Abstract
We report the present knowledge about RPHM21, a novel male-specific mitochondrial protein with a putative role in the paternal inheritance of sperm mitochondria in the Manila clam Ruditapes philippinarum, a species with doubly uniparental inheritance of mitochondria (DUI). We review all the available data on rphm21 transcription and translation, analyze in detail its female counterpart, RPHF22, discuss the homology with RPHM21, the putative function and origin, and analyze their polymorphism. The available evidence is compatible with a viral origin of RPHM21 and supports its activity during spermatogenesis. RPHM21 is progressively accumulated in mitochondria and nuclei of spermatogenic cells, and we hypothesize it can influence mitochondrial inheritance and sexual differentiation. We propose a testable model that describes how the acquisition of selfish features by a mitochondrial lineage might have been responsible for the emergence of DUI, and for the evolution of separate sexes (gonochorism) from hermaphroditism. The appearance of DUI most likely entailed the invasion of at least 1 selfish element, and the extant DUI systems can be seen as resolved conflicts. It was proposed that hermaphroditism was the ancestral condition of bivalves, and a correlation between DUI and gonochorism was documented. We hypothesize that DUI might have driven the shift from hermaphroditism to gonochorism, with androdioecy as transition state. The invasion of sex-ratio distorters and the evolution of suppressors can prompt rapid changes among sex-determination mechanisms, and DUI might have been responsible for one of such changes in some bivalve species. If true, DUI would represent the first animal sex-determination system involving mtDNA-encoded proteins.
Keywords: bivalve mollusc, doubly uniparental inheritance (DUI), endogenized virus, germ line, gonochorism, hermaphroditism
The common concept that animal mitochondrial genome (mtDNA) content and architecture are highly conserved is mostly due to a biased sampling: over 94% of the completely sequenced mtDNAs present in GenBank belongs to vertebrates (of which 85% are mammalian and 77% are human; Breton et al. 2014). Recently, the diversity of analyzed taxa has started to increase and several exceptions to the ‘usual’ gene content have been documented, showing that ‘non-canonical’ mtDNA genes are more common than previously imagined (see Breton et al. 2014 for a review). In most cases, the evidence of a protein product is still missing, so putative novel genes are predicted by: (1) the existence of an open reading frame (ORF), (2) sequence conservation, and (3) absence of nonsense mutations. Very often, ontology and function of these ORFs are unknown, so they are referred to as ORFans (Fischer and Eisenberg 1999).
Bivalve molluscs have an extremely fast-evolving mitochondrial genome, showing high-sequence polymorphism and divergence, variable architecture, high proportion of unassigned regions, and widespread presence of ORFans (Gissi et al. 2008; Ghiselli et al. 2013). That said, the most noteworthy feature of bivalve mitochondrial biology is the doubly uniparental inheritance (DUI) of mitochondria (Skibinski et al. 1994a, 1994b; Zouros et al. 1994a, 1994b) reported, so far, in about 50 species. In animals, DUI is the only known evolutionarily stable exception to the common strictly maternal inheritance (SMI) of mitochondria. Under DUI, mitochondria follow two distinct inheritance paths: 1 type of mitochondria is inherited through females (F type), the other through males (M type). The zygote receives both the mtDNA types then, if the embryo develops into a female, the M type is lost (degraded or diluted), and the adult will be homoplasmic for the F type. Conversely, if the embryo develops into a male, the M type is retained, it becomes the predominant mtDNA type in the gonad, and sperm will be homoplasmic for the M type. The somatic tissues of males show variable proportions of M and F type, depending on tissue and species (Ghiselli et al. 2011). It has to be noted that DUI is not a case of biparental inheritance, since each mtDNA lineage experiences a uniparental transmission. F and M mtDNAs are highly differentiated, with an amino acid divergence up to 50% (see Zouros 2013 for a thorough review of DUI). DUI represents an exceptional model for studying multiple aspects of mitochondrial biology (Passamonti and Ghiselli 2009; Milani et al. 2011; Breton et al. 2014), and since its discovery, the main focus has been to unveil the origin and the molecular mechanism behind it. The data gathered so far are consistent with DUI being originated from SMI by modification of the molecular machinery of SMI (Breton et al. 2007; Zouros 2013; Milani and Ghiselli 2015), but the underlying factors are still unknown. A fair amount of research was done on the mitochondrial genomes of DUI species to identify elements that could be responsible for, or involved in, the modification of the inheritance mechanism, and some good candidates have been spotted. Lineage specific mtORFans were identified in all the analyzed DUI species: fORFans specific to the F mtDNA, and mORFans specific to the M mtDNA have been found in the families Unionidae (Breton et al. 2009; Breton et al. 2011a), Mytilidae (Breton et al. 2011b), and Veneridae (Ghiselli et al. 2013; Milani et al. 2013a). Given the absence of identifiable homologs in databases, the function of these mtORFans has to be inferred by the presence of short regions showing similarity with known structural motifs. An in silico meta-analysis (Milani et al. 2013a) has highlighted some interesting patterns: features shared by MORFs (we refer to the translations of fORFs and mORFs as FORF and MORF, respectively) point to a role in the targeting of M-type mitochondria to germ line precursor blastomeres through interactions with the cytoskeleton. Moreover, both FORF and MORF features point to a role in nucleic acid binding and transcription regulation, and, most strikingly, all the analyzed mtORFans showed signatures that are consistent with a viral origin. Up to now, evidence for a protein product has been provided for only three mtORFans: RPHM21, specific to the M mtDNA of the Manila clam Ruditapes philippinarum, and the F and M mtORFans (here reported as Vel-FORF and Vel-MORF) specific to the freshwater mussel Venustaconcha ellipsiformis (Breton et al. 2009, 2011a). Immunoelectron microscopy and confocal microscopy showed that Vel-FORF and RPHM21 are localized in both mitochondria and nucleus (Breton et al. 2011a; Milani et al. 2014a). Milani et al. (2014a) found similarities between RPHM21 and MK3, a member of modulators of immune recognition (MIRs), which are viral proteins involved in immune recognition functioning as ubiquitin ligases (Coscoy and Ganem 2003). RPHM21 and MK3 show structural similarities, are both localized in mitochondria and nucleus, and share putative functions such as reorganization of cytoskeleton, cell migration, cell cycle control, chromatin remodeling, and transcriptional control (Kurz et al. 2002; Ronkina et al. 2007, 2008; Milani et al. 2013a, 2014a). Following these observations, Milani et al. (2014a) proposed that: (1) M-specific elements may have a role in preventing the recognition of male-transmitted mitochondria by the degradation machinery, thus allowing their survival in male embryos, and their preferential transmission to the progeny; (2) these elements might also function as transcriptional regulators.
