Abstract
Ruminants derived products have a prominent role in diets and economy worldwide; therefore, the capability to control the rumen microbial ecosystem, for ameliorating their quality, is of fundamental importance in the livestock sector. The aim of this study was to evaluate the effect of dietary supplementation with chestnut and quebracho tannins on microbial community and fatty acid profile, in the rumen fluid of dairy ewes. Multivariate analysis of PCR-DGGE profiles of rumen microbial communities showed a correlation among the presence of chestnut or quebracho in the diet, the specific Butyrivibrio group DGGE profiles, the increase in 18:3 cis9, cis12, and cis15; 18:2 cis9 and cis12; 18:2 cis9 and trans11; 18:2 trans11 and cis15; and 18:1 trans11 content, and the decrease in 18:0 concentration. Phylogenetic analysis of DGGE band sequences revealed the presence of bacteria representatives related to the genera Hungatella, Ruminococcus, and Eubacterium and unclassified Lachnospiraceae family members, suggesting that these taxa could be affected by tannins presence in the diets. The results of this study showed that tannins from chestnut and quebracho can reduce the biohydrogenation of unsaturated fatty acids through changes in rumen microbial communities.
1. Introduction
The manipulation of rumen microbial ecosystem is considered of primary importance in livestock sciences to improve the feed efficiency and to increase the quality of ruminant-derived products [1]. Recent findings have suggested that tannins, the second most abundant group of plant phenols after lignin, may be used as natural feed additives to modulate rumen fermentation through the inhibition of specific rumen microbial species [2, 3]. Tannins are chemically classified into two groups, hydrolysable and condensed tannins, both able to affect the growth and the metabolic activity of many species of rumen microorganisms [4]. Toxic effects of tannins have been attributed to different mechanisms, such as the inhibition of enzyme activities, the substrate or metal ion deprivation, and the detrimental action on biological membranes [1]. Moreover, tannin effects on rumen microorganisms appear to depend strongly on their chemical structure, their concentration in rumen liquor, and the microbial species involved [5]. For this reason, in vivo studies are requested to elucidate the effect of this class of polyphenols on rumen microbial communities and thus their actual employment in ruminant livestock.
Recently, many research studies have been attempted to understand how to increase the concentration of healthful fatty acids (FA) as rumenic acid (RA, 18:2 cis9, trans11) or vaccenic acid (VA, 18:1 trans11) and transient intermediate of the bacterial biohydrogenation (BH) of polyunsaturated fatty acids (PUFA), in ruminant milk and meat [6–8]. Among ruminal bacteria that appear to be involved in ruminal BH, the Butyrivibrio group is particularly sensitive to tannins [2, 3, 9–13]. A selective inhibition of Butyrivibrio proteoclasticus, involved in the last step of BH process of linoleic acid (LA, 18:2 cis9, cis12), may provide an accumulation of vaccenic acid (VA, 18:1 trans11) at the rumen level and, consequently, more RA in ruminant products [11, 14]. Tannins reduced in vitro activity and growth of B. proteoclasticus [11, 15, 16]. Moreover, literature [2] has shown that the inclusion of quebracho (Schinopsis lorentzii) tannins in ewe diet affected the Butyrivibrio group in a selective manner and enhanced RA content in dairy products. However, limited information is available on the in vivo effect of different sources of tannins on rumen microbiome and FA BH. Therefore, the aim of the present study was to investigate the effect of supplementing ewe diets with chestnut (CHT) or quebracho (QUE) tannin extracts (hydrolysable and condensed tannins, resp.) on rumen liquor FA profile, on the composition of the total rumen bacteria community, and, finally, on the composition of the Butyrivibrio group community, by using a polymerase chain reaction-denaturant gradient gel electrophoresis (PCR-DGGE) approach.
2. Materials and Methods
2.1. Animals and Experimental Design
The experiment was conducted at the Research Centre of the Department of Applied Biology, University of Perugia, Italy. Animals were handled according to the guidelines of the Italian law on animal welfare for experimental animals (Italian Ministry of Health, 2014) and of the University of Perugia Ethics Committee for animal use and care. Three nonlactating Bergamasca x Appenninica ewes (six years old, 60.5 ± 3.4 kg of body weight) equipped with a ruminal cannula of 10 cm internal diameter (Ankom Technology Corp., Macedon, NY, USA) were used. The animals were penned individually. The experiment was conducted as a 3 × 3 Latin square design. Each ewe was fed with the three diets in three consecutive experimental periods of 21 d, including 15 d of adaptation, before each one. At the 21st day the rumen liquor was sampled. The 3 × 3 Latin square was repeated twice with the aim of obtaining more replicates. During the whole experiment, the ewes had free access to water and hay, while the concentrates were administered twice daily (07:30 and 18:30). Orts were collected once daily.
2.2. Diets
The experimental diets were the same previously tested in an in vivo trial [3]. Diets were composed of chopped grass hay (particle size > 4 cm of length), administered ad libitum and by a concentrate (800 g/head/day), which contained 84.5 g kg−1 dry matter (DM) of soybean oil and 52.8 g kg−1 DM of bentonite (control, as an inert component to compensate the tannin introduction), or 52.8 g kg−1 DM of chestnut tannins (CHT) or 52.8 g kg−1 DM of quebracho tannins (QUE). The chemical composition of feeds and the ingredients of concentrates are presented in Table 1. The dose of tannins was chosen to obtain a diet tannin concentration of nearly 1.6% of expected DM intake. On the basis of results from previous studies in literature, this dose was considered as safe for the animal and practical for the farmers [7, 17, 18].
Table 1.
Ingredients, chemical composition, and fatty acids profile of the experimental concentrates and of the hay and rolled barley administered to the ewes.
| Grass hay | Rolled barley | Control diet | CHT diet | QUE diet | |
|---|---|---|---|---|---|
| Ingredients (g kg −1 of DM) | |||||
| Barley | 213.8 | 213.8 | 213.8 | ||
| Corn | 211.3 | 211.3 | 211.3 | ||
| Wheat bran | 158.5 | 158.5 | 158.5 | ||
| Soybean meal (44 CP) | 126.8 | 126.8 | 126.8 | ||
| Beet pulp | 89.8 | 89.8 | 89.8 | ||
| Soybean oil1 | 84.5 | 84.5 | 84.5 | ||
| Bentonite | 52.8 | - | - | ||
| Chestnut tannin extract2 | - | 52.8 | - | ||
| Quebracho tannin extract3 | - | - | 52.8 | ||
| Molasses | 41.3 | 41.3 | 41.3 | ||
| CaCO3 | 10.6 | 10.6 | 10.6 | ||
| Sodium bicarbonate | 5.3 | 5.3 | 5.3 | ||
| Dicalcium phosphate | 5.3 | 5.3 | 5.3 | ||
| Chemical composition (g kg −1 of DM) | |||||
| Organic matter | 847.0 | 859.9 | 816.9 | 858.1 | 869.6 |
| Crude protein | 111.2 | 121.0 | 165.6 | 173.7 | 170.3 |
| Ether extract | 12.0 | 16.1 | 109.4 | 105.4 | 102.4 |
| NDF | 636.4 | 134.1 | 174.7 | 181.4 | 172.1 |
| ADF | 501.3 | 54.2 | 77.6 | 72.4 | 74.3 |
| ADL | 105.7 | 14.9 | 10.6 | 13.3 | 8.7 |
| Ash | 69.6 | 21.0 | 84.6 | 39.9 | 39.4 |
| ME (MJ kg−1 DM) | 7.8 | 9.9 | 13.1 | 14.1 | 14.1 |
| NEl (Mcal kg−1 DM) | 0.9 | 1.2 | 2.0 | 2.1 | 2.1 |
| Fatty acids (g/100 g of total fatty acids) | |||||
| 16:0 | 35.5 | 18.2 | 14.0 | 14.4 | 14.9 |
| SA, 18:0 | 5.8 | 4.6 | 3.6 | 3.4 | 3.4 |
| 18:1 cis9 | 9.3 | 21.2 | 23.3 | 22.9 | 22.0 |
| LA, 18:2 cis9 cis12 | 28.5 | 45.0 | 51.4 | 51.7 | 51.8 |
| LNA, 18:3 cis9 cis12 cis15 | 2.8 | 6.0 | 5.8 | 5.6 | 5.8 |
| Other FA | 18.1 | 4.9 | 1.9 | 2.0 | 2.1 |
1Fatty acid profile of soybean oil (g/100 g of total fatty acids): C16:0, 11.01; C18:0, 3.6; C18:1 cis9, 22.09; C18:2 cis9 and cis12, 53.7; C18:3 cis9, cis12, and cis15, 7.2. 2Hydrolysable tannins extracted from chestnut wood (Castanea sativa Mill.) containing 750 g of equivalent tannic acid/kg DM (provided by Gruppo Mauro Saviola Srl Radicofani, Siena, Italy). 3Condensed tannins extracted from quebracho (Schinopsis lorentzii) containing 456 g of equivalent tannic acid/kg DM (provided by Guido Lapi SpA, Castel Franco di Sotto, Pisa, Italy).
