Abstract
The Warburg effect is a metabolic phenomenon characterized by increased glycolytic activity, decreased mitochondrial oxidative phosphorylation, and the production of lactate. This metabolic phenotype is characterized in rapidly proliferative cell types such as cancerous cells and embryonic stem cells. We hypothesized that a Warburg-like metabolism could be achieved in other cell types by treatment with pharmacological agents, which might, in turn, facilitate nuclear reprogramming. The aim of this study was to treat fibroblasts with CPI-613 and PS48 to induce a Warburg-like metabolic state. We demonstrate that treatment with both drugs altered the expression of 69 genes and changed the level of 21 metabolites in conditioned culture media, but did not induce higher proliferation compared to the control treatment. These results support a role for the reverse Warburg effect, whereby cancer cells induce cancer-associated fibroblast cells in the surrounding stroma to exhibit the metabolically characterized Warburg effect. Cancer-associated fibroblasts then produce and secrete metabolites such as pyruvate to supply the cancerous cells, thereby supporting tumor growth and metastasis. While anticipating an increase in the production of lactate and increased cellular proliferation, both hallmarks of the Warburg effect, we instead observed increased secretion of pyruvate without changes in proliferation.
Keywords: : metabolism, Warburg, glycolysis, tricarboxylic acid cycle, cell biology
Introduction
A hallmark of the “Warburg effect,” characterized by its namesake, Otto Warburg, is the predominate use of glycolysis for energy production as opposed to the use of pyruvate metabolism and mitochondrial oxidative phosphorylation typically employed in differentiated somatic cells. Warburg speculated that cancer was linked to the metabolic switch from respiration of oxygen to the fermentation of sugars for energy in normal body cells due to mitochondrial impairment (Koppenol et al., 2011; Warburg, 1956). Vander Heiden et al. (2009) illustrated the use of this seemingly atypical metabolism is attributed to be favorable for the production of biomass and reduction of reactive oxygen species (ROS). Indeed, this metabolic phenotype may likely be more resultant of mutations in tumor suppressors, oncogenes, and mitochondrial DNA that is causative of cancer [reviewed in Koppenol et al. (2011)]. Moreover, investigations finding that mitochondrial oxidative phosphorylation is active in cancer cells have stimulated an emerging hypothesis of the so-called “reverse Warburg effect” (Zheng, 2012).
In the reverse Warburg effect, epithelial cancer cells induce the Warburg effect in nearby stromal fibroblasts, thereby coercing them to produce pyruvate and lactate to support tumor growth and metastasis. We hypothesized that it may be possible to pharmacologically induce a Warburg-like effect metabolism in fibroblasts by inhibiting mitochondrial oxidative phosphorylation and promoting glycolytic activity by using pharmacological agents. In addition, we inquired whether programming of metabolism could lead to reprogramming of the nucleus, driving changes in gene expression that might be similar to Warburg effect-like cell types, and lead to increased cellular proliferation.
There is a correlation between increased glycolytic metabolism and pluripotency. Cytosolic acetyl-CoA produced by glycolysis promoted histone acetylation during pluripotency in stem cells and the inhibition of glycolysis caused deacetylation and differentiation of pluripotent cells (Moussaieff et al., 2015). Furthermore, before cells achieve pluripotency, there is upregulation of glycolysis early in induced pluripotent stem cell (IPSC) reprogramming (Folmes et al., 2011; Shyh-Chang and Daley, 2015). Increased signaling of the PI3K pathway is correlated with an increase in glucose metabolism within cancer cells and is consistent with the Warburg effect. The allosteric small molecule PS48 (5-(4-Chloro-phenyl)-3-phenyl-pent-2-enoic acid) activates phosphoinositide-dependent protein kinase 1 (Hindie et al., 2009; Stroba et al., 2009; Zorn and Wells, 2010).
Our goal was to utilize PS48 to enhance glycolysis and decrease mitochondrial oxidation. PS48 has been shown to enhance reprogramming efficiency of IPSCs by about 15-fold and is evidenced to promote glycolysis and lactate production (Zhu et al., 2010).
A second candidate to alter metabolism of donor cells to be more similar to that of an early embryo is a lipoate analog known as CPI-613 (6, 8-bis(benzylthio)octanoic acid: hereafter called CPI). CPI is a mitochondrial disrupter by inhibition of the mitochondrial enzymes pyruvate dehydrogenase (PDH) and α-ketoglutarate dehydrogenase (Stuart et al., 2014; Zachar et al., 2011). Inhibiting precursors for entry into the tricarboxylic acid (TCA) cycle should decrease use of the TCA cycle activity and promote the glycolytic pathway to meet metabolic requirements.
In this study, we investigated effects of PS48 and CPI dosages on cell viability, longevity, and cell cycle length. Conditioned media metabolites and gene expression were analyzed to determine if metabolic pathways consistent with the Warburg effect were induced in fibroblasts.
Materials and Methods
All materials were purchased from Sigma-Aldrich, St. Louis, MO, unless otherwise specified.
Compliance with ethical standards
This article does not contain any study with human participants performed by any of the authors. All procedures performed in studies involving animals were in accordance with the ethical standards of the University of Missouri Institutional Animal Care and Use Committee at the University of Missouri in Columbia, MO.
Fetal-derived fibroblast cell culture and trypan blue exclusion
A gilt was artificially inseminated and humanely euthanized by ear vein infusion of Euthasol at 35 days postinsemination. The reproductive tract was excised from the cervix to ovaries and opened along the entire length of uterine horns for fetal collection. Tissue was excised from the posterior region of a single eviscerated fetus and washed in 1 × phosphate-buffered saline (PBS). Genotyping was used to confirm this fetus was female. Cells were liberated by brief homogenization and tissue pieces were then placed in digestion media (Dulbecco's modified Eagle's medium [DMEM] containing 1 g/L and d-glucose and l-glutamine; Corning, Corning, NY) with 200 units/mL collagenase and 25 Kunitz units/mL DNaseI for 3 hours. Fibroblast cells were pelleted, then replated in DMEM (15% fetal bovine serum [FBS]; Corning, Manassas, VA), and cultured to confluence.
Afterward, they were dissociated, pelleted, resuspended, and diluted to 1.5 million/mL in media containing 85% FBS and 15% DMSO, and then frozen in liquid nitrogen (0.5 mL aliquots; ∼80 cryogenic vials). A separate vial of fibroblasts was thawed for each replicate of each experiment; the same cell line was used throughout all experiments. During the experiments, incubators were maintained at 38.5°C with a humidified atmosphere of 5% oxygen, 5% carbon dioxide, and 90% nitrogen. In both experiments, all cells were thawed and cultured in DMEM (1 g/L glucose, glutamine, and pyruvate with phenol red; supplemented with 15% FBS [Corning, Manassas, VA]) for 7 days in T25 flasks (Corning, Corning, NY) with or without the addition of respective treatment concentrations of CPI-613 or PS48 (Stemgent, Cambridge, MA).
Media were changed daily in all flasks, that is, those which received PS48 or CPI had new drugs applied daily (24 ± 2 hours). In a separate experiment, cells were grown in T75 flasks and drugs were spiked in during the week of treatment (no media change), and cell growth was measured by trypan blue staining (1:1 ratio of cell suspension and stain volumes) by using an automated cell counter.