The aim of this article is to report the present knowledge about RPHM21, proposed to be one of the factors responsible for paternal inheritance of sperm mitochondria in R. philippinarum. The evidence collected so far strongly supports the activity of this protein during spermatogenesis, and we hypothesize that the viral sequence provided infected mitochondria with the ability to escape the degradation process in early embryos, invading the germ line, and being preferentially transmitted to the progeny. In the present work we: (1) review all the available data on rphm21 transcription and translation, providing new in situ hybridization and confocal data; (2) analyze in detail the F-type specific mtORFan RPHF22, discussing its homology with RPHM21, its putative function, and its origin; (3) analyze the M-type and F-type polymorphism; (4) propose a testable model that describes how the acquisition of selfish features by a mitochondrial lineage (through endogenization of a viral sequence in its mtDNA) might have been responsible for the emergence of an aberrant mitochondrial inheritance system, and for the evolution of separate sexes (condition usually referred to as gonochorism in animals and dioecy in plants) from hermaphroditism.
Materials and Methods
Real-Time qPCR data
A comparative analysis of rphm21 transcript amount obtained with Real-Time qPCR on young and adult male specimens was performed to highlight possible differences between tissues and developmental stages. The target (129 bp sequence) was quantified using the primers Rph_SYBR_orf21_ forward TCTGTGAAAGGAAACCCATGTGAG and Rph_SYBR_orf21_ reverse ACTAATAATAATTGGAGCCGAATAAACTTG (Milani et al. 2014b, 2015). In total, data from 112 male samples were used: 28 juveniles (whole bodies), 16 gonads from gametogenic adults (adults during gonad maturation), 20 gonads of ripe adults (adults with mature gametes), 24 adductor muscles, and 24 mantles. Details about experimental design and quantification method are available in Milani et al. (2014b, 2015). Statistical analyses and plots were performed with R 3.1.1.
Microscopy
Mature adult specimens (gonads and surrounding tissues of 5 individuals, 3 males, and 2 females) were used for in situ hybridization with rphm21 riboprobes and immunological localization of RPHM21 in tissues to document the presence and localization of transcript and protein in tissues. A digoxigenin (DIG)-labeled riboprobe antisense to rphm21 transcript was obtained using the Roche in vitro transcription labeling protocol (Roche DIG RNA labeling kit). Cryosections of gonads of 10–20 µm were used for in situ hybridization, following the method in Milani et al. (2013b, 2014a). Samples were observed with a Nikon Eclipse 80i microscope and images were captured using NIS-Elements D3.2 software.
An anti-RPHM21 antibody was used to immunolocalize the male-specific mitochondrial protein, following the method in Milani et al. (2014a). Nuclei were stained with 1 mM TO-PRO3 (Molecular Probes). Some sections were incubated with a monoclonal anti-alpha-tubulin (clone DM1A; Sigma) (method as in Milani et al. 2011) to stain microtubules and to better show the structure of both male and female acinus (i.e., the structural unit of bivalve gonad; a gonad is formed by thousands of acini). Imaging was recorded by a confocal laser scanning microscope (Leica confocal SP2 microscope), using Leica software.
In silico analyses on RPHM21 and RPHF22
We performed in silico analyses to better characterize the novel mitochondrial elements RPHM21 and RPHF22 because they were reported not to show a clear sequence similarity with known proteins. The tertiary structure of RPHM21 and RPHF22 was predicted using I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER; Zhang 2008). The best model in pdb format was used as input for the Chimera 1.8.1 software (Pettersen et al. 2004) to obtain the 3D structure.
Alignments between RPHM21 and RPHM22 were performed with TM-COFFEE (http://tcoffee.crg.cat/apps/tcoffee/do:tmcoffee; Chang et al. 2012), which is specifically designed to align transmembrane proteins, and with HMMER 3.1b2 (Finn et al. 2011). HMMER uses profile hidden Markov models (profile HMMs; Eddy 1998) to perform alignments. Profile HMM converts an alignment into a position-specific scoring system that is particularly efficient in searches for remote homology. To obtain specific-profile HMMs for RPHM21 and RPHF22, we obtained an amino acid sequence alignment for each protein using Clustal Omega (Sievers et al. 2011), and then we used the hmmbuild tool of the HMMER 3.1b2 package. We then aligned the 2 proteins using hmmalign bidirectionally (RPHM21 profile HMM vs. RPHF22 and vice versa).
Interproscan5 (http://www.ebi.ac.uk/Tools/pfa/iprscan5; Jones et al. 2014) was used to search InterPro signatures in RPHM21 and RPHF22.
Polymorphism analysis was performed on F and M mitochondrial genes for which the number of sequences available was ≥10, namely: atp6, cox3, nd3, nd4, nd5, nd6 (F type n = 12; M type n = 11), rphm21 (n = 16), rphf22 (n = 20). We combined data obtained by Sanger sequencing (F type: AB065375, AF484332-36, KC243324-31; M type: AB065374, AF484337-40, KC243347-53) with data obtained from a single nucleotide polymorphism (SNP) analysis performed on RNA-Seq data (BioProject PRJNA68513, Ghiselli et al. 2012). SNPs were called from gonad transcriptomes of 6 males and 6 females. Details about SNP calling procedure are in Ghiselli et al. (2013). To obtain alignable FASTA sequences from the VCF file containing the SNP informations, we first prepared the reference FASTA by building an index with the Samtools faidx tool (Li et al. 2009), and creating a sequence dictionary with the CreateSequenceDictionary program from PicardTools (http://broadinstitute.github.io/picard). Then we used the FastaAlternateReferenceMaker command from the program GenomeAnalysisTK of the GATK distribution 3.4-46 (McKenna et al. 2010). Later, we used ClustalW to align the FASTA files containing the alternative alleles obtained from the SNP calling with the Sanger sequences. Nucleotide and amino acid p-distances, number of synonymous substitutions for synonymous sites (synonymous polymorphism, pS), and number of nonsynonymous substitutions per nonsynonymous sites (nonsynonymous polymorphism, pN) were calculated using MEGA 5 (Tamura et al. 2011) with the following settings: 1,000 bootstrap replicates, transitions+transversions, complete deletion, Kumar method.