2.3. Tannin Sources
Chestnut tannins (750 g kg−1 DM of equivalent tannic acid) were provided by Gruppo Mauro Saviola Srl (Radicofani, Siena, Italy), while extract of QUE (456 g kg−1 DM of equivalent tannic acid) was provided by Guido Lapi SpA (Castel Franco di Sotto, Pisa, Italy).
Both the extracts were titrated according to Burns [19] to evaluate the equivalent tannic acid. The chemical composition and gas chromatographic profile of CHT were published by Campo et al. [20] and the characteristics of QUE were reported by Vasta et al. [2].
2.4. Feed Sampling and Analysis
Samples of feeds were collected daily and stored at −80°C until further analysis. Samples were then ground for chemical analysis by mill Cyclotec 1093 (PBI International, Milan, Italy), using a mesh size of one mm. Concentrations of crude protein (CP), ether extract (EE), and ash were determined according to the AOAC methods 976.06, 920.39, and 942.05, respectively [21]. Neutral detergent fibre (NDF), acid detergent fibre (ADF), and lignin (ADL) contents were determined according to van Soest et al. [22], using heat stable amylase and sodium sulphite, and expressed inclusive of residual ash. Metabolizable energy (ME) and net energy for lactation (NEL) were calculated according to Cannas et al. [23]. Feed FA were extracted according to Folch et al. [24], esterified according to Christie [25] with 19:0 (Sigma Chemical Co., St Louis, MO, USA) as the internal standard, and identified using the same procedure described below for FA of rumen samples.
2.5. Rumen Sample Collection and Fatty Acid Profile
Rumen content was sampled from each ewe before morning feeding from two different sites of the rumen and after 21 days of trials on each diet and immediately frozen at −80°C until further analysis.
The FA were extracted according to Folch et al. [24] and methylated according to Buccioni et al. [3]. The FA methyl ester (FAME) composition was carried out by gas-chromatography, according to Buccioni et al. [3]. Individual FAMEs were quantified using valeric acid (5:0) and nonadecanoic acid (19:0) methyl esters (cod W275204 and cod N5377, resp.; Sigma Chemical Co., St. Louis, MO, USA) as internal standards and identified by the comparison of the relative retention times of FAME peaks from samples, with those of the standard mixture 37 Component FAME Mix (C4:0-C24:0, cod 18919-1AMP, Supelco, Bellefonte, PA, USA), individual 18:1 trans9 and 18:1 trans11 (cod 46903 and v1381, resp., Sigma-Aldrich, St. Louis, MO, USA), individual 18:2 cis9, trans11 (cod 1255, Matreya Inc. Pleasant GAP, PA, USA), CLA mix standard (cod 05632; Sigma-Aldrich, St. Louis, MO, USA), and published isomeric profile [26–28]. The 18:1 isomers elution sequence was performed according to Kramer et al. [29]. Moreover, standard mix of linolenic acid (LNA) isomers (cod 47792, Supelco, Chemical Co., St. Louis, MO, USA) and of LA isomers (cod 47791, Supelco, Chemical Co., St. Louis, MO, USA) and published isomeric profiles [30] were used to identify the isomers of interest (conjugated α-linolenic acid, CALNA, 18:3 cis9, trans11, cis15; vaccelenic acid, VLA, 18:2 trans11, cis15). Two bacterial acid methyl ester mixes (cod 47080-U Supelco, Chemical Co., St. Louis, MO; GLC110, Matreya, Pleasant Gap, PA) and individual standard for methyl ester of 14:0 iso, 14:0 anteiso, 15:0 iso and 17:0 anteiso (cods 21-1211-11, 21-1210-11, 21-1312-11, and 21-1415-11, Larodan Malmo, Sweden) were used to identify branched FA profile. Inter- and intra-assay coefficients of variation were calculated by using a reference standard butter (CRM 164, Community Bureau of Reference, Bruxelles, Belgium) and detection threshold of FA was 0.001 g kg−1 of FA [31]. All FA composition results are expressed as g/100 g of FA.
2.6. DNA Extraction and PCR Amplification
Total DNA was extracted from 1 mL of each frozen rumen fluid, using the Fast DNA SPIN kit for soil (MP Biomedicals, Santa Ana, CA, USA) with some modifications. Briefly, each sample was thawed and transferred to a 15 mL tube, containing 4.5 mL of a buffer consisting of 150 mM NaCl, 10 mM−1 Tris-HCl, pH 8.0, and 10 mM EDTA, vortexed vigorously, and centrifuged at 200g at 4°C for 5 min. One mL of supernatant was transferred to a 2 mL centrifuge tube and centrifuged at 14,600g at 4°C for 5 min. The pellet was dissolved in 978 μL of buffer sodium phosphate and 122 μL of MT buffer (both solutions are supplied by the Fast DNA SPIN kit for soil, MP Biomedicals, Santa Ana, CA, USA) and then processed, according to the manufacturer's guidelines. DNA yield was quantified by its absorbance at 260 nm and DNA quality was verified using agarose gel electrophoresis (1% w/v).
The extracted DNA was used as a template for PCR amplification of the V6–V8 region of 16S rRNA genes of total bacteria or for the Butyrivibrio group. PCR amplifications were conducted using the following primer pairs: F968GC and R1401 for total bacteria (fragment size ~470 bp), according to Nübel et al. [32], and F968GC and B fib for the Butyrivibrio group (fragment size ~470 bp), according to Kim et al. [33]. Reactions were carried out using an iCycler Thermal Cycler (Bio-Rad Laboratories, Hertfordshire, UK) in 25 μL volumes containing 1X PCR buffer (67 mM Tris-HCl, pH 8.8, 1.66 mM (NH4)2SO4, 0.1% Tween-20), 1.5 mM MgCl2, 250 μM deoxynucleotide triphosphates (dNTPs), 400 nM of each primer, 1U of Polytaq (Polymed, Florence, Italy), and 10 ng of DNA. PCR reactions were performed under the following conditions: initial denaturation of 94°C for 5 min, followed by 35 cycles of 94°C for 20 s, 56°C for 30 s and 72°C for 45 s, and a final extension of 72°C for 10 min. PCR products were verified by agarose gel (1.2% w/v) ecectrophoresis.
