[14] human rotavirus strain from a child with diarrhea in Egypt. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2011;11:1436–1442. doi: 10.1016/j.meegid.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Estes MK, Kapikian A. Rotaviruses. In: Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Fields Virology. 5. Kluwer/Lippincott, Williams and Wilkins; Philadelphia, PA: 2007. pp. 1917–1974. [Google Scholar]
- Fukai K, Onoda H, Itou T, Sato M, Miura Y, Sakai T. Genetic and serological characterization of novel serotype G8 bovine group A rotavirus strains isolated in Japan. J Vet Med Sci/Jpn Soc Vet Sci. 2004;66:1413–1416. doi: 10.1292/jvms.66.1413. [DOI] [PubMed] [Google Scholar]
- Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch JR, Laird AR, Bielfelt B, Griffin DD, Banyai K, Ramachandran M, Jain V, Cunliffe NA, Nakagomi O, Kirkwood CD, Fischer TK, Parashar UD, Bresee JS, Jiang B, Glass RI. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J Infect Dis. 2005;192(Suppl 1):S146–159. doi: 10.1086/431499. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Varghese V, Samajdar S, Sinha M, Naik TN, Kobayashi N. Evidence for bovine origin of VP4 and VP7 genes of human group A rotavirus G6P[14] and G10P[14] strains. J Clin Microbiol. 2007;45:2751–2753. doi: 10.1128/JCM.00230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomara MI, Green J, Gray J. Methods of rotavirus detection, sero- and genotyping, sequencing, and phylogenetic analysis. Methods Mol Med. 2000;34:189–216. doi: 10.1385/1-59259-078-0:189. [DOI] [PubMed] [Google Scholar]
- Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Holmes JL, Kirkwood CD, Gerna G, Clemens JD, Rao MR, Naficy AB, Abu-Elyazeed R, Savarino SJ, Glass RI, Gentsch JR. Characterization of unusual G8 rotavirus strains isolated from Egyptian children. Arch Virol. 1999;144:1381–1396. doi: 10.1007/s007050050594. [DOI] [PubMed] [Google Scholar]
- Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull JJ, Teel EN, Kerin TK, Freeman MM, Esona MD, Gentsch JR, Cortese MM, Parashar UD, Glass RI, Bowen MD National Rotavirus Strain Surveillance System. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. Pediatr Infect Dis J. 2011;30:S42–47. doi: 10.1097/INF.0b013e3181fefd78. [DOI] [PubMed] [Google Scholar]
- Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. Rotavirus VP8*: phylogeny, host range, and interaction with histoblood group antigens. J Virol. 2012;86:9899–9910. doi: 10.1128/JVI.00979-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes P, Matthijnssens J, Rahman M, Van Ranst M. RotaC: a web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009;9:238. doi: 10.1186/1471-2180-9-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda T, Nagai M, Yamasato H, Tsuchiaka S, Okazaki S, Katayama Y, Oba M, Nishiura N, Sassa Y, Omatsu T, Furuya T, Koyama S, Shirai J, Taniguchi K, Fujii Y, Todaka R, Katayama K, Mizutani T. Identification of novel bovine group A rotavirus G15P[14] strain from epizootic diarrhea of adult cows by de novo sequencing using a next-generation sequencer. Vet Microbiol. 2014;171:66–73. doi: 10.1016/j.vetmic.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthijnssens J, De Grazia S, Piessens J, Heylen E, Zeller M, Giammanco GM, Banyai K, Buonavoglia C, Ciarlet M, Martella V, Van Ranst M. Multiple reassortment and interspecies transmission events contribute to the diversity of feline, canine and feline/canine-like human group A rotavirus strains. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2011;11:1396–1406. doi: 10.1016/j.meegid.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Matthijnssens J, Potgieter CA, Ciarlet M, Parreno V, Martella V, Banyai K, Garaicoechea L, Palombo EA, Novo L, Zeller M, Arista S, Gerna G, Rahman M, Van Ranst M. Are human P[14] rotavirus strains the result of interspecies transmissions from sheep or other ungulates that belong to the mammalian order Artiodactyla? J Virol. 2009;83:2917–2929. doi: 10.1128/JVI.02246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici MC, Abelli LA, Martinelli M, Dettori G, Chezzi C. Molecular characterization of VP4, VP6 and VP7 genes of a rare G8P[14] rotavirus strain detected in an infant with gastroenteritis in Italy. Virus Res. 2008;137:163–167. doi: 10.1016/j.virusres.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Midgley S, Bottiger B, Jensen TG, Friis-Moller A, Person LK, Nielsen L, Barzinci S, Fischer TK. Human group A rotavirus infections in children in Denmark: detection of reassortant G9 strains and zoonotic P[14] strains. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2014;27:114–120. doi: 10.1016/j.meegid.2014.07.008. [DOI] [PubMed] [Google Scholar]
- Midgley SE, Hjulsager CK, Larsen LE, Falkenhorst G, Bottiger B. Suspected zoonotic transmission of rotavirus group A in Danish adults. Epidemiol Infect. 2012;140:1013–1017. doi: 10.1017/S0950268811001981. [DOI] [PubMed] [Google Scholar]
- Mijatovic-Rustempasic S, Frace MA, Bowen MD. Cost-effective paramagnetic bead technique for purification of cycle sequencing products. Sequencing. 2012;2012:4. [Google Scholar]
- Muller H, Johne R. Rotaviruses: diversity and zoonotic potential – a brief review. Berl Munch Tierarztl Wochenschr. 2007;120:108–112. [PubMed] [Google Scholar]
- Mullick S, Mukherjee A, Ghosh S, Pazhani GP, Sur D, Manna B, Nataro JP, Levine MM, Ramamurthy T, Chawla-Sarkar M. Genomic analysis of human rotavirus strains G6P[14] and G11P[25] isolated from Kolkata in 2009 reveals interspecies transmission and complex reassortment events. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2013;14:15–21. doi: 10.1016/j.meegid.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Parreno V, Bok K, Fernandez F, Gomez J. Molecular characterization of the first isolation of rotavirus in guanacos (Lama guanicoe) Arch Virol. 2004;149:2465–2471. doi: 10.1007/s00705-004-0371-2. [DOI] [PubMed] [Google Scholar]
- Posada D. JModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol. 2005;15:29–56. doi: 10.1002/rmv.448. [DOI] [PubMed] [Google Scholar]
- Steyer A, Sagadin M, Kolenc M, Poljsak-Prijatelj M. Whole genome sequence analysis of bovine G6P[11] rotavirus strain found in a child with gastroenteritis. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2013;13:89–95. doi: 10.1016/j.meegid.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Swiatek DL, Palombo EA, Lee A, Coventry MJ, Britz ML, Kirkwood CD. Detection and analysis of bovine rotavirus strains circulating in Australian calves during 2004 and 2005. Vet Microbiol. 2010;140:56–62. doi: 10.1016/j.vetmic.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Tam KI, Roy S, Esona MD, Jones S, Sobers S, Morris-Glasgow V, Rey-Benito G, Gentsch JR, Bowen MD. Full genomic characterization of a novel genotype combination, G4P[14], of a human rotavirus strain from barbados. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2014 doi: 10.1016/j.meegid.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FT, Banyai K, Wu HS, Yang DC, Lin JS, Hsiung CA, Huang YC, Hwang KP, Jiang B, Gentsch JR. Identification of a G8P[14] rotavirus isolate obtained from a Taiwanese child: evidence for a relationship with bovine rotaviruses. Jpn J Infect Dis. 2012;65:455–457. [PMC free article] [PubMed] [Google Scholar]