Results
Real-Time qPCR
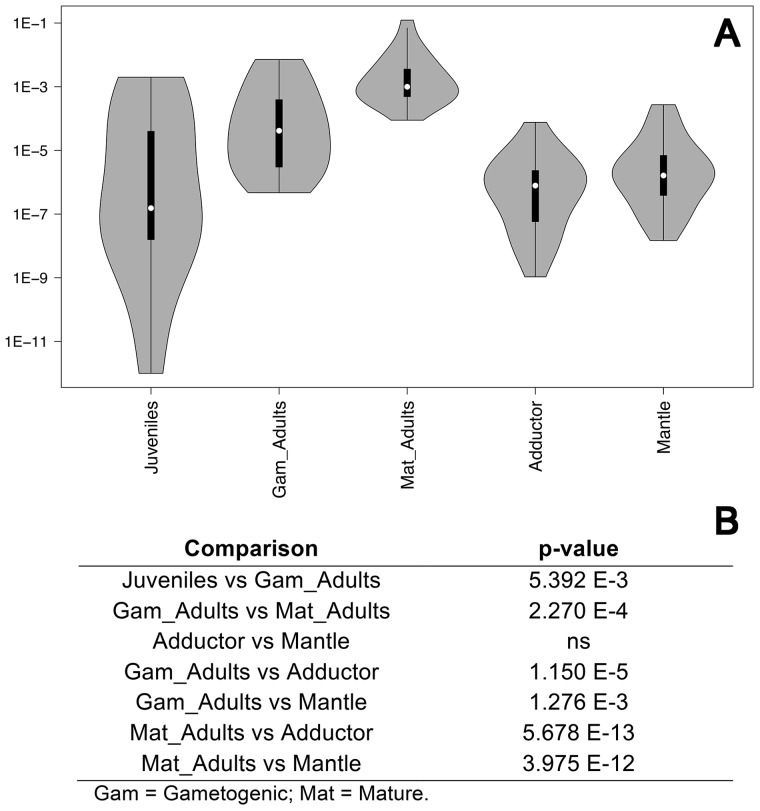
This analysis was performed in male individuals only; sex in juveniles was assigned by the presence of the M-type genome, while in adults by microscope examination of gonadic tissue. Real-time qPCR data indicate that rphm21 transcript abundance increases from the time of gonad formation (juveniles) to the fully mature gonad of ripe adults (Figure 1). rphm21 transcript amount is lower and uniform in the 2 somatic tissues analyzed (Figure 1).
Figure 1.
rphm21 transcript amount. (A) Violin plots of rphm21 transcript amount obtained by Real-Time qPCR. Sample sizes: 28 juveniles, 16 gonads from gametogenic adults, 20 gonads of mature adults, 24 adductor muscles, and 24 mantles. Y-axis: log10 copy number relative to the nuclear endogenous control (18S) (see Milani et al. 2014b, 2015 for details). (B) Statistical significance of transcript amount differences; p-values obtained with Wilcoxon Rank-Sum test.
In situ hybridization
Figure 2 shows the morphology of male and female acini. Consistently with what was previously described (Milani et al. 2014a), rphm21 transcripts are localized in male gonads (Figure 3). Many more spermatozoa are produced in male acini compared to eggs in female acini (50:1, spermatozoa:eggs per z-section at the confocal microscope; Figure 2). In male acini, the transcript is present in early spermatogenic cells along the acinus wall (Figure 3A,B), then, during spermiogenesis, it accumulates in spermatozoa that fill the acinus lumen when gonads are mature. The transcript is not detected in female acini (Figure 3C).
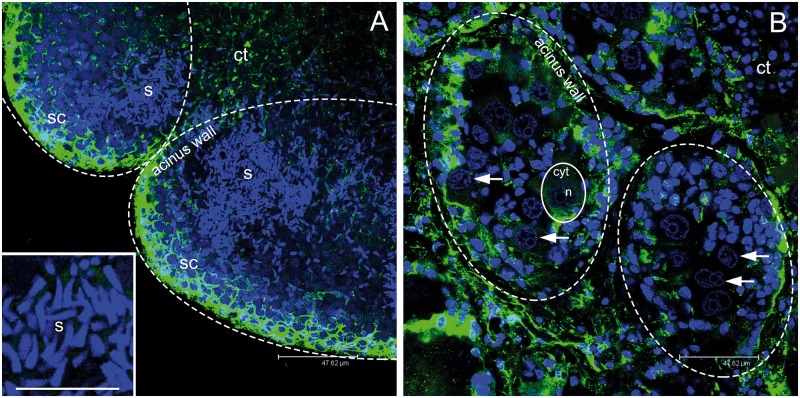
Figure 2.
Gonadic structure of Ruditapes philippinarum. (A) Two adjacent male acini at confocal microscope showing many spermatozoa (s; around 500 gametes per z-section). Magnification of sperm nuclei, in light blue, in the inset. (B) Two adjacent female acini (dashed ovals) at confocal microscope showing up to 10 eggs per section (1 egg is lined with a solid oval). The nucleus of some eggs is indicated by an arrow. (A, B scale bar = 47.62 µm; A inset scale bar = 16.26 µm). s = spermatozoa; sc = spermatogenic cells along the acinus wall; n = egg nucleus; cyt = egg cytoplasm; ct = connective tissue. Microtubules in green; TO-PRO3 nuclear dye in blue.
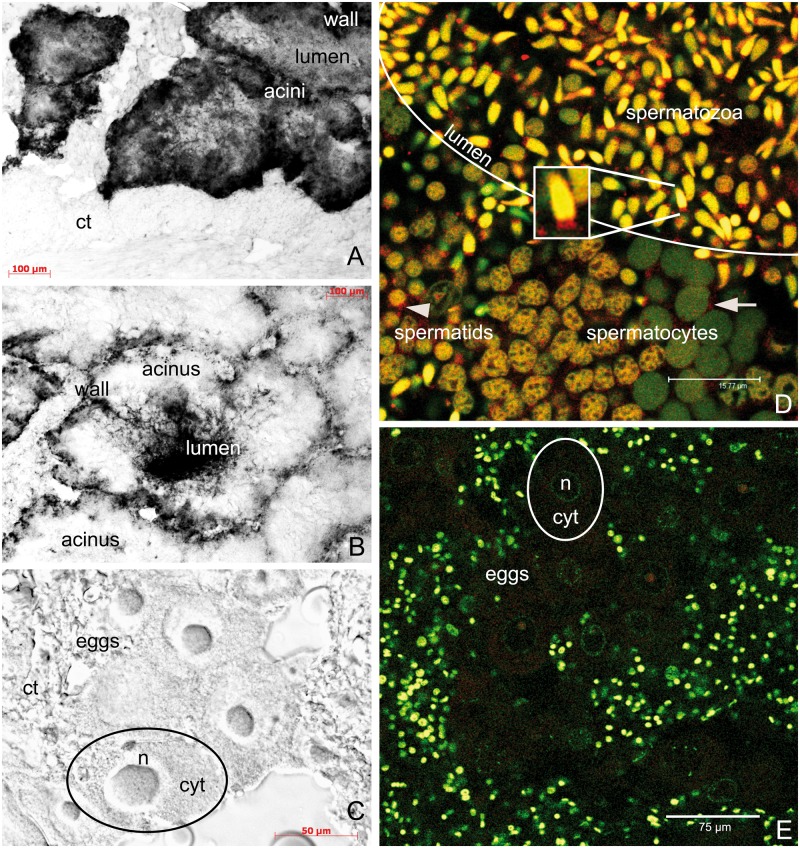
Figure 3.