2.7. PCR-DGGE Analysis of Total Bacteria and Butyrivibrio Group Communities
The PCR amplicons were electrophoretically separated by DGGE on a 6% polyacrylamide gel (acrylamide/bis 37.5:1) in 1X TAE Buffer (40 mM Tris base; 20 mM glacial acetic acid; 1 mM EDTA) using a 50–60% denaturant gradient obtained with a 100% denaturant solution, consisting of 40% v/v deionized formamide, 7 M urea. The gels were run at 60°C and 75 V for 17 h in a Phor-U system (Ingeny International, Goes, NL) and at the end of electrophoretic runs, the gels were stained with SYBR® Gold (Molecular Probes, Eugene, OR) and gel images digitalized using the ChemiDoc XRS apparatus (Bio-Rad Laboratories, Hertfordshire, UK).
2.8. Sequence Analysis of PCR-DGGE Fragments
The middle portion of 16 bands selected from Butyrivibrio group DGGE profiles was aseptically excised and placed in 20 μL distilled water. The PCR products were eluted through freezing and thawing [34] and reamplified using F968/B fib primer pairs without GC clamp, as previously described. PCR products were checked by DGGE gel electrophoresis and then subjected to directly sequencing by Macrogen Service [35]. The sequence chromatograms were edited using Chromas Lite Software [36] to verify the absence of ambiguous peaks and convert them to FASTA format. The Decipher Find Chimera Web tool [37] was used to uncover chimeras hidden in the 16S rDNA sequences. The BLASTN program [38] available at the NCBI website [39] was used to find taxonomic closely related nucleotide sequences. To increase the accuracy of the assignments, different sequence similarity thresholds were used for different taxonomic levels: a similarity of ≥97% for a species level identification and 95%, 90%, 85%, 80%, and 75% for assignment at the genus, family, order, class, and phylum level, respectively [40].
A phylogenetic dendrogram was constructed to display the apparent relatedness of the partial 16S rRNA gene sequences to each other and to other sequences of equivalent length retrieved from the GenBank database using the software ClustalX 2.0.11 [41] to perform sequence alignment and the software TREECON 1.3b [42] for the construction of the phylogenetic tree using the neighbor-joining method [43]. Bootstrap analysis was performed based on 1000 resamplings.
2.9. Statistical Analysis
Statistical analysis was performed using the mixed procedure of SAS [44]. Data were analyzed with the following model:
| (1) |
where Y is the dependent variable, calculated as the mean of the measurements during each sampling period, μ is the overall mean, Ai is the random animal effect (i = 1 to 3), Pj is the period effect (j = 1 to 3), Dk is the diet effect (k = 1 to 3), Dk × Pi is their interaction, Rz is the random replicates of the Latin square (z = 1 to 2), and eijkz is the residual error.
Least squares means estimates are reported. For all statistical analyses, significance was declared at P ≤ 0.05.
DGGE profiles were normalized and analyzed using GelCompar II software v 4.6 (Applied Maths, Sint-Martens-Latem, Belgium). The number of bands (species richness) and their relative abundance were used as a proxy of richness and diversity (Shannon index, H′, and Simpson index, D) of rumen microbial communities, as described by Pastorelli et al. [45]. The banding profiles of DGGEs, extracted as presence/absence matching tables, were imported into PAST software [46] for multivariate statistical analysis as previously described by Lagomarsino et al. [47]. One-way analysis of similarity (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA) were performed to determine significance differences in the microbial community structure due to the different dietary regimes. In order to find potential connection between community composition and ruminal FA profile and to evidence how these connections may be influenced by the different diets two different canonical correspondence analyses (CCA) were carried out: in the first, the FA assumed to be mainly implicated in BH process (i.e., 18:0, stearic acid, SA; VA; VLA; LA; LNA; RA) were selected; in the second, the FA assumed to be markers of rumen microbial metabolism according to Fievez et al. [48] were considered (i.e., 15:0 iso; 15:0 ante; 17:0 iso; 17:0 ante). The length and the angle of vectors indicate the relative importance of that FA in discriminating the bacterial community of the different rumen liquors [49]. To identify taxa that mainly contribute to separation of microbial communities, according to the different diets, DGGE band scores were also plotted in the CCA diagrams. The Butyrivibrio group DGGE profiles that were mainly related to different FA profile were sequenced.
3. Results
3.1. Fatty Acids (FA) Composition of Rumen Liquor
The presence of tannins in the diets induced changes in FA profile of rumen liquor. Tannins lowered BH of PUFA leading to an accumulation of linoleic acid (LA), linolenic acid (LNA), and their BH intermediates, reducing the accumulation of stearic acid (SA) (Table 2). In particular, vaccenic acid (VA) and rumenic acid (RA) percentage was significantly higher in rumen liquor from ewes fed with CHT and QUE than in samples from animals fed control diet. Other 18:1 isomers such as cis15, cis9, cis11, trans5, trans6–8, trans9, and trans10 showed a similar trend. QUE diet was also associated with an increase of 18:2 trans10, cis12 content in rumen liquor.
Table 2.
Fatty acid profile of rumen liquor from sheep fed with 800 g/head/day of a concentrate containing 84 g kg−1 DM of soybean oil plus 0 (control) or 52.8 g kg−1 DM of a chestnut tannin extract (CHT) or 52.8 g kg−1 of DM of quebracho tannin extract (QUE).
| FA g/100 g of total fatty acids | Control | CHT | QUE | SEM1 | P value2 |
|---|---|---|---|---|---|
| 10:0 | 0.197 b | 0.186 b | 0.439 a | 0.077 | 0.0473 |
| 12:0 ante | 0.237 b | 0.387 a | 0.261 b | 0.041 | 0.0402 |
| 12:0 | 0.297 b | 0.318 b | 0.656 a | 0.087 | 0.0213 |
| 13:0 | 0.305 c | 0.453 b | 0.542 a | 0.024 | <0.0001 |
| 14:0 iso | 0.317 b | 0.613 a | 0.404 b | 0.068 | 0.0184 |
| 14:0 | 0.485 b | 0.556 b | 1.438 a | 0.076 | <0.0001 |
| 15:0 iso | 0.116 c | 0.185 b | 0.239 a | 0.020 | 0.0030 |
| 15:0 ante | 0.378 b | 0.376 b | 0.507 a | 0.039 | 0.0499 |
| 15:0 | 0.776 b | 0.605 c | 1.054 a | 0.017 | <0.0001 |
| 16:0 iso | 0.175 b | 0.178 b | 0.276 a | 0.029 | 0.0353 |
| 16:0 | 10,428 c | 17.423 b | 23.108 a | 0.712 | <0.0001 |
| 17:0 iso | 7.544 a | 7.418 a | 6.135 b | 0.288 | 0.0039 |
| 17:0 ante | 0.189 b | 0.154 c | 0.247 a | 0.010 | <0.0001 |
| 17:0 | 0.327 b | 0.342 b | 0.532 a | 0.037 | 0.0013 |
| SA, 18:0 | 50.447 a | 44.616 b | 32.770 c | 1.307 | <0.0001 |
| 18:1 trans5 | 0.051 c | 0.313 a | 0.132 b | 0.055 | 0.0122 |
| 18:1 tran6–8 | 0.476 c | 0.682 b | 1.134 a | 0.131 | 0.0051 |
| 18:1 trans9 | 0.334 c | 0.602 b | 0.665 a | 0.016 | <0.0001 |
| 18:1 trans10 | 0.681 b | 1.254 a | 1.359 a | 0.147 | 0.0071 |
| VA, 18:1 trans11 | 1.922 c | 6.304 b | 7.589 a | 0.244 | <0.0001 |
| 18:1 trans12 | 0.535 c | 0.792 b | 1.299 a | 0.033 | <0.0001 |
| 18:1 cis5 | 0.374 b | 0.697 a | 0.799 a | 0.069 | 0.0007 |
| 18:1 cis7 | 0.510 c | 1.297 b | 1.498 a | 0.064 | <0.0001 |
| 18:1 cis9 | 2.337 c | 3.879 b | 5.340 a | 0.111 | <0.0001 |
| 18:1 cis11 | 0.414 c | 0.702 b | 0.892 a | 0.014 | 0.0137 |
| 18:1 cis12 | 0.258 | 0.315 | 0.363 | 0.042 | 0.1927 |
| VLA, 18:2 trans11, cis15 | 0.123 | 0.181 | 0.179 | 0.049 | 0.5901 |
| LA, 18:2 cis9, cis12 | 0.845 c | 1.096 b | 1.926 a | 0.075 | <0.0001 |
| LNA, 18:3 cis9, cis12, and cis15 | 0.305 c | 0.363 b | 0.467 a | 0.023 | 0.0004 |
| RA, 18:2 cis9, trans11 | 0.651 c | 2.137 b | 2.600 a | 0.042 | <0.0001 |
| CLA trans10, cis12 | 0.163 b | 0.162 b | 0.258 a | 0.017 | 0.0008 |
| 20:0 | 0.230 b | 0.323 b | 0.637 a | 0.025 | <0.0001 |
| 20:4 | 0.738 | 1.016 | 0.959 | 0.110 | 0.1855 |
| 22:0 | 0.144 c | 0.226 b | 0.399 a | 0.021 | <0.0001 |
1Standard error mean; 2probability of significant effect (a, b, and c for P < 0.05).