In a preliminary experiment, porcine fetal fibroblasts were treated with CPI (25, 50, or 100 μM), PS48 (1, 5, or 10 μM), or as controls (0 μM) for 7 days of culture to determine which doses of CPI and PS48 to utilize. In the main study, porcine fetal fibroblasts were treated with PS48 (10 μM), CPI (100 μM), a mixture of PS48 and CPI (10 and 100 μM), or treated as controls (0 μM) for 7 days of culture. Stock aliquots of CPI were diluted to 100 mM and PS48 stocks were diluted to 10 mM to eliminate potential confounding of DMSO quantity between the two pharmaceutical treatments (CPI and PS48). Cells were initially plated at 1 × 105 cells/mL in T25 flasks. After 72 and 120 hours, cells were passaged to new T25 flasks. At 120 hours, cells in all treatments were plated (density: 5 × 105 cells) to achieve ∼80% confluence at 168 hours.
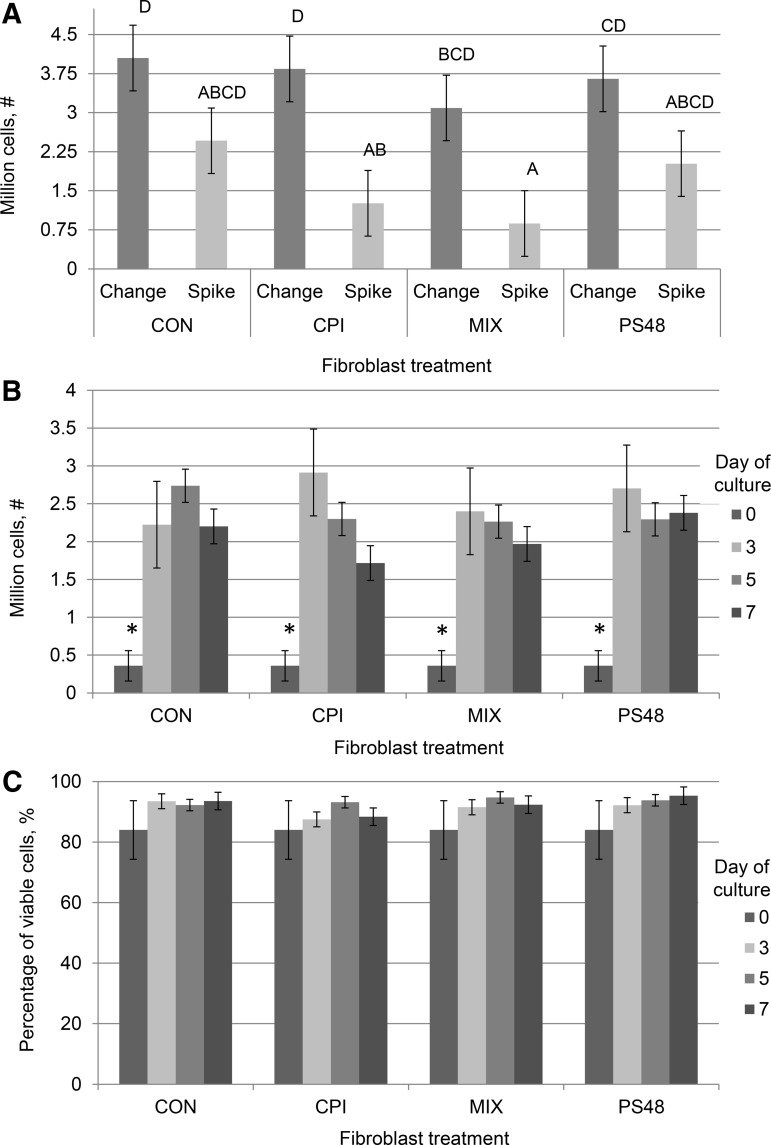
At initial thawing and subsequent passages, cell proliferation and percentage of live cells were measured by trypan blue exclusion staining by an automated cell counter (TC20 automated cell counter; Bio-Rad). At passage, cells were briefly rinsed with PBS 0.01 M EDTA and dissociated by brief incubation (37°C) from flasks using 1 × TrypLE Express (Gibco, Denmark). Cells were pelleted (5 minutes at 500 g) and suspended in 5 mL DMEM containing 15% FBS; for every treatment, 200 μL of cell suspension was mixed in a 1:1 ratio with 1 × trypan blue stain and 10 μL of the mixture was quantified in counting chamber slides. As per the vertical axes in Figure 1A and B, samples measured for viability contained at least 250,000 cells. After trypan measures were taken, treatments were diluted as needed to achieve an even cell number across treatments and replated with respective treatment media.
FIG. 1.
Growth and viability of cells cultured for 0, 3, 5, and 7 days with the pharmaceutical agents CPI (100 μM), PS48 (10 μM), the mixture of the two (MIX), or without drugs (CON; 0 μM); error bars reflect standard error. (A) Cell proliferation numbers from cells grown for 5 days with pharmaceutical agents either spiked in daily without media change (Spike) or when drugs were added in fresh culture media daily (Change). Means with different letters are significantly different (p < 0.05). (B) Cell proliferation numbers from cells cultured for 0, 3, 5, and 7 days with pharmaceutical agents. *Significant at p < 0.01. (C) Percentage of viable cells from cells cultured for 0, 3, 5, and 7 days with pharmaceutical agents.
Life span was determined for PS48-treated cells; fibroblasts treated daily with 0, 5, or 10 μM PS48 were continuously passaged after reaching ∼80% confluence and considered dead at the point where after 24 hours of passaging the cells did not replate and grow. Normality of data was assessed by Univariate procedure in SAS 9.4 (SAS, Cary, NC) using the option for generation of normality test p-values, which included Shapiro–Wilk, Kolmogorov–Smirnov, Anderson–Darling, and Cramér–von Mises tests. Log transformations were made, where appropriate, before statistical analysis. All trypan blue exclusion proliferation data were analyzed for effects of treatment, day, and the interaction of treatment by day using the generalized linear model procedure of SAS 9.4. For ease of interpretation, all figures depict SAS generated least squared means and standard error values, which are not transformed.
Flow cytometry: cell preparation and staining
On day 7 (168 hours), media were removed, flasks were briefly rinsed with PBS 0.01 M EDTA, and fibroblast cells were dissociated from flasks by brief incubation (37°C) with 1 × TrypLE Express (Gibco) and pelleted (5 minutes at 500 g). Fibroblasts used for live cell mitochondrial staining were immediately suspended in 1 × DMEM without phenol red (Corning, Corning, NY) and appropriate drug concentrations were added for each treatment. For viability staining, fibroblasts were suspended in 1 × binding buffer, then stained, and incubated with annexin-V-FITC and propidium iodide (PI) (50 μg/mL, 37°C, 5 minutes; BioVision, Milpitas, CA). In viability cytometry acquisitions, fibroblast cells received their respective treatments during staining and pharmaceutical agents were present in media during data collection.