Localization of rphm21 products. (A–C) rphm21 transcript localization with in situ hybridization in male (A, B) and female (C) gonadic tissue of the Manila clam Ruditapes philippinarum. (A) Male immature acinus in which rphm21 riboprobe labels spermatogenic cells along the acinus wall (positive signal in black). (B) Male mature acinus in which spermatozoa stored in the acinus lumen are deeply stained with rphm21 riboprobe. (A, B scale bars = 100 µm). (C) No staining is present in eggs (C scale bar = 50 µm). n = egg nucleus; cyt = egg cytoplasm; ct = connective tissue. (D, E) RPHM21 protein localization in male and female gonadic tissue, respectively. (D) In the male gonad, anti-RPHM21 (in red) labeled a clear spot at 1 side of the nucleus of spermatocytes and spermatids (arrow and arrowhead, respectively). Anti-RPHM21 staining is strong in mature spermatozoa in the acinus lumen in both mitochondrial midpiece and nucleus (yellow due to the colocalization of the nuclear dye, in green, and the antibody, in red). Initially, in spermatocytes the nuclei are visible in green, and no RPHM21 appears to be stored in the nucleus; in spermatids the nuclei are brownish as indicating RPHM21 storing (clearly seen in the bottom of the image going from the right to the left). (D scale bar = 15.77 µm). (E) Eggs do not show anti-RPHM21 detectable staining. The smaller nuclei of somatic cells surrounding acini are also visible. (E scale bars = 75 µm). RPHM21 immunolabeling in red; TO-PRO3 nuclear dye in green.
A-C: optical microscope. D, E: confocal microscope.
Immunohistochemistry
In the male gonad, anti-RPHM21 labels a clear spot at one side of the nucleus of spermatocytes and spermatids (Figure 3D). Anti-RPHM21 staining is also present in spermatozoa that fill the acinus (Figure 3D), showing the staining both in the sperm mitochondrial midpiece and in the nucleus (Figure 3D, box). The nuclei of spermatogenic cells appear with different color, depending on the different proportion of colocalizing fluorescence signals (TO-PRO3, in green, and anti-RPHM21, in red). RPHM21 protein is not detected in female acini and eggs (Figure 3E).
In silico analyses
I-TASSER proposed 5 models for RPHM21 and RPHF22. The C-score for the inferred proteins was in the range [−4.31, −4.96] and [−1.96, −5. 00], respectively. The 10 threading templates used by I-TASSER to model RPHF22 and RPHM21 are reported in Table 1. The results of InterProScan 5, TM-COFFEE, and HMMER are shown in Figure 4A–D. 3D models of RPHM21 and RPHF22 obtained using structures predicted by I-TASSER are shown in Figure 4E,F.
Table 1.
Threading templates used by I-TASSER to model RPHF22 and RPHM21
| Template | RCSB protein data bank | Description | Classification |
|---|---|---|---|
| RPHF22 | |||
| 1 | 3J5P | TRPV1 ion channel | Transport protein |
| 2 | 3DM8 | Putative Isomerase | Unknown function |
| 3 | 3UJM | NTF2-like domain (Rasputin protein) | Signaling protein |
| 4 | 1U5O | Nuclear transport carrier NTF2 | Transport protein |
| 5 | 2JNE | YfgJ modeled with 2 Zn+2 bound | Metal binding protein |
| 6 | 4TPS | Sporulation inhibitor of DNA replication (SirA) | Replication |
| 7 | 3UJM | NTF2-like domain (Rasputin protein) | Signaling protein |
| 8 | 2VXR | Botulinum neurotoxin serotype G-binding domain | Toxin |
| 9 | 3VNE | Ebolavirus protein VP24 | Viral protein |
| 10 | 4TPS | Sporulation inhibitor of DNA replication (SirA) | Replication |
| RPHM21 | |||
| 1 | 2ACW | UGT71G1 complexed with UDP-glucose | Transferase |
| 2 | 2NPI | Clp1-ATP-Pcf11 complex | Transcription |
| 3 | 1MKF | Viral chemokine binding protein (gammaherpesvirus 68) | Immune system |
| 4 | 3DFR | Dihydrofolate reductase | Oxido-reductase |
| 5 | 3TU5 | Actin complex with gelsolin segment 1 | Structural protein/actin-binding protein |
| 6 | 3DBA | cGMP-bound GAF a domain | Hydrolase |
| 7 | 4H51 | Putative aspartate aminotransferase | Transferase |
| 8 | 3FL7 | Ephrin A2 ectodomain | Transferase signaling protein |
| 9 | 4EE6 | Novel phenazine prenyltransferase EpzP | Transferase |
| 10 | 4CR4 | 26S proteasome | Hydrolase |
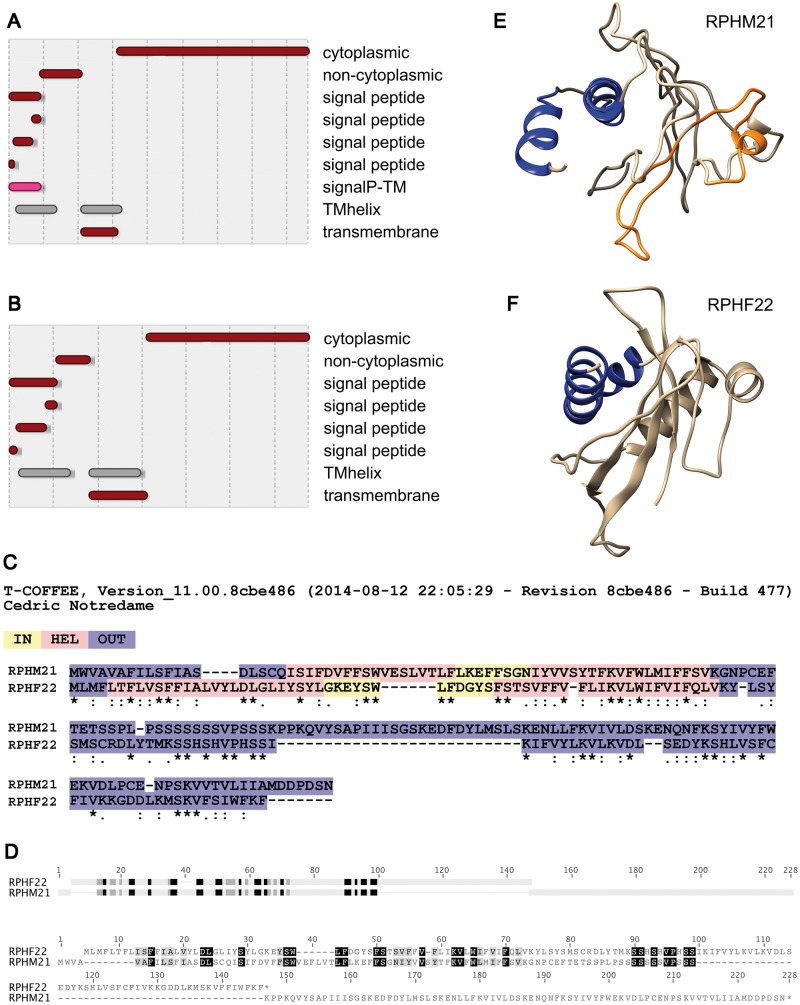
Figure 4.