Considering the odd and even branched fatty acids, 14:0 iso content increased only in rumen liquor samples from ewes fed CHT, whereas the content of 15:0 iso increased in rumen liquor of both tannin-rich diets. However, the content of 15:0 iso was higher in rumen liquor from ewes fed with QUE. Rumen liquor samples from ewe fed with control and CHT diet had the highest concentration of 17:0 iso (Table 2). Considering the ante/iso FA, the content of 12:0 ante was significantly higher in CHT samples, whereas 15:0 ante and 17:0 ante content was higher in QUE samples.
3.2. Effect of Chestnut and Quebracho Tannins on Rumen Microbial Communities
The DGGE banding profiles obtained for total bacteria (Supplementary Material 1) showed a number of bands ranging from 16 to 28. The profiles generated with Butyrivibrio group primers were less complex, with a band number of 4–16 (Supplementary Material 2). Richness was not affected by the presence of tannins in the diet in rumen liquor bacterial (P = 0.324) and Butyrivibrio group (P = 0.206) communities. H′ index obtained from the DGGE analysis of bacteria (P = 0.352) and Butyrivibrio group (P = 0.117) was similar among treatments and D index did not change significantly in relation to diet in bacterial (P = 0.383) or Butyrivibrio group communities (P = 0.071).
The ANOSIM test applied to 16S rDNA PCR-DGGE profiles showed that the different dietary regimens significantly separated the rumen bacterial communities and that bacterial banding profiles of replicates (6 animal samples × 3 diets) for each diet were more similar to each other (ANOSIM global test R = 0.233; P < 0.05) than those find when the Butyrivibrio group (ANOSIM global test R = 0.4216; P < 0.01) was analyzed. PERMANOVA analysis confirmed that diet significantly affected the microbial community structure (PERMANOVA global test: bacteria F = 2.446, P < 0.05; Butyrivibrio group F = 4.276, P < 0.01). PERMANOVA pair-wise test established that bacterial communities under CHT and QUE were significantly different from that of control diet (Table 3) and that for Butyrivibrio group communities under CHT were significantly different from the others, whereas the control community was not significantly different to QUE (Table 3).
Table 3.
P values from PERMANOVA pair-wise comparison of band profiles from 16S rDNA bacterial DGGE (in boldface, upper right side) and from 16S rDNA Butyrivibrio group DGGE (in italics, lower left side).
| Diet | Control | CHT | QUE |
|---|---|---|---|
| Control | 0.0450 a | 0.0272 a | |
| CHT | 0.0196 a | 0.1310 | |
| QUE | 0.0737 | 0.0046 a |
aSignificant value (P < 0.05).
3.3. Bacterial Community Composition in relation to Diet
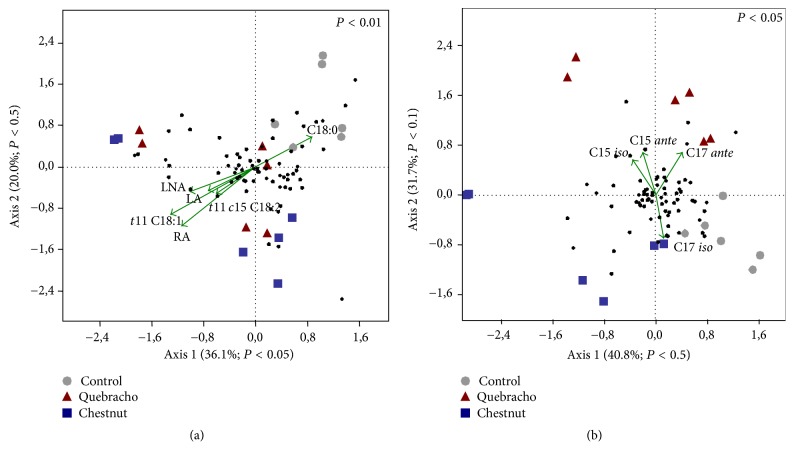
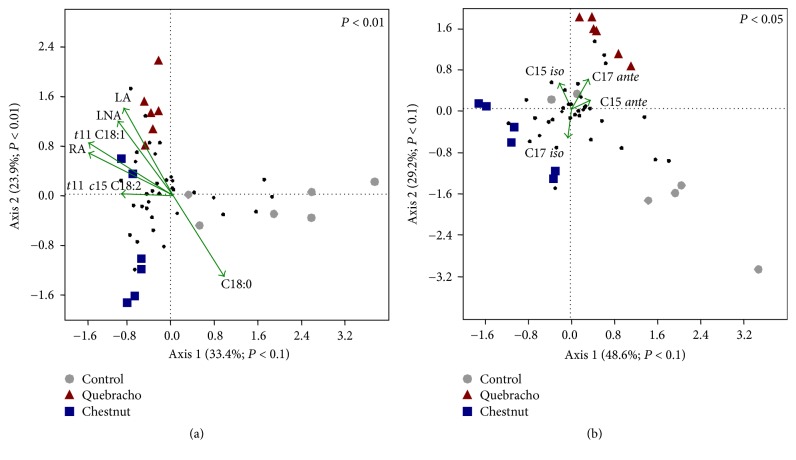
Canonical correspondence analysis carried out between total bacteria or Butyrivibrio group DGGE profiles and the FA assumed to be mainly implicated in BH process (SA; VA, VLA, and LA; LNA; RA) showed that ruminal communities under tannin dietary treatments were separated from the control, Figures 1(a) and 2(a). Similarly, CCA carried out between total bacteria or Butyrivibrio group DGGE profiles and the FA assumed to be markers of rumen microbial metabolism (15:0 iso; 15:0 ante; 17:0 iso; 17:0 ante) according to Fievez et al. [48] indicated that ruminal community under QUE was separated from the control and CHT, Figures 1(b) and 2(b).
Figure 1.
Canonical correspondence analysis (CCA) ordination diagram of ruminal bacterial communities and ruminal fatty acids variables [(a) FA assumed to be mainly implicated in BH process (SA, 18:0; VA, 18:1 trans11; VLA, 18:1 trans11, cis15; LA; LNA; RA); (b) FA assumed to be markers of rumen microbial metabolism (15:0 iso; 15:0 ante; 17:0 iso; 17:0 ante)] (vectors) defined by the first and second axes. DGGE band scores were also plotted (black filled circle). For each diagram significance (global test) is reported in upper right side.