For cell cycle analysis, pelleted cells were suspended in ice-cold 1 × PBS and 2.5 mL of cold (4°C) 100% ethanol was slowly dripped into the PBS suspension while under constant slow vortex. After fixation, fibroblast cells were rested on ice for 30 minutes. Fibroblast cells were subsequently pelleted and incubated in a 500 μL PBS mixture with 50 μg/mL of PI, 0.05% Triton X-100, and 0.1 mg/mL RNase A for 45 minutes. After staining cells, 3 mL of 1 × PBS was added, cells were pelleted (500 g for 5 minutes), and 3 mL of media was removed and pellet resuspended leaving PI in excess during the cytometry run.
Flow cytometry: data acquisition and analysis
For cytometry data collected, gating protocols were applied for analysis of a population free of debris and doublet cells by using plots of side scatter height × forward scatter height, forward scatter height × forward scatter area, and forward scatter height × forward scatter width. In all flow cytometry experiments, three biological replicates were collected for analysis; in each, we used three technical samples with at least 50,000 cells in the “single cell” gate using the protocol described above for each treatment. Data were assessed for normality by using the Univariate procedure in SAS 9.4 (SAS), by using the option for generation of normality test p-values, which included Shapiro–Wilk, Kolmogorov–Smirnov, Anderson–Darling, and Cramér–von Mises tests.
Log and square root transformations were made, where appropriate, before statistical analysis. Data were analyzed by using the generalized linear model procedure of SAS 9.4 for main effect of treatment for flow cytometry acquisitions. For ease of interpretation, all figures depict least squared means and standard error values generated by using SAS, which are not transformed.
Annexin-V-FITC and PI cell viability
Fibroblast viability was measured based on positive fluorescence in an FL × FL2 (530/40 × 575/25 filters) channel plot by using a Beckman Coulter CyAN ADP Analyzer cytometer (Beckman Coulter, Inc., Fullerton, CA). Axis of plots were determined by using fibroblasts singly stained with either annexin-V (FITC conjugated) or PI, as well as by using unstained control cells. Fibroblasts were defined as healthy and viable if they did not stain positively for annexin-V or PI.
PI cell cycle cytometric acquisition
Intensity of PI staining in ethanol fixed fibroblasts was measured by using a BD LSR Fortessa X-20 cytometer (Becton Dickinson, Franklin Lakes, NJ) equipped with a 552 nm laser and 610/20 dichroic filter. Stage of the cell cycle was determined by using a Watson pragmatic model and curvilinear analysis in FlowJo version 7 software (FlowJo, LLC, Ashland, OR).
Conditioned media metabolite gas chromatography and mass spectroscopy analysis
Conditioned media samples were collected from three replicates of CPI (100 μM)-, PS48 (10 μM)-, MIX (CPI 100 μM+PS48 10 μM)-, and CON (0 μM)-treated flasks on days 3, 5, and 7 of culture and were spun at 600 g for 6 minutes to remove any cellular debris, and then frozen at −20°C. The number of cells growing on flasks from which media was collected are reported in Figure 1A and B. Samples were thawed, vortexed, and 1 mL was used for metabolite analysis. Chloroform (1 mL) and high-performance liquid chromatography (HPLC)-grade water containing internal standard 25 μg/mL ribitol (1 mL) were added to media samples. The samples were then vortexed and centrifuged at 2900 g for 30 minutes at 4°C to separate the layers. The upper aqueous layer (1 mL) was collected and transferred to individual 2.0 mL autosampler vials and dried under nitrogen at 45°C.
Dried polar compounds were methoximated in pyridine with 120 μL of 15.0 μg/mL methoxyamine-HCl, briefly sonicated, and incubated at 50°C until the residue was resuspended. Metabolites were then derivatized with 120 μL of MSTFA +1% TMCS for 1 hour at 50°C. The samples were subsequently transferred to a 300 μL glass insert and analyzed using an Agilent 6890 gas chromatographer coupled to a 5973 MSD scanning from m/z 50 to 650. Samples were injected at a 15:1 split ratio, and the inlet and transfer line were held at 280°C. Separation was achieved on a 630 m DB-5MS column (0.25 mm ID, 0.25 μm film thickness; J&W Scientific) with a temperature gradient of 5°C/min from 80°C to 315°C and held at 315°C for 12 minutes, and a constant helium flow of 1.0 mL/min.
The raw data were processed using AMDIS software (Automated Mass spectral Deconvolution and Identification System, http://chemdata.nist.gov/mass-spectra/amdis/). Derivatized metabolites were identified by matching retention time and mass spectra to those in a custom library of authentic compounds. Abundances of the metabolites were extracted with MET-IDEA (Broeckling et al., 2006; Lei et al., 2012), and then normalized to the abundance of the internal standard ribitol for statistical analyses.
Conditioned media were analyzed using the program SAS. The model for each of the metabolites included treatment effect (CON, CPI, MIX, or PS48) and day effect (3, 5, or 7) as fixed effects, and the replicate as a random effect. The interaction between treatment and day was included when significant. The heterogeneous autoregressive (1) covariance structure was used to model the correlations among the repeated measures at different days. To meet the normality assumption in the linear regression models, the metabolites were either modeled at original scale or transformed to log-scale or square root scale. The studentized residual plot and normal quantile plot were used for checking model fitting. For the pairwise comparisons, the Tukey-Kramer method for multiple test adjustment was used.
For day 5 comparisons between culture media with drugs spiked in (Spike; no daily media changes) or added daily in a media change (Change), cells proliferated differently (Fig. 2). To account for this, cell numbers of treatments were used to normalize data relative to the Change CON treatment before further analysis. The model for each response variable consisted of the treatment effect (CON, CPI, MIX, or PS48), change effect (Spike or Change), and the interaction between these two, as fixed effects. The replicate was considered a random effect. The data were normalized by using quantile normalization.
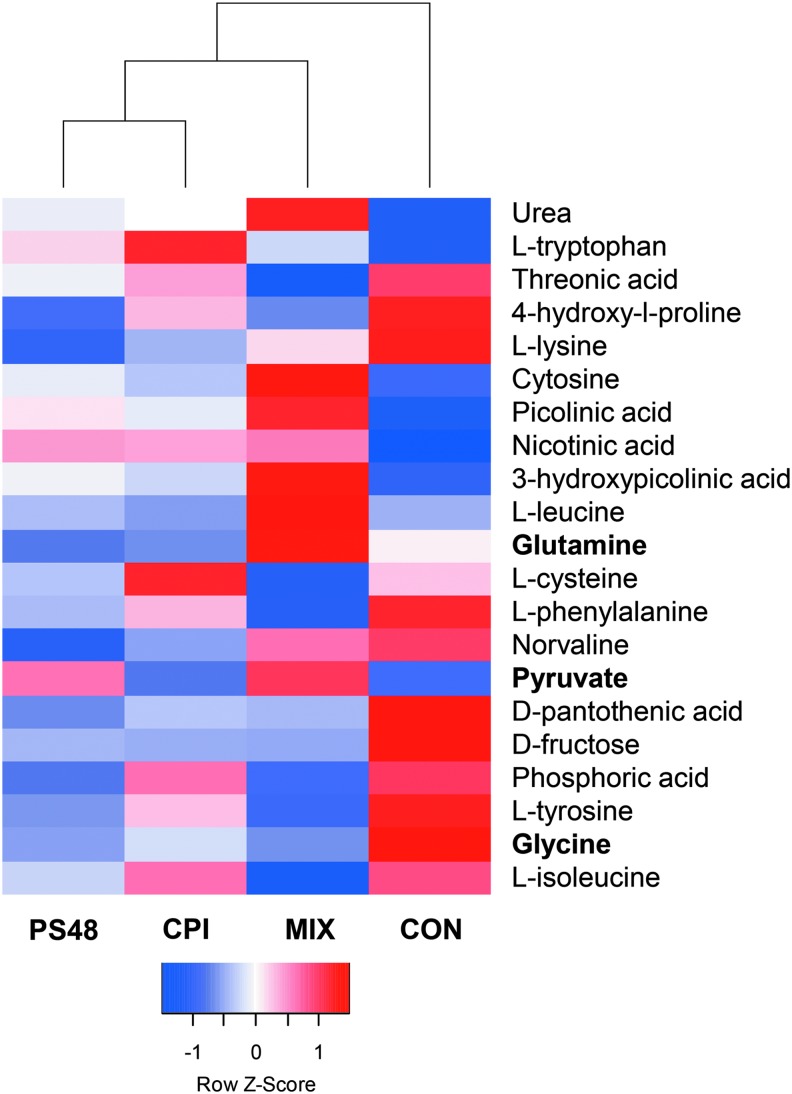
FIG. 2.