Structural analysis of RPHM21 and RPHF22. (A, B) Protein domains detected with InterProScan 5: RPHM21 (A) and RPHF22 (B) both show transmembrane domains in their N-terminus, whereas the C-terminus is cytoplasmic. (C) Similarities in domain localization detected with TM-COFFEE alignment. (D) HMMER detected good alignment of profile HMMs in correspondence of the transmembrane domains. (E, F) 3D models of RPHM21 (E) and RPHF22 (F) obtained using structures predicted by I-TASSER.
Polymorphism analysis (Table 2) shows that both rphm21 and rphf22 are quite conserved, and that, compared with other mitochondrial genes, their synonymous and nonsynonymous polymorphism is low. rphm21 has the lowest nonsynonymous polymorphism of all (both F and M mtDNAs) the analyzed genes (pN = 0.003, 95% CI [0.000, 0.007]).
Table 2.
Polymorphism of M-type and F-type mtDNAs
| gene | pD nt | 95% CI | pD nt | 95% CI | pD aa | 95% CI | pD aa | 95% CI |
|---|---|---|---|---|---|---|---|---|
| F type | M type | F type | M type | |||||
| atp6 | 0.016 | 0.012–0.020 | 0.019 | 0.013–0.025 | 0.019 | 0.011–0.027 | 0.020 | 0.010–0.030 |
| cox3 | 0.018 | 0.014–0.022 | 0.021 | 0.015–0.027 | 0.025 | 0.015–0.035 | 0.026 | 0.016–0.036 |
| nd3 | 0.013 | 0.007–0.019 | 0.006 | 0.002–0.010 | 0.021 | 0.009–0.033 | 0.013 | 0.000–0.027 |
| nd4 | 0.015 | 0.011–0.019 | 0.013 | 0.009–0.017 | 0.022 | 0.016–0.028 | 0.015 | 0.009–0.021 |
| nd5 | 0.008 | 0.006–0.010 | 0.016 | 0.012–0.020 | 0.012 | 0.008–0.016 | 0.014 | 0.008–0.020 |
| nd6 | 0.019 | 0.013–0.025 | 0.018 | 0.012–0.024 | 0.025 | 0.013–0.037 | 0.025 | 0.011–0.039 |
| rphf22 | 0.009 | 0.003–0.015 | NA | NA | 0.014 | 0.004–0.024 | NA | NA |
| rphm21 | NA | NA | 0.006 | 0.002–0.010 | NA | NA | 0.007 | 0.001–0.013 |
| gene | pS | 95% CI | pS | 95% CI | pN | 95% CI | pN | 95% CI |
| F type | M type | F type | M type | |||||
| atp6 | 0.029 | 0.015–0.043 | 0.031 | 0.017–0.045 | 0.011 | 0.005–0.017 | 0.012 | 0.006–0.018 |
| cox3 | 0.031 | 0.017–0.045 | 0.037 | 0.021–0.053 | 0.013 | 0.007–0.019 | 0.015 | 0.009–0.021 |
| nd3 | 0.016 | 0.004–0.028 | 0.005 | 0.000–0.013 | 0.011 | 0.003–0.019 | 0.006 | 0.000–0.012 |
| nd4 | 0.027 | 0.017–0.037 | 0.021 | 0.013–0.029 | 0.012 | 0.008–0.016 | 0.008 | 0.004–0.012 |
| nd5 | 0.013 | 0.005–0.021 | 0.034 | 0.024–0.044 | 0.005 | 0.003–0.007 | 0.008 | 0.004–0.012 |
| nd6 | 0.037 | 0.019–0.055 | 0.020 | 0.006–0.034 | 0.012 | 0.006–0.018 | 0.013 | 0.005–0.021 |
| rphf22 | 0.020 | 0.002–0.038 | NA | NA | 0.007 | 0.001–0.013 | NA | NA |
| rphm21 | NA | NA | 0.011 | 0.001–0.021 | NA | NA | 0.003 | 0.000–0.007 |
Note: pD nt = nucleotide p-distance; pD aa = amino acid p-distance; 95% CI = 95% confidence interval; pS = synonymous polymorphism (number of synonymous substitutions per synonymous site); pN = nonsynonymous polymorphism (number of nonsynonymous substitutions per nonsynonymous site); NA = not available. Only genes with ≥ 10 sequences available were utilized in the analyses.