Figure 2.
Canonical correspondence analysis (CCA) ordination diagram of ruminal Butyrivibrio-related communities (symbols) and ruminal fatty acids variables [(a) FA assumed to be mainly implicated in BH process (SA, 18:0; VA, 18:1 trans11; VLA, 18:1 trans11, cis15; LA; LNA; RA); (b) FA assumed to be markers of rumen microbial metabolism (15:0 iso; 15:0 ante; 17:0 iso; 17 ante)] (vectors) defined by the first and second axes. DGGE band scores were also plotted (black filled circle). For each diagram significance (global test) is reported in upper right side.
Both total bacterial and Butyrivibrio group communities under tannin extract diets were positively correlated to LA, LNA, RA, and VA production, Figures 1(a) and 2(a), whereas only those under QUE were positively correlated to C15 ante and C17 ante, Figures 1(b) and 2(b). Total bacterial and Butyrivibrio group communities of control samples were positively correlated to SA production, Figures 1(a) and 2(a).
3.4. Association and Identification of Butyrivibrio Group 16S rDNA PCR-DGGE Bands with Key Fatty Acids in the Biohydrogenation Pathway
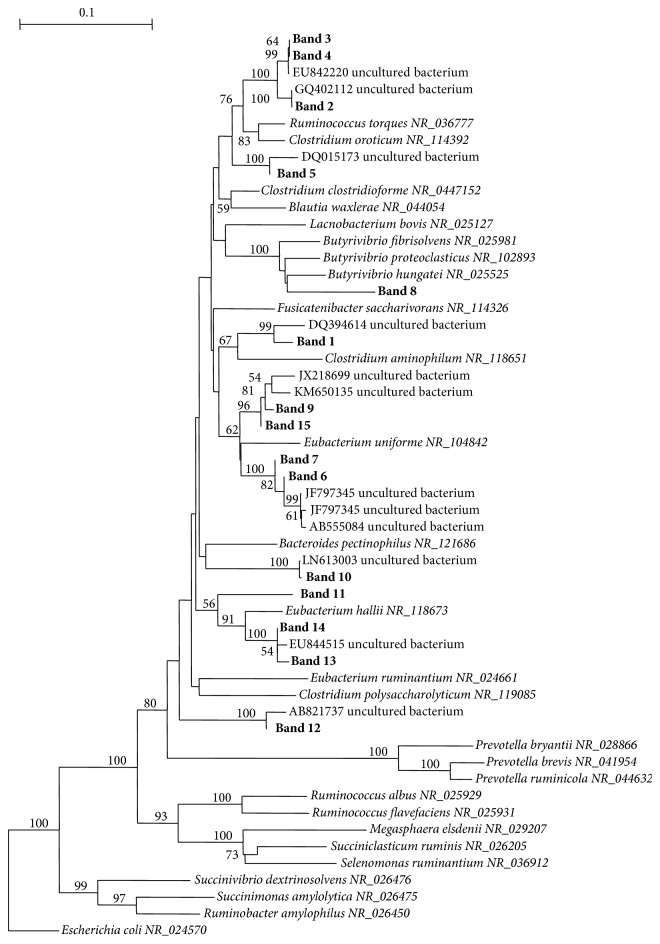
Multivariate CCA analysis of data generated from Butyrivibrio group DGGE allowed identifying bacterial species or groups mainly correlated to a specific FA; thus, bacteria identified by sequencing DGGE bands 1, 2, 3, 4, 5, 6, and 7 (Table 4) were significantly associated with LA, LNA, RA, VA, and VLA (data not shown), whereas bacteria corresponding to bands 8, 9, 10, 11, 12, 13, 14, and 15 (Table 4) were significantly linked with SA (data not shown).
Table 4.
Identification of the selected polymerase chain reaction denaturing gradient gel electrophoresis (16S rDNA PCR-DGGE) fragments.
| PCR-DGGE band |
Nearest match (GenBank accession number; % sequence similarity) |
Relative taxonomic classification |
|---|---|---|
| (1) | Clostridium aminophilum (NR_118651.1; 93%) | Unclassified Lachnospiraceae |
| (2) | Ruminococcus torques (NR_036777.1; 95%) | Ruminococcus spp. |
| (3) | Ruminococcus torques (NR_036777.1; 95%) | Ruminococcus spp. |
| (4) | Ruminococcus torques (NR_036777.1; 96%) | Ruminococcus spp. |
| (5) | Clostridium clostridioforme (NR_0447152; 95%) | Hungatella spp. |
| (6) | Fusicatenibacter saccharivorans (NR_114326.1; 93%) | Unclassified Lachnospiraceae |
| (7) | Eubacterium uniforme (NR_104842.1; 94%) | Unclassified Lachnospiraceae |
| (8) | Butyrivibrio proteoclasticus (NR_102893.1; 94%) | Unclassified Lachnospiraceae |
| (9) | Clostridium oroticum (NR_114392.1; 94%) | Unclassified Lachnospiraceae |
| (10) | Blautia waxlerae (NR_044054.1; 93%) | Unclassified Lachnospiraceae |
| (11) | Roseburia intestinalis (NR_027557.1; 92%) | Unclassified Lachnospiraceae |
| (12) | Bacteroides pectinophilus (NR_121686.1; 90%) | Unclassified Lachnospiraceae |
| (13) | Eubacterium hallii (NR_118673.1; 93%) | Unclassified Lachnospiraceae |
| (14) | Eubacterium hallii (NR_118673.1; 93%) | Unclassified Lachnospiraceae |
| (15) | Clostridium oroticum (NR_114392.1; 94%) | Unclassified Lachnospiraceae |
Putative taxonomic identification of DNA bands associated with LA, LNA, RA, and VA revealed that they were related to genera Hungatella (band 5), Ruminococcus (bands 2, 3, and 4), and unclassified Lachnospiraceae (bands 1, 6, and 7; Table 4; Figure 3). Moreover, putative taxonomic identification of bands associated with SA revealed that they were related to unclassified Lachnospiraceae (bands 8, 9, 10, 11, 12, 13, 14, and 15; Table 4; Figure 3).
Figure 3.
Phylogenetic analysis of Butyrivibrio partial 16S rRNA sequences obtained from PCR-DGGE bands using primers F968/B fib and identified species. Sequences obtained in this study are shown in boldface. Bootstrap values of >50% based on 1000 replications are indicated at the nodes. The 16S rRNA gene sequence of Escherichia coli (NR_024570) was selected as the outgroup.
4. Discussion
The BH process of dietary PUFA was strongly lowered by both the tannin-rich diets, regardless of the type of tannin. However, results showed that QUE tannins had a stronger ability to favor the accumulation of BH-intermediate, such as VA and RA, and to reduce the SA concentration in rumen liquor, if compared to CHT tannins. Recently, we found similar results by feeding lactating dairy ewes with diets containing soybean oil supplemented or not with chestnut and quebracho tannins [3]. In the present trial, in rumen liquor trans10 isomers of 18:1 and 18:2 also significantly were accumulated in response to tannin supplementation. Previously, in an in vivo trial on lactating dairy ewes [3] the administration of the same amount of soybean oil and tannins in the concentrate feed did not result in significant effects on trans10 isomers of 18:1 and 18:2, suggesting that in the present trial the rumen environment was more favorable to alternative BH pathway of LA from soybean oil.
The presence of condensed tannins in QUE diet induced the increase of 15:0 ante and 17:0 ante content and the decrease of 17:0 iso content in rumen liquor. Since the first two FA were associated with the growth of cellulolytic strains and the latter with the growth of amylolytic bacteria [48], this pattern or branched FA suggested a detrimental effect of condensed tannins on cellulolytic bacteria. In contrast, the pattern of branched chain FA between rumen liquor samples from CHT and control diets was quite similar, suggesting that CHT tannins did not perturb the growth of cellulolytic bacteria. Indeed, the previous in vivo experiment on lactating ewes [3] demonstrated that QUE tannins were more efficient than CHT in limiting cellulolytic bacteria proliferation.