Differentially expressed genes between MIX- and CON-treated fibroblasts after 7 days with the pharmaceuticals CPI (100 μM), PS48 (10 μM), the mixture of the two (MIX), or without drugs (CON; 0 μM). Metabolites in bold type are important in Warburg effect-like metabolism. The Z-score of differential metabolite quantities are shown.
The response variables were either modeled at original scale or transformed to log-scale or square root scale, to fulfill the normality and homogeneity of variance assumptions in the linear regression models. The studentized residual plot and normal quantile plot were used for checking model fitting. For pairwise comparisons between the main effects, the Tukey-Kramer method for multiple test adjustment was used. In the case of the interaction, the comparison Change versus Spike was considered in each of the treatments.
Extraction of RNA and sequencing
Fibroblasts were collected for RNA after 7 days of respective culture treatments for four biological replicates. The number of cells in a flask collected for sequencing is reflected in Figure 1A and B. The cells were dissociated from culture flasks by brief incubation (37°C) with 1 × TrypLE Express (Gibco) by the same method used for cytometry. Cells were pelleted (5 minutes at 500 g), rinsed with 1 × PBS, again pelleted, and cells were plunged into liquid nitrogen and stored at −80°C. Extraction of RNA was performed as per specifications using Qiagen RNeasy mini kits (Qiagen, Germantown, MD). Total RNA quality was assessed at the University of Missouri DNA core facility by using the Advanced Analytical Fragment Analyzer and RNA quality scores were assigned based on (1) the presence of discrete 18S and 28S rRNA bands, (2) the mass ratio between the 28S and 18S rRNA, (3) the absence of fragments in the pre-18S and 28S regions, and (4) absence of contaminating high molecular weight fragments.
All RNA utilized in this study had scores of 10 (the highest score possible). RNAseq libraries were prepared at the University of Missouri Core facility by using standard Illumina protocol and sequenced on an Illumina HiSeq 2000 platform as single-end reads with read depth of 50 million reads/sample. The raw sequences (FASTQ) were subjected to quality check by FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/). The program fqtrim (https://ccb.jhu.edu/software/fqtrim/) was used to remove adapters, perform quality trimming (phred score >30) by sliding window scan (6 nucleotides), and select read length 30 nucleotides or longer after trimming. The reads obtained from the quality control step were mapped to the Sus Scrofa (v10.2) reference genome using Hisat2 aligner, which is a fast and sensitive alignment program of next-generation sequencing data (Kim et al., 2015).
The program Feature Counts (Liao et al., 2014) was used to quantify read counts using the sequences alignment files of each sample. The differentially expressed (DE) genes between sample groups, representing the culture treatment, were determined by fitting the read counts to a generalized linear model implemented in edgeR-robust (Zhou et al., 2014). The false discovery rate (FDR) <0.05 was used as threshold for statistically significant differential expression of genes.
Results
Impact of pharmacological treatment on fibroblast viability and proliferation
Without daily media changes, proliferation of cells was decreased in some cases to 50% of the cell number achieved with media changes (CPI and MIX; Fig. 1A); therefore in our further experiments, new media and pharmaceutical agents were applied daily, as opposed to being spiked into the media. Initial experiments were conducted to determine the effect of CPI and PS48 dosage on cell viability by using annexin-V-conjugated FITC and PI staining. Fibroblasts that did not stain positively for either annexin-V-conjugated FITC or PI staining were considered to be healthy nonapoptotic cells. Treatment with CPI dosages of 50 and 100 μM did not significantly alter (p = 0.40) the percentage of cells considered healthy from untreated fibroblasts (all dosages ≥88.4% appeared healthy; Table 1).
Table 1.
Viability of Fibroblasts Treated with CPI-613 for 7 Days
| Treatmenta | |||||
|---|---|---|---|---|---|
| Population | 0 μM | 50 μM | 100 μM | SE | p |
| Healthyb, % | 91.2 | 88.4 | 91.0 | 1.6 | 0.39 |
| Early apoptoticc, % | 0.7A | 0.3B | 0.4B | 0.1 | 0.05 |
| Late apoptoticd, % | 3.8A | 9.5B | 7.3B | 1.5 | 0.03 |
| Necrotice, % | 4.5A | 1.8B | 1.3B | 0.4 | <0.01 |
Fibroblasts treated with 0, 50, or 100 μM CPI-613 by daily media changes at 24 ± 2 hours.
Percentage of fibroblast population that stained negative for annexin-V-FITC and PI.
Percentage of fibroblast population that stained positive for annexin-V-FITC and negative for PI.
Percentage of fibroblast population that stained positive for PI and negative for annexin-V-FITC.
Percentage of fibroblast population that stained positive for annexin-V-FITC and PI.
PI, propidium iodide.
Means with capitalized letters are significantly different (p < 0.05).
Treatment with 50 and 100 μM CPI did increase (p = 0.03) the percentage of cells that were PI positive compared to untreated cells (9.5 and 7.3 vs. 3.8% ± 1.5%). However, untreated control cells in initial CPI dosage experiments had higher proportions of cells that were annexin-V positive, as well as double PI and annexin-V positive compared to 50 and 100 μM CPI (p ≤ 0.05; Table 1). Inversely, treatment with PS48 did not affect cell viability compared to controls (p > 0.4; Table 2). After determining that the highest concentration of CPI and PS48 that would still yield high percentages of viable cells, we then tested the combination of the two drugs.
Table 2.