Discussion
Lineage-specific mitochondrial elements and DUI
In the absence of proper functional studies, the role of RPHM21 and of other DUI mtORFans can be inferred only through in silico predictions and bioinformatics analyses. In the last years, several works have provided new insights about these novel mitochondrial elements (Breton et al. 2011a, 2011b, 2014; Ghiselli et al. 2013; Milani et al. 2013a, 2014a, 2015), but clear evolutionary and functional patterns have not emerged yet. The difference among these elements might be the result of divergent evolution after a common origin, or it might be indicative of an independent origin; in the latter case the genes involved in DUI would be taxon-specific. Accordingly, and alternatively to what hypothesized earlier (see Zouros 2013), it was recently proposed that DUI might have evolved multiple times through the endogenization of different viral elements in the mitochondrial genome of some bivalve species (Milani et al. 2013a), thus explaining the scattered distribution of DUI in the class Bivalvia (Zouros 2013) and the higher difference in amino acid sequence among the above-mentioned lineage-specific mitochondrial elements when compared to standard mitochondrial proteins (see Figure 5 in Milani et al. 2013a). If so, DUI in different species could be achieved by different modifications of SMI, and the genes involved could be nonhomologous.
M-type specific RPHM21
The sequence of rphm21 was reported to be quite conserved (Ghiselli et al. 2013), and the new analysis performed on additional sequences (both from Sanger sequencing and RNA-Seq SNP calling) confirms this observation. Both the nucleotide and amino acid p-distances are among the lowest of all the analyzed mitochondrial genes (see Table 1) and the synonymous–nonsynonymous polymorphism indicates negative selection (pS = 0.011, 95% CI [0.001, 0.021]; pN 0.003, 95% CI [0.000, 0.007]). All the data collected so far show that rphm21 transcript is more abundant in gonads of ripe adults, when compared with juveniles and gametogenic adults (Figure 1), and that rphm21 transcript is localized in gametes, as the protein RPHM21 (Figure 3). The transcript is already present in early spermatogenic cells, alongside the acinus wall where spermatogonia are located, and it remains detectable in spermatozoa, as shown by the riboprobe labeling the acinus lumen (Figure 3A–C). The protein appears to be produced and stored in the cytoplasm portion in which the mitochondrial midpiece forms, as shown by the red spot at one side of the nucleus visible in some spermatocytes/spermatids (Figure 3). Initially the protein is not detectable in the nucleus (as shown by the green nuclear labeling in spermatocytes in Figure 3D bottom right), but as soon as the spermatogenic cells proceed with the differentiation process, RPHM21 appears to be progressively accumulated in the nucleus (as shown by the brown–green color of nuclei, that eventually turn to yellow in spermatozoa). RPHM21 storing in nuclei may indicate an increasing activity during spermatogenic cell differentiation, and the protein accumulation in spermatozoa might indicate a function after fertilization, during embryonic development. Indeed, given the mentioned dual localization (nuclear and mitochondrial), RPHM21 may be also involved in transcriptional regulation, enhancing, or repressing nuclear genes involved in mitochondrial inheritance and sexual differentiation. In this concern, recent analyses in R. philippinarum documented the presence of RPHM21 in a subset of male primordial germ cells (Milani et al. 2015). The fact that all the spermatozoa analyzed so far contain RPHM21 led to the hypothesis that only the fraction of germ cells expressing the male-specific mitochondrial element can differentiate in mature gametes, or are greatly advantaged in the process. It is also noteworthy that RPHM21 protein appears to be expressed since the first stage of embryo development: as observed, it is localized along the first cleavage furrow in 2-blastomere embryos, in the same position in which M-type mitochondria and germ line determinants are located (Milani et al. 2011, 2014a). The process of sperm mitochondria elimination/maintenance might include the activation/repression of nucleases that might act in mtDNA degradation. These nucleases were demonstrated to be responsible for depleting Drosophila melanogaster mtDNA from developing sperm, thus promoting maternal inheritance of mtDNA (DeLuca and O’Farrell 2012). Interestingly, viral nucleases mainly found in the nucleus, but also targeted to mitochondria, appear to participate with mitochondrial endonucleases in mtDNA degradation (Duguay and Smiley 2013).
F-type specific RPHF22
Ruditapes philippinarum female-specific mitochondrial ORF rphf22 is not deeply analyzed until now. Previous analyses using an algorithm for detecting remote homology produced a fairly good amino acid alignment between RPHM21 and the predicted protein RPHF22 (Milani et al. 2013a). Indeed, the 2 sequences, despite not showing high identity, are more similar to each other than to other sex-specific mtORFans found in other bivalves. rphf22 is shorter and with a lower amount of transcripts in female gonads in respect to the quantity of rphm21 transcripts in male gonads (Ghiselli et al. 2013), but its amino acid sequence appears to be quite conserved in the females analyzed so far (Table 2), and never shows any nonsense mutation. If we assume that rphm21 and rphf22 are actually the same endogenized element that diverged following the separation that occurred between the 2 mitochondrial genomes, we can speculate that in the 2 sexes the element progressively diverged and may be acquired of different sex-specific functions. Data consistent with the origin from the same endogenized element for RPHM21 and RPHF22 are: (1) the predicted structure: C-terminus containing helices and a N-terminus cytoplasmic region (Figure 4); (2) sequence similarity especially in the predicted transmembrane region (Figure 4 and Milani et al. 2013a); (3) presence of a polyserine domain in the same protein position (the domain is absent in the other analyzed mtORFans). Instead of a polyserine, in the species of the Mytilus edulis complex a poly-lysine domain is present. Both the domains support the possibility of association with membranes (Howard et al. 2004; Bouaouina et al. 2012). Interestingly, polyserine domains were proposed to be involved in the targeting of proteins to the nucleus (Wolf et al. 2013), and are also required for a normal viral gene expression (Bates and DeLuca 1998).
A model for the evolution of DUI and gonochorism
In the present article, first, we focused on the characterization, origin, and possible function of RPHM21 and RPHM22 and their likely derivation from the same selfish element. Starting from the assumption that the 2 elements have the same origin, we formulate a model on how selection on selfish mitochondria may have resulted in the appearance of DUI.
There is a strict correlation between the presence of DUI and gonochorism, indeed, hermaphrodites closely related to DUI species do not have DUI and show extensive mutations in the mitochondrial fORFs; as a consequence of this observation, a role of DUI lineage-specific mtORFs in the maintenance of gonochorism was hypothesized by Breton et al. (2011a). However, we still do not know if the strict correlation between mitochondrial inheritance and sex is actually causal or coincidental. Here we propose a model that describes a scenario under which DUI and gonochorism might have evolved together in some bivalve species.
Step 1
In a population of hermaphrodite bivalves, a virus infects some mitochondria and confers them the ability to avoid degradation in embryos, and to be preferentially transmitted through generations (segregation distortion).