As regards the composition of the whole rumen bacterial community, the diversity indices did not change. As a consequence, changes in the BH pattern could be due to rearrangements of the bacterial species. Indeed, the toxic effect of tannins on specific strains could be compensated by an increase of tannin resistant bacteria in total population. These data are in accordance with previous findings about the selective inhibition of plant extracts containing tannins on the growth and the activity of specific bacterial species representative of rumen microbial populations [2, 11, 50, 51]. Vasta et al. [2], by means of a T-RFLP analysis, demonstrated that dietary supplementation of QUE tannins affected total bacteria community structure in the rumen of lambs fed with QUE supplemented diets. However, it is worth noting that the percentage of tannins employed in the present study was much lower (<2% DM) than that used by Vasta et al. [2]. Thus, putting together data from the present and previous in vivo trials on dairy ewes, the addition of practical doses of tannins in the diet is able to cause shifts in the rumen total bacterial community favoring the accumulation of LA and its BH intermediates in rumen liquor.
Bacteria involved in the BH process have been categorized traditionally into two distinct groups: those belonging to group A converting PUFA as LA or LNA into VA and those belonging to group B hydrogenating VA into SA [14]. Group A comprises many known species, among which Butyrivibrio spp. are the most important [52, 53], whereas B. proteoclasticus is the only known cultivable rumen bacterium belonging to group B [54, 55]. However, recent studies demonstrate that other microorganisms as-yet-uncultivated bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales, and Ruminococcaceae might be involved in BH processes with a relevant role [10, 12, 13, 56]. Our data from DGGE analysis showed an effect of 2% DM of QUE or CHT tannins on the composition of the Butyrivibrio group. Butyrivibrio species are particularly sensitive to condensed tannins [16]. Indeed, these phytochemicals can penetrate the cell wall of Butyrivibrio and other Gram positive bacteria and selectively inhibit the cell wall biosynthesis [50]. However, at the same time, remarkable differences among Butyrivibrio species were observed in the level of their sensitivity to tannins [11, 50]. The persistence and the appearance of some bands but not of other ones in the Butyrivibrio group DGGE gel are consistent with these previous findings.
Multivariate statistics allowed the selection of DGGE bands representative of Butyrivibrio group that are putatively involved in the BH process in vivo. Seven bands disappeared when ewes were fed with QUE enriched diet, and this effect was associated with reduced rumen SA concentrations. Therefore, it is possible that this group of DNA bands might be representative of other bacteria that play a role in the BH of VA into SA, confirming the findings of several studies [10, 12, 13, 56]. Indeed, the bands here identified are highly related to uncultured rumen bacteria belonging to the family Lachnospiraceae. Interestingly, no sequences were identified as B. proteoclasticus, which according to literature is the only known bacterial species able to efficiently biohydrogenate PUFA into SA in the rumen. However, in vivo studies have shown contrasting results, since a clear relationship between the reduced amount of B. proteoclasticus and a decreased production of SA in rumen liquor was found only in a limited number of trials [2, 3]. A possible explanation to our data may be that the concentration of tannins used in this study was not able to modify B. proteoclasticus growth in the rumen but only lowered its capacity to hydrogenate 18:1 trans FA, as previously suggested by Boeckaert et al. [10]. Otherwise, it is possible that B. proteoclasticus has a limited contribution to SA formation in vivo and that other yet not known species may have a more important role in this step of the BH pathway. Since only a limited number of rumen species is presently known, this hypothesis is likely. Moreover, it is in agreement with the opinion of other authors who evaluated the effect of marine algae [10] and fish oil [56] on rumen bacterial diversity, evidencing a possible relation between the disappearance of many uncultivated Lachnospiraceae strains, genetically distant from B. proteoclasticus, and a significant decrease of rumen SA concentration.
Our study evidenced also that the two types of tannins induced an increase in LA, LNA, RA, and VA and 18:2 trans11 and cis15 in rumen liquor and this was associated with a higher intensity of seven bands in the Butyrivibrio group DGGE profiles. Phylogenetic analysis revealed that these sequences were representative of species belonging to genera Hungatella, Ruminococcus, Eubacterium and to unclassified Lachnospiraceae. Once more, these data confirm that other Butyrivibrio groups may be involved in the BH pathway and that their increased amount may promote the accumulation of 18:1 intermediates in the rumen. The employment of other and more powerful molecular techniques, such as functional metagenomics, could be useful to clear the role of these uncultured bacteria in the BH pathway.
5. Conclusions
The use of chestnut and quebracho tannins in the diet of dairy ewes at a level below 2% DM reduced the extent of ruminal BH process, lowering SA concentration and enhancing the percentage of LA, LNA, VA, RA, and other 18:1 isomers. The changes observed in the FA profile were associated with changes in total bacteria and Butyrivibrio group communities, even if they were more evident in presence of quebracho. Bands that disappeared or increased in presence of tannins in the Butyrivibrio group DGGE profiles were related to many uncultivated species of Lachnospiraceae, suggesting that these yet not known species may play a role in BH of PUFA. Our study indicates that chestnut and quebracho tannins offer an interesting possibility of modulating favorably rumen bacterial lipid metabolism toward precursors of healthful FA, which are produced in mammary tissues of lactating ewes during milk fat synthesis.
Acknowledgments
The authors would like to thank the following staff of the University of Florence: Francesca Decorosi, Doria Benvenuti, and Antonio Pezzati for technical assistance. This work has been funded by means of the financial support of University of Florence (Fondi di Ricerca di Ateneo, 2013/2014/2015) and Gruppo Mauro Saviola Srl, Radicofani, Siena, Italy.
Abbreviations
- ADF:
Acid detergent fibre
- BH:
Biohydrogenation
- CLNA:
Conjugated linolenic acid
- CHT:
Chestnut tannins
- DM:
Dry matter
- CP:
Crude protein
- EE:
Ether extract
- FA:
Fatty acids
- FAME:
Fatty acid methyl esters
- LA:
Linoleic acid
- LNA:
Linolenic acid
- ME:
Metabolizable energy
- NEL:
Net energy for lactation
- NDF:
Neutral detergent fibre
- PUFA:
Polyunsaturated fatty acids
- QUE:
Quebracho tannins
- RA:
Rumenic acid
- SA:
Stearic acid
- VA:
Vaccenic acid
- VLA:
Vaccelenic acid.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Arianna Buccioni and Grazia Pallara are equal contributors.
Supplementary Materials
Supplementary Material 1: DGGE profiles of 16S rDNA PCR products obtained from DNA extracted from rumen liquor using primer for the total bacteria (F968GC-1401R). CTR, control diet (84 g kg−1 DM of soybean oil); CHT, chestnut tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a chestnut tannin extract); QUE, quebracho tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a quebracho tannin extract); M, marker used for normalization of bands. Supplementary Material 2: DGGE profiles of 16S rDNA PCR products obtained from DNA extracted from rumen liquor using primer for the Butyrivibrio group (F968GC-B fib). CTR, control diet (84 g kg−1 DM of soybean oil); CHT, chestnut tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a chestnut tannin extract); QUE, quebracho tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a quebracho tannin extract); M, marker used for normalization of bands.