Viability of Fibroblasts Treated with PS48 Dosages for 7 Days
| Treatmenta | |||||
|---|---|---|---|---|---|
| Population | 0 μM | 5 μM | 10 μM | SE | p |
| Healthyb, % | 95.3 | 93.6 | 93.1 | 1.3 | 0.48 |
| Early apoptoticc, % | 0.8 | 0.8 | 0.8 | 0.1 | 0.99 |
| Late apoptoticd, % | 1.1 | 1.3 | 1.2 | 0.4 | 0.88 |
| Necrotice, % | 2.9 | 4.2 | 4.4 | 1.0 | 0.55 |
Fibroblasts treated with 0, 5, or 10 μM PS48 by daily media changes at 24 ± 2 hours.
Percentage of fibroblast population that stained negative for annexin-V-FITC and PI.
Percentage of fibroblast population that stained positive for annexin-V-FITC and negative for PI.
Percentage of fibroblast population that stained positive for PI and negative for annexin-V-FITC.
Percentage of fibroblast population that stained positive for annexin-V-FITC and PI.
Proliferation of cells as well as the percentage of cells alive at the time of passage (as measured by trypan blue staining) were not augmented with treatment (p = 0.4; see Fig. 1C). All treatments had increased proliferation from day 0 to 3 (p < 0.01; Fig. 1B); however in subsequent passages (days 3, 5, and 7), numbers did not differ between treatments (p > 0.05). There was no interaction of treatment by day of culture among treatments (p = 0.8; Fig. 1). Lifespan of cells treated daily with PS48 (0 μM = 61.3; 5 μM = 64.7; 10 μM = 68.0 ± 4.1 days) was measured, as this compound has been demonstrated to have pluripotency-inducing properties, but was not different (p = 0.5). The stage of cell cycle as measured by PI staining in ethanol-fixed fibroblasts was not impacted by the 7-day treatments (p ≥ 0.4; Table 3).
Table 3.
Cell Cycle Phase of Fibroblasts After 7-Day Pharmacological Treatment
| Treatmenta | ||||||
|---|---|---|---|---|---|---|
| Measure | CON | CPI | MIX | PS48 | SE | p |
| G1 phase, % | 65.9 | 69.2 | 69.3 | 67.7 | 3.0 | 0.84 |
| S phase, % | 22.5 | 21.1 | 20.3 | 22.1 | 2.6 | 0.92 |
| G2 phase, % | 8.4 | 7.1 | 7.3 | 7.0 | 0.6 | 0.41 |
Fibroblast treatments applied during daily media changes at 24 ± 2 hours; CON (0 μM), PS48 (10 μM PS48), CPI (100 μM CPI-613), or MIX (10 μM PS48 and 100 μM CPI-613).
Conditioned cell culture media metabolites
Using tandem gas chromatography and mass spectroscopy, 55 metabolites were detected in the conditioned media from culture-treated fibroblast cells. The same metabolites were present in analyzed media, which had never been used in cell culture (unconditioned media). Of the metabolites detected, 21 had significantly different quantities (p < 0.05) among treatments where media were changed daily (Fig. 2). Of note was increased pyruvate in the MIX treatment, whereas other treatments had decreased values compared with the quantity in day 0 media.
Comparatively, all metabolites had higher quantities (p < 0.05) when comparing day 5 media from cells that did not have media changes (drugs spiked in) with day 5 media from cells that had daily media changes (drugs added during media change). The quantity of PS48 and CPI was detectible in conditioned media by their respective treatments; PS48: MIX and PS48 treatments were both 14.7 SE = 2.80 AU; CPI: MIX and CPI treatments had 6.15 and 4.57; SE = 1.74 AU. The quantity of PS48 did not differ between day 5 media samples where media were changed daily or spiked in (p > 0.12). Inversely, the quantity of CPI in conditioned media was higher (p = 0.006) when it was spiked in daily compared to when media were changed (22.2 vs.4.57; SE = 2.64 AU), and this also occurred in MIX-treated cells (15.96 vs. 6.15; SE = 2.64).
Gene expression
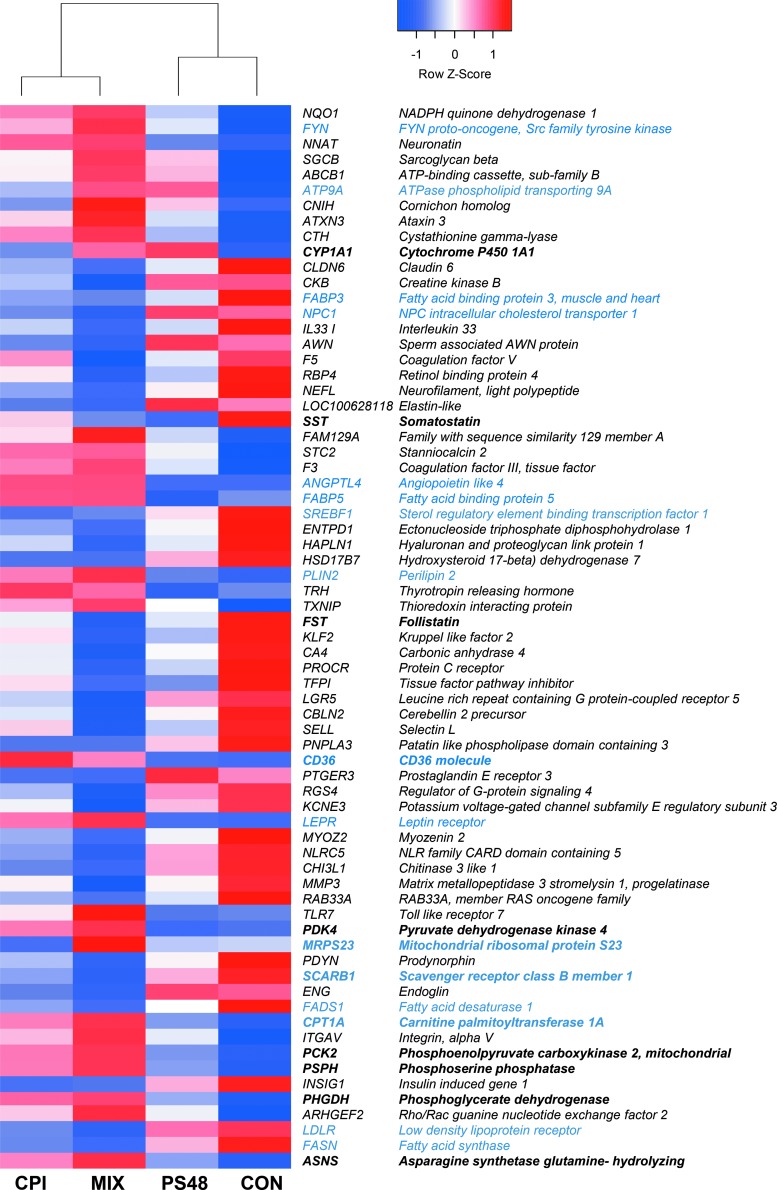
Sequencing of mRNA revealed 69 DE genes between MIX and CON treatments (Fig. 3). Of these genes, 16 play roles in lipid synthesis and beta-oxidation of fatty acids (Fig. 3, blue font). Compared to CON, MIX cells had downregulation of twofold change or greater in 18 genes and upregulation of twofold change or greater in expression of 8 genes. Among CON and CPI, there were 27 DE genes. All, but six of these genes were found DE between MIX and CON; those genes are ribosomal protein S16, ADP ribosylation factor-like GTPase 4C, kruppel-like factor 9, oxytocin receptor, lysine demethylase and nuclear receptor corepressor, and tumor necrosis factor alpha induced protein 6. All these genes were downregulated, except ribosomal protein S16. There was only one gene DE between PS48 and CON, which was a decreased expression of Somatostatin.