Step 2
Infected mitochondria end up both in male and female germ lines, but we could expect that they spread more efficiently through sperm for the following reasons:
males produce more gametes than females. Actually, in R. philippinarum, as visible in cross sections of male and female acini, many more spermatozoa are produced in male acini compared to eggs in female acini (spermatozoa:eggs ratio indicatively 50:1; Figure 2). According to some authors, the majority of retroviral insertions are acquired by the male germ line due to the relatively high number of cell divisions involved in the production of sperm in comparison to eggs (Katzourakis et al. 2007);
even though eggs contain more mitochondria than sperm, the mtDNA copy number in R. philippinarum gonads is much higher in males than in females (at least 1 order of magnitude, see Ghiselli et al. 2011);
the integration of viral elements is easier in presence of a high replication rate, a condition that well matches mitochondrial proliferation during spermatogenesis. In fact, proliferating cells usually experience higher mitochondrial replication, with the only notable exception of the early stages of embryo development (Yamano and Youle 2011; Milani et al. 2012; Mishra and Chan 2014). This mitochondrial proliferation may be even stronger for germ cells, when a proper distribution and quality control of mitochondria is needed more;
the high incidence of mitochondrial fusion and fission during cell replication can be an additional mechanism allowing the element to spread in the mitochondrial population (Mitra 2013; Mishra and Chan 2014).
All that considered, given its specific feature, the spermatozoon appears to be more suited to incorporate and transmit mtDNA carrying the integrated element.
Step 3
The frequency of infected sperm increases in the population. If we assume that the viral sequence allows mitochondria containing it to survive and to be preferentially transported to the germ line (a behavior clearly visible for sperm mitochondria in R. philippinarum, Milani et al. 2012, and in other DUI species, Zouros 2013) and transmitted to the progeny, in the subsequent generations the viral element can widely spread in the population.
Step 4
The infection allowed both maternal and paternal inheritance of mitochondria. In this condition, mitochondria are under selection also for male functions, such as spermatogenesis and—most importantly for a broadcast spawning species—sperm swimming. So now mutations that increase male fitness have a high selection coefficient and can spread quickly in the population.
Step 5
Emergence of males and transition from hermaphroditism to androdioecy. Gonochorism can evolve when a unisexual mutant (i.e., carrying a female- or male-sterility mutation) invades the population and the remaining hermaphrodites specialize in the complementary unisexual type (Charlesworth and Charlesworth 1978). Theoretical models show that this transition can occur through 2 intermediate transitory stages: gynodioecy (females + hermaphrodites), and androdioecy (males + hermaphrodites). In animals, the transition from hermaphroditism to androdioecy is much more common (Weeks 2012). Unisexual mutants can be maintained in hermaphrodite populations if they gain a strong fitness advantage over the hermaphrodites (Charlesworth and Charlesworth 1978, Equation 10), and this can be achieved either by avoiding self-fertilization (thus inbreeding depression), or through a more efficient resource allocation: spermatozoa are cheaper to produce, so the reproductive success of males is size-independent, namely given the same energy allocated in reproduction, male relative fitness is higher (Charlesworth and Charlesworth 1978; Weeks et al. 2006; Weeks 2012).
Going back to our model, we think that in R. philippinarum 2 main factors contributed to the increase in male fitness that allowed the establishment of androdioecy: (1) bivalves show the kind of resource allocation discussed above, for example, in most hermaphrodite species, younger/smaller individuals are males, and they switch to females when they grow older/bigger (Wright 1998); (2) considering what we mentioned in Step 2, we expect a longer permanence of the selfish element in male germ cells than in female germ cells, so infected mtDNA will coevolve mostly with male-specific alleles and male-biased genes. This suggests that males with infected mitochondria (now under selection for male functions, see Step 4) may have an even higher fitness in respect to other males. That said, the evolution of androdioecy and eventually separate sexes can occur even in the absence of resource reallocation, if segregation distortion is present (Billiard et al. 2015).
Step 6
Evolution of gonochorism from androdioecy. The proportion of males increases in the population, until their initial fitness advantage is neutralized by the disadvantage of a male-biased population. In this condition, the selective pressure on egg production would be strong, and male-sterility mutation in hermaphrodites highly favored.
Step 7
Evolution of one or more restorer genes that counteract the action of RPHM21. The coevolutionary interactions of the nuclear genome with cytoplasmic sex-ratio distorters can lead to the evolution of compensatory genes, to restore a balanced sex ratio (Hurst 1992). Such genes might induce monogeny, which is a condition where all offspring of each individual female are either exclusively male or exclusively female (Werren and Beukeboom 1998). Interestingly, DUI species show sex-biased lineages (Kenchington et al. 2002; Ghiselli et al. 2012; Machordom et al. 2015): independent of the sire, there are females producing exclusively or almost exclusively female offspring, females producing almost exclusively male offspring, and females producing intermediate proportions of the 2 sexes. In the literature about genomic conflicts, the restorer genes are usually nuclear, and that might be the case also for R. philippinarum, but then this would not explain the persistence of 2 lineage-specific elements (RPHM21 and RPHF22). Indeed, if RPHM21 was the segregation distorter, and a restorer system evolved from nuclear genes, most likely RPHF22 would have pseudogenized, remaining a genic relict. This is not what we observe, since RPHF22 sequence is well conserved, as suggested by polymorphism data (pS = 0.020, 95% CI [0.002, 0.038]; pN = 0.007, 95% CI [0.001, 0.013], see also Table 2). RPHF22 might have become part of the restorer mechanism, interacting with nuclear elements to counteract RPHM21.
We propose 3 different scenarios.
The viral element also kept infecting mtDNA transmitted through eggs, but the slower replication in the female germ line led to a less efficient insertion ratio and a consequently slower diffusion in the population, so the process of differentiation from the original sequence would have been slowed down as well. Interestingly, I-TASSER generated a better model for RPHF22 compared with RPHM21 (−1,96 versus less than −4, respectively, see ‘Results’ section), because of a higher structural similarity found with known protein sequences.
The viral element was first endogenized in sperm-transmitted mtDNA, starting DUI, and then, by recombination (an event that was documented multiple times in DUI species, see Zouros 2013), it invaded the original mitochondrial lineage that was still transmitted maternally. At this point the 2 elements diverged by evolving sex-specific features.
RPHF22 originated from a different viral element, but with similar features.
Given the fast-evolving nature of viral sequences, the fast-evolving nature of mtDNA in bivalves, and also given the long divergence time between F and M mtDNAs in R. philippinarum, it will be very difficult (if not impossible) to resolve this issue.