References
- 1.Patra A. K., Saxena J. Exploitation of dietary tannins to improve rumen metabolism and ruminant nutrition. Journal of the Science of Food and Agriculture. 2011;91(1):24–37. doi: 10.1002/jsfa.4152. [DOI] [PubMed] [Google Scholar]
- 2.Vasta V., Yáñez-Ruiz D. R., Mele M., et al. Bacterial and protozoal communities and fatty acid profile in the rumen of sheep fed a diet containing added tannins. Applied and Environmental Microbiology. 2010;76(8):2549–2555. doi: 10.1128/AEM.02583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buccioni A., Pauselli M., Viti C., et al. Milk fatty acid composition, rumen microbial population, and animal performances in response to diets rich in linoleic acid supplemented with chestnut or quebracho tannins in dairy ewes. Journal of Dairy Science. 2015;98(2):1145–1156. doi: 10.3168/jds.2014-8651. [DOI] [PubMed] [Google Scholar]
- 4.Goel G., Puniya A. K., Aguilar C. N., Singh K. Interaction of gut microflora with tannins in feeds. Naturwissenschaften. 2005;92(11):497–503. doi: 10.1007/s00114-005-0040-7. [DOI] [PubMed] [Google Scholar]
- 5.Wanapat M., Kongmun P., Poungchompu O., et al. Effects of plants containing secondary compounds and plant oils on rumen fermentation and ecology. Tropical Animal Health and Production. 2012;44(3):399–405. doi: 10.1007/s11250-011-9949-3. [DOI] [PubMed] [Google Scholar]
- 6.Serra A., Mele M., La Comba F., Conte G., Buccioni A., Secchiari P. Conjugated Linoleic Acid (CLA) content of meat from three muscles of Massese suckling lambs slaughtered at different weights. Meat Science. 2009;81(2):396–404. doi: 10.1016/j.meatsci.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Buccioni A., Serra A., Minieri S., et al. Milk production, composition, and milk fatty acid profile from grazing sheep fed diets supplemented with chestnut tannin extract and extruded linseed. Small Ruminant Research. 2015;130:200–207. doi: 10.1016/j.smallrumres.2015.07.021. [DOI] [Google Scholar]
- 8.Correddu F., Gaspa G., Pulina G., Nudda A. Grape seed and linseed, alone and in combination, enhance unsaturated fatty acids in the milk of Sarda dairy sheep. Journal of Dairy Science. 2016;99(3):1725–1735. doi: 10.3168/jds.2015-10108. [DOI] [PubMed] [Google Scholar]
- 9.Paillard D., McKain N., Rincon M. T., Shingfield K. J., Givens D. I., Wallace R. J. Quantification of ruminal Clostridium proteoclasticum by real-time PCR using a molecular beacon approach. Journal of Applied Microbiology. 2007;103(4):1251–1261. doi: 10.1111/j.1365-2672.2007.03349.x. [DOI] [PubMed] [Google Scholar]
- 10.Boeckaert C., Vlaeminck B., Fievez V., Maignien L., Dijkstra J., Boon N. Accumulation of trans C18:1 fatty acids in the rumen after dietary algal supplementation is associated with changes in the Butyrivibrio community. Applied and Environmental Microbiology. 2008;74(22):6923–6930. doi: 10.1128/AEM.01473-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durmic Z., McSweeney C. S., Kemp G. W., Hutton P., Wallace R. J., Vercoe P. E. Australian plants with potential to inhibit bacteria and processes involved in ruminal biohydrogenation of fatty acids. Animal Feed Science and Technology. 2008;145(1-4):271–284. doi: 10.1016/j.anifeedsci.2007.05.052. [DOI] [Google Scholar]
- 12.Belenguer A., Toral P. G., Frutos P., Hervás G. Changes in the rumen bacterial community in response to sunflower oil and fish oil supplements in the diet of dairy sheep. Journal of Dairy Science. 2010;93(7):3275–3286. doi: 10.3168/jds.2010-3101. [DOI] [PubMed] [Google Scholar]
- 13.Castro-Carrera T., Toral P. G., Frutos P., et al. Rumen bacterial community evaluated by 454 pyrosequencing and terminal restriction fragment length polymorphism analyses in dairy sheep fed marine algae. Journal of Dairy Science. 2014;97(3):1661–1669. doi: 10.3168/jds.2013-7243. [DOI] [PubMed] [Google Scholar]
- 14.Harfoot C. G., Hazlewood G. P. Lipid metabolism in the rumen , In, The rumen microbial. In: ecosystem., Hobson P. N., Stewart C. S., editors. The rumen microbial ecosystem. Springer Netherlands; 1997. [Google Scholar]
- 15.Min B. R., Attwood G. T., Reilly K., et al. Lotus corniculatus condensed tannins decrease in vivo populations of proteolytic bacteria and affect nitrogen metabolism in the rumen of sheep. Canadian Journal of Microbiology. 2002;48(10):911–921. doi: 10.1139/w02-087. [DOI] [PubMed] [Google Scholar]
- 16.Sivakumaran S., Molan A. L., Meagher L. P., et al. Variation in antimicrobial action of proanthocyanidins from Dorycnium rectum against rumen bacteria. Phytochemistry. 2004;65(17):2485–2497. doi: 10.1016/j.phytochem.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 17.Hervás G., Pérez V., Giráldez F. J., Mantecón A. R., Almar M. M., Frutos P. Intoxication of sheep with quebracho tannin extract. Journal of Comparative Pathology. 2003;129(1):44–54. doi: 10.1016/S0021-9975(02)00168-8. [DOI] [PubMed] [Google Scholar]
- 18.Hervás G., Frutos P., Giráldez F. J., Mantecón Á. R., Álvarez Del Pino M. C. Effect of different doses of quebracho tannins extract on rumen fermentation in ewes. Animal Feed Science and Technology. 2003;109(1-4):65–78. doi: 10.1016/S0377-8401(03)00208-6. [DOI] [Google Scholar]
- 19.Burns R. E. Methods of tannin analysis for forage crop evaluation. Technical Bulletin N. S. Georgia Agricultural Experiment Stations. Athens, GA, USA, 1963. [Google Scholar]
- 20.Campo M., Pinelli P., Romani A. HPLC/DAD/MS characterization and antioxidant activity of sweet chestnut (Castaneasativa M.) fractions. Proceedings of the XXVI International Conference on Polyphenols; 2012; Florence Italy. pp. 135–136. [Google Scholar]
- 21. AOAC International,Official Methods of Analysis, International, Gaitersburg, MD, 1995.