FIG. 3.
Spent media metabolite quantities of fibroblasts treated for 7 days with the pharmaceuticals CPI (100 μM), PS48 (10 μM), the mixture of the two (MIX), or without drugs (CON; 0 μM). Genes in bold type may play a role in influencing a Warburg effect-like metabolism. Genes in blue are involved in beta-oxidation or lipid synthesis. The Z-score of gene differential expression is shown.
Discussion
Our experimental goal was to determine if pharmacological treatment can be used to induce fibroblasts to have a metabolism that is characteristic of the Warburg effect, wherein there is a greater degree of glycolysis rather than TCA cycle activity. In this study, we determined concentrations of both CPI and PS48 that would provide us with a high percentage of viable cells (Tables 1 and 2). We show that, when PS48 and CPI are used in combination, gene expression and metabolite flux are augmented. To our knowledge, these drugs have not been used as a treatment combination in any type of cell line to promote a Warburg effect-type metabolic phenotype previously.
Contrary to our proposed Warburg proliferation hypothesis, there was no difference in proliferation between treatments throughout the week. At passages 3 and 5, all cell treatments were diluted so that fibroblasts across treatments would get similar daily drug exposure (see Fig. 1B for cell growth); this may contribute to the reason treatments yielded similar numbers and were not different overall. The percentage of viable cells (as measured by trypan blue) did not differ among treatments throughout the week if media were changed daily (Fig. 1C). In addition, passaging on days 3 and 5 likely contribute to the similarity in cell cycle states on day 7 among treatments (Table 3).
Previous CPI studies have focused on investigating the efficacy of this drug in anti-cancer treatment as a tumor-specific antimitochondrial mediator that has been tested in a number of cancerous cell lines as well as in vivo (Lee et al., 2011a, 2011b; 2014; Pardee et al., 2014; Perera et al., 2012; Senzer et al., 2012; Zachar et al., 2011). A comparison of CPI treatment between cancerous and non-cancerous cell lines from lung, breast, and kidney demonstrated that CPI reduced cell survival in all of the cancerous lines compared to the non-cancerous lines, implying non-cancerous cells are less sensitive to CPI (Zachar et al., 2011). The mechanism of cell death in cancerous cells by CPI may be, in part, through redox activity induced within mitochondria of tumor cells (Stuart et al., 2014), and cell death can be induced through apoptotic and nonapoptotic pathways (Zachar et al., 2011).
In this study, CPI decreased the number of cells after 5 days of growth (Fig. 1A) and is detected at twofold or higher concentration in conditioned media from flasks it was “spiked into” (CPI- and MIX-treated cells) compared to those with daily media changes. While the half-life in cell culture is not reported to our knowledge, in clinical cancer trials, the half-life is reported to be 1.3 hours in patients (Pardee et al., 2014). We speculate that either CPI has a long enough half-life duration that changing the media removes some of the CPI alleviating toxic ROS-inducing effects, or perhaps that when CPI is metabolized those derivatives may build up and become toxic. To detect CPI and PS48 through tandem liquid chromatography–mass spectroscopy (LC/MS), pure samples were analyzed and added to the existing metabolite library at the University of Missouri Metabolomics Core facility; the metabolites of CPI and PS48 are not in the library for us to identify.
Comparatively, the PS48 concentration was similar in conditioned media when spiked in daily or changed daily. The concentration of CPI cells were exposed to was 10 times higher than PS48 in this study (100 μM vs.10 μM), yet the quantity (relative to the internal standard ribitol) of PS48 detected in the media was higher than CPI, implying differences in the rate of metabolism and stability of the two drugs. The small molecule PS48, which stimulates glycolysis, enhances in vitro reprogramming efficiency by about 15-fold in human somatic cells (Zhu et al., 2010).
Experimentation using PS48 has been largely focused in the area of cellular reprogramming and pluripotency induction. In this experiment, however, we utilized it in an effort to stimulate the PI3K pathway and promote higher glycolytic activity. Lack of altered gene expression in addition to a normal length of life for fibroblasts treated with PS48 (at the concentration tested; 10 μM) do not indicate increased pluripotency in this experiment. While expression of 27 genes was impacted by CPI, only expression of SST (somatostatin) was augmented by treatment of PS48 alone.
The nucleus controls and regulates gene expression for products such as enzymes involved in metabolic processes. In this study, we sought to determine if metabolic regulation and feedback could be pharmacologically manipulated in such a way as to “reprogram” nuclear gene expression to a more Warburg-like state. Some of the DE genes in MIX-treated fibroblasts have been identified to play roles in various cancers and proliferation. For example, mitochondrial ribosomal protein S23 was increased 28-fold and carnitine palmitoyltransferase 1A was increased 1.95-fold in MIX-treated fibroblasts; these were two of eight genes identified as drivers of proliferation in luminal subtype breast tumors in humans (Gatza et al., 2014).
Expression of the gene angiopoietin-like 4 (ANGPTL4) was increased ninefold in MIX cells. This gene is evidenced to be induced by multiple factors, including hypoxia, presence of free fatty acids, and expression of tumor necrosis factor β (González-Muniesa et al., 2011; Kim et al., 2011). The ANGPTL4 protein targets peroxisome proliferator-activated receptors and is involved in the regulation of cancer metastasis, evasion of anoikis, and lipid metabolism (Kersten, 2005; Kim et al., 2011; Semenza, 2012).
In the reverse Warburg effect, it is proposed that stroma surrounding cancerous cells provide nutrients through autophagy, mitophagy, and aerobic glycolysis. The stromal catabolites such as pyruvate, lactate, glutamine, and free fatty acids support the growth of cancer (Yoshida, 2015). Compared to CON-, CPI-, and PS48-treated fibroblasts, MIX produced glutamine, whereas others consumed it as per comparison to unused media quantity (Fig. 2).
Many of the DE genes between MIX and CON were related to lipid regulation, including fatty acid synthesis, uptake, and transport (Fig. 3, blue font). The changes in gene expression related to lipid synthesis and metabolism may be due to decreased de novo lipid synthesis through the action of CPI. Mitochondrial citrate production is needed for de novo lipogenesis; in order for acetyl-CoA to be used for the synthesis of fatty acid, cholesterol, and isoprenoids, acetyl-CoA must be converted to citrate to travel to the cytosol [reviewed in Ward et al. (2012)]. The drug CPI is a lipoate derivative that activates the E1α subunit of PDH, preventing the flux of pyruvate into the TCA cycle, therefore the conversion of pyruvate to acetyl-CoA (Zachar et al., 2011). The compound CPI is inhibitory to α-ketoglutarate dehydrogenase, which also facilitates the eventual production of citrate (Stuart et al., 2014; Zachar et al., 2011).
Pyruvate dehydrogenase kinase (PDK) phosphorylates PDH and thus inactivates the PDH enzyme. In the MIX treatment, PDK4 expression was upregulated 7.4-fold from CON. Hypoxic inducible factor-1 can facilitate the expression of PDK1 as a cellular adaptation to hypoxia, which aids in the prevention of ROS production and apoptosis (Kim et al., 2006). Research evidences that “PDK4 is a ‘lipid status’ responsive PDK isoform” whereby it aids in mitochondrial fatty acid oxidation (Sugden et al., 2001; 2003). Increased pyruvate is demonstrated to inhibit the PDKs; however, pyruvate was increased in the MIX treatment compared to day 0 controls, whereas all other groups had negative values indicative of pyruvate consumption (Bao et al., 2004; Saunier et al., 2016).
Glycine was decreased in the conditioned media of MIX-treated fibroblasts compared to CON fibroblasts (Fig. 2). Compared with day 0 media quantities, MIX had a value of −781.4 and CON had a value of 478.4. We speculate based on previously described mitochondrial stress induced by CPI (Stuart et al., 2014) that, ROS may likely be generated and some of the glycine might be used for glutathione synthesis. A review from Amelio et al. (2014) highlights the roles of serine and glycine metabolism in cancer, whereby they are necessary to provide precursors for proteins, nucleic acids, and lipids. In this review, the coauthors detail the de novo synthesis of serine from glycolysis. Two genes within this pathway phosphoglycerate dehydrogenase (PHGDH) and phosphoserine phosphatase (PSPH) were upregulated from CON expression in the MIX treatment.
Serine serves as an allosteric regulator of pyruvate kinase M2 isoform. Pyruvate kinase catalyzes the conversion of phosphoenolpyruvate (Pepino et al.) to pyruvate. The M2 variant has a weaker enzymatic activity and lowers the PEP-to-pyruvate conversion, thus favoring accumulation of glycolytic intermediates for use in other pathways inadvertently promoting glycolytic activity [reviewed in Amelio et al. (2014)]. This could, in part, contribute to an increased production of pyruvate in MIX-treated fibroblasts. In addition, expression of phosphoenolpyruvate carboxykinase 2, mitochondrial (PCK2) was upregulated with MIX treatment; this enzyme catalyzes the conversion of oxaloacetate to phosphoenolpyruvate in the presence of guanosine triphosphate and could also be contributing to the production of pyruvate.
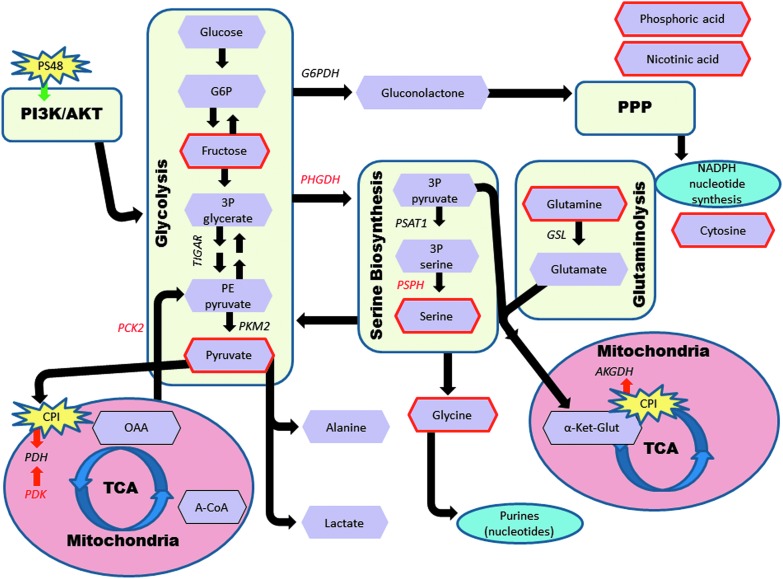
While more research is warranted for verification, our best interpretation of what metabolites and genes are influencing Warburg effect-related metabolic pathways in MIX-treated cells is represented in Figure 4. In this figure, significant genes are in red font and metabolites (purple hexagons), which were significantly impacted, are outlined in red.
FIG. 4.
Influence of CPI and PS48 treatment on gene expression and metabolite quantities in Warburg effect-related metabolic pathways in porcine fetal fibroblasts. Enzymes are in italic font and metabolites are in purple hexagons. The genes that were significantly impacted in fibroblasts treated with CPI and PS48 are in red font. Metabolites that were significantly impacted are outlined in red. This figure was modeled and adapted based on figures created by Vander Heiden et al. (2009) and Amelio et al. (2014). Black arrows represent forward possible directions for metabolites and roles of enzymes. Red arrows indicate inhibitory roles of enzymes and CPI. Green arrow depicts PS48 stimulation of the PI3K/AKT pathway.
Our study is novel in its approach to augment fibroblast cell metabolism pharmacologically by promoting glycolysis and inhibiting TCA activity in an attempt to achieve a more Warburg effect-like state. The pharmaceutical agents CPI and PS48 can be applied at concentrations used on other cell lines without harmfully impacting viability or proliferation in a porcine fetal cell line. While there is evidence of changes in gene expression that are implicated in the Warburg effect and expressed in cancerous cells, instead of the production of lactate and alanine, the MIX-treated cells increase the concentration of pyruvate in media to a level above what that media originally contained. We propose that this phenomenon, whereby we attempted to induce a Warburg effect in fibroblasts, minics the role of hypoxia-coerced fibroblasts in supporting the growth cancer stroma proposed by other researchers known as the Reverse Warburg effect.