- 22.van Soest P. J., Robertson J. B., Lewis B. A. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. Journal of Dairy Science. 1991;74(10):3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 23.Cannas A., Tedeschi L. O., Fox D. G., Pell A. N., Van Soest P. J. A mechanistic model for predicting the nutrient requirements and feed biological values for sheep. Journal of Animal Science. 2004;82(1):149–169. doi: 10.2527/2004.821149x. [DOI] [PubMed] [Google Scholar]
- 24.Folch J., Lees M., Sloane Stanley G. H. A simple method for the isolation and purification of total lipides from animal tissues. The Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 25.Christie W. W. A simple procedure for rapid transmethylation of glycerolipids and cholesteryl esters. Journal of Lipid Research. 1982;23(7):1072–1075. [PubMed] [Google Scholar]
- 26.Kramer J. K. G., Fellner V., Dugan M. E. R., Sauer F. D., Mossoba M. M., Yurawecz M. P. Evaluating acid and base catalysts in the methylation of milk and rumen fatty acids with special emphasis on conjugated dienes and total trans fatty acids. Lipids. 1997;32(11):1219–1228. doi: 10.1007/s11745-997-0156-3. [DOI] [PubMed] [Google Scholar]
- 27.Kramer J. K. G., Cruz-Hernandez C., Deng Z., Zhou J., Jahreis G., Dugan M. E. R. Analysis of conjugated linoleic acid and trans 18:1 isomers in synthetic and animal products. American Journal of Clinical Nutrition. 2004;79(6):1137S–1145S. doi: 10.1093/ajcn/79.6.1137S. [DOI] [PubMed] [Google Scholar]
- 28.Cruz-Hernandez C., Kramer J. K. G., Kraft J., et al. Systematic analysis of trans and conjugated linoleic acids in the milk and meat of ruminants. Advances in Conjugated Linoleic Acid Research. 2006;3:45–933. [Google Scholar]
- 29.Kramer J. K. G., Hernandez M., Cruz-Hernandez C., Kraft J., Dugan M. E. R. Combining results of two GC separations partly achieves determination of all cis and trans 16:1, 18:1, 18:2 and 18:3 except CLA isomers of milk fat as demonstrated using ag-ion SPE fractionation. Lipids. 2008;43(3):259–273. doi: 10.1007/s11745-007-3143-4. [DOI] [PubMed] [Google Scholar]
- 30.Destaillats F., Trottier J. P., Galvez J. M. G., Angers P. Analysis of α-linolenic acid biohydrogenation intermediates in milk fat with emphasis on conjugated linolenic acids. Journal of Dairy Science. 2005;88(9):3231–3239. doi: 10.3168/jds.S0022-0302(05)73006-X. [DOI] [PubMed] [Google Scholar]
- 31.Contarini G., Povolo M., Pelizzola V., Monti L., Lercker G. Interlaboratory evaluation of milk fatty acid composition by using different GC operating conditions. Journal of Food Composition and Analysis. 2013;32(2):131–140. doi: 10.1016/j.jfca.2013.08.008. [DOI] [Google Scholar]
- 32.Nübel U., Engelen B., Felsre A., et al. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. Journal of Bacteriology. 1996;178(19):5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim E. J., Huws S. A., Lee M. R. F., et al. Fish oil increases the duodenal flow of long chain polyunsaturated fatty acids and trans-11 18:1 and decreases 18:0 in steers via changes in the rumen bacterial community. Journal of Nutrition. 2008;138(5):889–896. doi: 10.1093/jn/138.5.889. [DOI] [PubMed] [Google Scholar]
- 34.Throbäck I. N., Enwall K., Jarvis Å., Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiology Ecology. 2004;49(3):401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 35. Macrogen Ltd, http://www.macrogen.co.
- 36.v2.1.1Technelysium Pty Ltd. http://www.technelysium.com.au/chromas_lite.htm.
- 37. http://decipher.cec.wisc.edu.
- 38.Zhang Z., Schwartz S., Wagner L., Miller W. A greedy algorithm for aligning DNA sequences. Journal of Computational Biology. 2000;7(1-2):203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 39. http://www.ncbi.nlm.nih.gov/BLAST.
- 40.Webster N. S., Taylor M. W., Behnam F., et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environmental Microbiology. 2010;12(8):2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larkin M. A., Blackshields G., Brown N. P., et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Van De Peer Y., De Wachter R. Treecon for windows: a software package for the construction and drawing of evolutionary trees for the microsoft windows environment. Bioinformatics. 1994;10(5):569–570. doi: 10.1093/bioinformatics/10.5.569. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.SAS Institute. SAS User’s Guide: Statistics Version 8.0. SAS Institute Inc, Cary, NC, USA, 1999.
- 45.Pastorelli R., Landi S., Trabelsi D., et al. Effects of soil management on structure and activity of denitrifying bacterial communities. Applied Soil Ecology. 2011;49(1):46–58. doi: 10.1016/j.apsoil.2011.07.002. [DOI] [Google Scholar]
- 46. http://folk.uio.no/ohammer/past.
- 47.Lagomarsino A., Agnelli A. E., Pastorelli R., Pallara G., Rasse D. P., Silvennoinen H. Past water management affected GHG production and microbial community pattern in Italian rice paddy soils. Soil Biology & Biochemistry. 2016;93:17–27. doi: 10.1016/j.soilbio.2015.10.016. [DOI] [Google Scholar]
- 48.Fievez V., Colman E., Castro-Montoya J. M., Stefanov I., Vlaeminck B. Milk odd- and branched-chain fatty acids as biomarkers of rumen function-an update. Animal Feed Science and Technology. 2012;172(1-2):51–65. doi: 10.1016/j.anifeedsci.2011.12.008. [DOI] [Google Scholar]
- 49.Zhang N., Wan S., Li L., Bi J., Zhao M., Ma K. Impacts of urea N addition on soil microbial community in a semi-arid temperate steppe in northern China. Plant and Soil. 2008;311(1-2):19–28. doi: 10.1007/s11104-008-9650-0. [DOI] [Google Scholar]
- 50.Jones G. A., McAllister T. A., Muir A. D., Cheng K.-J. Effects of sainfoin (Onobrychis viciifolia Scop.) condensed tannins on growth and proteolysis by four strains of ruminal bacteria. Applied and Environmental Microbiology. 1994;60(4):1374–1378. doi: 10.1128/aem.60.4.1374-1378.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Min B. R., Attwood G. T., McNabb W. C., Molan A. L., Barry T. N. The effect of condensed tannins from Lotus corniculatus on the proteolytic activities and growth of rumen bacteria. Animal Feed Science and Technology. 2005;121(1-2):45–58. doi: 10.1016/j.anifeedsci.2005.02.007. [DOI] [Google Scholar]
- 52.Polan C. E., McNeill J. J., Tove S. B. Biohydrogenation of unsaturated fatty acids by rumen bacteria. Journal of Bacteriology. 1964;88:1056–1064. doi: 10.1128/jb.88.4.1056-1064.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harfoot C. G., Noble R. C., Moore J. H. Factors influencing the extent of biohydrogenation of linoleic acid by rumen micro‐organisms in vitro. Journal of the Science of Food and Agriculture. 1973;24(8):961–970. doi: 10.1002/jsfa.2740240814. [DOI] [PubMed] [Google Scholar]
- 54.John Wallace R., Chaudhary L. C., McKain N., et al. Clostridium proteoclasticum: A ruminal bacterium that forms stearic acid from linoleic acid. FEMS Microbiology Letters. 2006;265(2):195–201. doi: 10.1111/j.1574-6968.2006.00487.x. [DOI] [PubMed] [Google Scholar]
- 55.Moon C. D., Pacheco D. M., Kelly W. J., et al. Reclassification of Clostridium proteoclasticum as Butyrivibrio proteoclasticus comb. nov., a butyrate-producing ruminal bacterium. International Journal of Systematic and Evolutionary Microbiology. 2008;58(9):2041–2045. doi: 10.1099/ijs.0.65845-0. [DOI] [PubMed] [Google Scholar]
- 56.Huws S. A., Kim E. J., Lee M. R. F., et al. As yet uncultured bacteria phylogenetically classified as Prevotella, Lachnospiraceae incertae sedis and unclassified Bacteroidales, Clostridiales and Ruminococcaceae may play a predominant role in ruminal biohydrogenation. Environmental Microbiology. 2011;13(6):1500–1512. doi: 10.1111/j.1462-2920.2011.02452.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material 1: DGGE profiles of 16S rDNA PCR products obtained from DNA extracted from rumen liquor using primer for the total bacteria (F968GC-1401R). CTR, control diet (84 g kg−1 DM of soybean oil); CHT, chestnut tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a chestnut tannin extract); QUE, quebracho tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a quebracho tannin extract); M, marker used for normalization of bands. Supplementary Material 2: DGGE profiles of 16S rDNA PCR products obtained from DNA extracted from rumen liquor using primer for the Butyrivibrio group (F968GC-B fib). CTR, control diet (84 g kg−1 DM of soybean oil); CHT, chestnut tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a chestnut tannin extract); QUE, quebracho tannins diet (84 g kg−1 DM of soybean oil plus 52.8 g kg−1 DM of a quebracho tannin extract); M, marker used for normalization of bands.