Acknowledgments
The authors would like to thank Daniel Jackson and Kathy Schieber at the University of Missouri Flow Cytometry Core for their assistance and training in flow cytometry data collection. The authors also thank Dr. Zhentian Lei, Dr. Barbara Sumner, and Dr. Llyod Sumner from the University of Missouri Metabolomics Core for their training and assistance in processing and analysis of conditioned media. Funding for this project was provided by the National Institutes of Health (R01HD080636), Food for the 21st Century at the University of Missouri, the National Science Foundation (Award 1615789), and the University of Missouri Molecular Life Science Fellowship.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Amelio I., Cutruzzolá F., Antonov A., Agostini M., and Melino G. (2014). Serine and glycine metabolism in cancer. Trends Biochem Sci 39, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao H., Kasten S.A., Yan X., Hiromasa Y., and Roche T.E. (2004). Pyruvate dehydrogenase kinase isoform 2 activity stimulated by speeding up the rate of dissociation of ADP. Biochemistry 43, 13442–13451 [DOI] [PubMed] [Google Scholar]
- Bodart V., et al. (2002). CD36 mediates the cardiovascular action of growth hormone-releasing peptides in the heart. Circ Res 90, 844–849 [DOI] [PubMed] [Google Scholar]
- Briest F., and Grabowski P. (2014). PI3K-AKT-mTOR-signaling and beyond: the complex network in gastroenteropancreatic neuroendocrine neoplasms. Theranostics 4, 336–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeckling C.D., Reddy I R., Duran A.L., Zhao X., and Sumner L.W. (2006). MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78, 4334–4341 [DOI] [PubMed] [Google Scholar]
- Bull H.A., Brickell P.M., and Dowd P.M. (1994). Src-related protein tyrosine kinases are physically associated with the surface antigen CD36 in human dermal microvascular endothelial cells. FEBS Lett 351, 41–44 [DOI] [PubMed] [Google Scholar]
- Carlson H.E., Mariz I.K., and Daughaday W.H. (1974). Thyrotropin-releasing hormone stimulation and somatostatin inhibition of growth hormone secretion from perfused rat adenohypophyses*. Endocrinology 94, 1709–1713 [DOI] [PubMed] [Google Scholar]
- Dörsam B., and Fahrer J. (2016). The disulfide compound α-lipoic acid and its derivatives: a novel class of anticancer agents targeting mitochondria. Cancer Lett 371, 12–19 [DOI] [PubMed] [Google Scholar]
- Folmes Clifford D.L., et al. (2011). Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14, 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatza M.L., Silva G.O., Parker J.S., Fan C., and Perou C.M. (2014). An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet 46, 1051–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Muniesa P., de Oliveira C., Pérez de Heredia F., Thompson M.P., and Trayhurn P. (2011). Fatty acids and hypoxia stimulate the expression and secretion of the adipokine ANGPTL4 (angiopoietin-like protein 4/fasting-induced adipose factor) by human adipocytes. J Nutrigenet Nutrigenomics 4, 146–153 [DOI] [PubMed] [Google Scholar]
- Hankinson O. (1995). The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol 35, 307–340 [DOI] [PubMed] [Google Scholar]
- Hindie V., et al. (2009). Structure and allosteric effects of low-molecular-weight activators on the protein kinase PDK1. Nat Chem Biol 5, 758–764 [DOI] [PubMed] [Google Scholar]
- Ibrahimi A., and Abumrad N.A. (2002). Role of CD36 in membrane transport of long-chain fatty acids. Curr Opin Clin Nutr Metab Care 5, 139–145 [DOI] [PubMed] [Google Scholar]
- Kersten S. (2005). Regulation of Lipid Metabolism via Angiopoietin-like Proteins. Biochem Soc Transactions 33, 1059–1062 [DOI] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.-W., Tchernyshyov I., Semenza G.L., and Dang C.V. (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3, 177–185 [DOI] [PubMed] [Google Scholar]
- Kim S.-H., et al. (2011). ANGPTL4 induction by prostaglandin E2 under hypoxic conditions promotes colorectal cancer progression. Cancer Res 71, 7010–7020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppenol W.H., Bounds P.L., and Dang C.V. (2011). Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 11, 325–337 [DOI] [PubMed] [Google Scholar]
- Lee K., et al. (2011a). Long-term stable disease of stage IV pancreatic neuroendocrine tumors and without significant adverse effect by CPI-613, an investigational novel anti-cancer agent. Case Study Case Rep 1, 137–145 [Google Scholar]
- Lee K., Shorr R., Maturo C., Boteju L.W., and Sheldon A. (2011b). Formation and anti-tumor activity of uncommon in vitro and in vivo metabolites of CPI-613, a novel anti-tumor compound that selectively alters tumor energy metabolism. Drug Metab Lett 5, 163–182 [DOI] [PubMed] [Google Scholar]
- Lee K.C., et al. (2014). Translational assessment of mitochondrial dysfunction of pancreatic cancer from in vitro gene microarray and animal efficacy studies, to early clinical studies, via the novel tumor-specific anti-mitochondrial agent, CPI-613. Ann Transl Med 2, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Z., Li H., Chang J., Zhao P.X., and Sumner L.W. (2012). MET-IDEA version 2.06; improved efficiency and additional functions for mass spectrometry-based metabolomics data processing. Metabolomics 8, 105–110 [Google Scholar]
- Liao Y., Smyth G.K., and Shi W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 [DOI] [PubMed] [Google Scholar]
- Moussaieff A., et al. (2015). Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21, 392–402 [DOI] [PubMed] [Google Scholar]
- Pardee T.S., et al. (2014). A phase I study of the first-in-class antimitochondrial metabolism agent, CPI-613, in patients with advanced hematologic malignancies. Clin Cancer Res 20, 5255–5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepino M.Y., Kuda O., Samovski D., and Abumrad N.A. (2014). Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr 34, 281–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera C.N., Rodriguez R., and Shorr R. (2012). Regulation of pancreatic, gliosarcoma and non-small cell lung cancer via CPI-613, a novel anticancer therapeutic agent. Cancer Res 72, 3807–380722710435 [Google Scholar]
- Saunier E., Benelli C., and Bortoli S. (2016). The pyruvate dehydrogenase complex in cancer: an old metabolic gatekeeper regulated by new pathways and pharmacological agents. Int J Cancer 138, 809–817 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. (2012). Molecular mechanisms mediating metastasis of hypoxic breast cancer cells. Trends Mol Med 18, 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senzer N., et al. (2012). CPI-613, an investigational novel anti-cancer agent, provides long-term stable disease without significant adverse effects in a patient with stage IV relapsed hepatocellular carcinoma. Case Study Case Rep 2, 38–45 [Google Scholar]
- Shyh-Chang N., and Daley G.Q. (2015). Metabolic switches linked to pluripotency and embryonic stem cell differentiation. Cell Metab 21, 349–350 [DOI] [PubMed] [Google Scholar]
- Sidis Y., Schneyer A.L., and Keutmann H.T. (2005). Heparin and activin-binding determinants in follistatin and FSTL3. Endocrinology 146, 130–136 [DOI] [PubMed] [Google Scholar]
- Stroba A., et al. (2009). 3,5-Diphenylpent-2-enoic acids as allosteric activators of the protein kinase PDK1: structure-activity relationships and thermodynamic characterization of binding as paradigms for PIF-binding pocket-targeting compounds. J Med Chem 52, 4683–4693 [DOI] [PubMed] [Google Scholar]
- Stuart S.D., et al. (2014). A strategically designed small molecule attacks alpha-ketoglutarate dehydrogenase in tumor cells through a redox process. Cancer Metab 2, 10.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden M., Bulmer K., and Holness M. (2001). Fuel-sensing mechanisms integrating lipid and carbohydrate utilization. Biochem Soc Trans 29, 272–278 [DOI] [PubMed] [Google Scholar]
- Sugden M.C., and Holness M.J. (2003). Recent advances in mechanisms regulating glucose oxidation at the level of the pyruvate dehydrogenase complex by PDKs. Am J Physiol Endocrinol Metab 284, E855–E862 [DOI] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C., and Thompson C.B. (2009). Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- Ward P.S., and Thompson C.B. (2012). Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell 21, 297–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida G.J. (2015). Metabolic reprogramming: the emerging concept and associated therapeutic strategies. J Exp Clin Cancer Res 34, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z., et al. (2011). Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Med 89, 1137–1148 [DOI] [PubMed] [Google Scholar]
- Zheng J. (2012). Energy metabolism of cancer: glycolysis versus oxidative phosphorylation (review). Oncol Lett 4, 1151–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Lindsay H., and Robinson M.D. (2014). Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 42, e91–e91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., et al. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell stem cell 7, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn J.A., and Wells J.A. (2010). Turning enzymes ON with small molecules. Nat Chem Biol 6, 179–188 [DOI] [PubMed] [Google Scholar]