How to test the model
In follow-up work, we will test our working hypotheses using mathematical models and simulations. This approach will show which biological and evolutionary parameters are compatible with our model (e.g., number of male and female gametes, number of cell divisions in gametogenesis, mtDNA copy number, selection coefficient on infected mitochondria, relative fitness, etc.), and will greatly help us to understand if it is realistic and consistent with the available data. The ongoing increase of bivalve genomic resources will also help to better understand the mechanisms of germ line establishment, gametogenesis and mitochondrial inheritance, thus providing fundamental knowledge to test our suppositions. Another approach that would answer the question of the role of RPHM21 and RPHF22 would be the manipulation of their transcription/expression (e.g., over/under expression, knock-out, RNAi), but this is a medium-term goal, since such techniques still need to be developed for bivalve molluscs. Last, to answer more general questions about the evolution of DUI, a more comparative approach will be necessary, but this will require the overcoming of the intrinsic challenges of comparative analyses (see, for example, Dunn et al. 2013; Roux et al. 2015).
Conclusions
The origin of DUI most likely entailed the invasion of at least one selfish genetic element, and the extant DUI systems can be seen as resolved conflicts (Passamonti and Ghiselli 2009). Specifically, we think that the acquisition of selfish features by a mitochondrial lineage could have started the genetic conflict that originated DUI, and that this process could have happened in independent events for single bivalve species, as described by the model in R. philippinarum. Under this light it is interesting to point out the similarities between the DUI system and one of the most studied cases of genomic conflicts, the cytoplasmic male sterility (CMS) in plants (reviewed in Chase 2007). CMS is responsible for gynodioecy, it consists of a condition under which a plant is unable to produce functional pollen, and it is determined by mitochondrial ORFs derived either from mitochondrial gene-coding and gene-flanking sequences, or from sequences of unknown origin. The 2 most striking CMS features that could be applied also to the model for DUI origin proposed here are the selfish nature of mitochondria, and the influence of a mitochondrial ORF on germ line development. Also, CMS is a clear example of nuclear–cytoplasmic sex determination, a scenario hypothesized also for DUI (Breton et al. 2007; Passamonti and Ghiselli 2009; Zouros 2000; Yusa et al. 2013). It was proposed that hermaphroditism was the ancestral condition of bivalves (Davison 2006), and a correlation between DUI and gonochorism was documented (Breton et al. 2011a). The invasion of sex-ratio distorters and the evolution of suppressors can prompt rapid transitions among sex-determination mechanisms (Bachtrog et al. 2014), and DUI might have been responsible for the shift from hermaphroditism to gonochorism in some bivalve species. If true, DUI would represent the first animal sex-determination system involving mtDNA-encoded proteins, paralleling CMS in plants.
But why do only bivalves have DUI? Is bivalve mitochondrial genome more prone to foreign sequence insertion/endogenization? Or is the nuclear genome more permissive to such kind of biological novelties? First of all, it is possible that modifications of SMI are present in other organisms, but they have not been discovered yet. That said, we think that bivalve molluscs have several features that could favor the emergence of a system like DUI. First, it is known that bivalve mitochondria can function in absence of oxygen, by switching to an anaerobic energy metabolism. This “anaerobically functioning mitochondria” appear to be widespread in species that often have to face hypoxic or anoxic conditions (Müller et al. 2012). For example, the blue mussel Mytilus edulis (actually a DUI species) can enter an oxygen-independent cytosolic energy metabolism pathway, producing adenosine triphosphate (ATP) with only Complex I and V (Müller et al. 2012). It is tempting to speculate that a more relaxed selection, at least on some complexes, allows a greater mtDNA polymorphism and even heteroplasmy. Another feature we think might have favored the emergence of DUI in bivalves is the absence of sexual chromosomes and of any sex-specific phenotype (if not the presence of eggs or spermatozoa in the gonads at maturity) which may be key feature for the evolution of this mitochondrial inheritance mode.
At the moment it is not known whether in DUI species the presence of lineage-specific mitochondrial proteins (or RNAs) in germ cells can drive the gonad development toward 1 specific sex, or if their segregation into male (female) germ cells is simply a consequence of being a male (female). If the scenario proposed here will be proved correct, DUI will became another example of how a selfish element (in this particular case a segregation distorter of viral origin) can profoundly affect the biology of an organism, and eventually its evolution.
Funding
This study was supported by the Italian Ministry of Education, University and Research MIUR - SIR Programme no. RBSI14G0P5 funded to LM, by the Italian Ministry of Education, University and Research MIUR - FIR Programme no. RBFR13T97A funded to FG, and by the Canziani bequest funded to MP.
References
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, et al. , 2014. Sex determination: why so many ways of doing it? PLoS Biol 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates PA, DeLuca NA, 1998. The polyserine tract of herpes simplex virus ICP4 is required for normal viral gene expression and growth in murine trigeminal ganglia. J Virol 72:7115–7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billiard S, Husse L, Lepercq P, Godé C, Bourceaux A, et al. , 2015. Selfish male-determining element favors the transition from hermaphroditism to androdioecy. Evolution 69:683–693. [DOI] [PubMed] [Google Scholar]
- Bouaouina M, Goult BT, Huet-Calderwood C, Bate N, Brahme NN, et al. , 2012. A conserved lipid-binding loop in the kindlin FERM F1 domain is required for kindlin-mediated αIIbβ3 integrin coactivation. J Biol Chem 287:6979–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Doucet-Beaupré H, Stewart DT, Hoeh WR, Blier PU, 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough? Trends Genet 23:465–474. [DOI] [PubMed] [Google Scholar]
- Breton S, Doucet-Beaupré H, Stewart DT, Piontkivska H, Karmakar M, et al. , 2009. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: gender-specific open reading frames and putative origins of replication. Genetics 183:1575–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, et al. , 2011a. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol Biol Evol 28:1645–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, et al. , 2011b. Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine mussels (Bivalvia: Mytilidae) PLoS ONE 6:e19365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton S, Milani L, Ghiselli F, Guerra D, Stewart DT, et al. , 2014. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet 30:555–564. [DOI] [PubMed] [Google Scholar]
- Chang JM, Di Tommaso P, Taly JF, Notredame C, 2012. Accurate multiple sequence alignment of transmembrane proteins with PSI-Coffee. BMC Bioinform 13(4 Suppl): S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D, 1978. A model for the evolution of dioecy and gynodioecy. Am Nat 112:975–997. [Google Scholar]
- Chase CD, 2007. Cytoplasmic male sterility: a window to the world of plant mitochondrial-nuclear interactions. Trends Genet 23:81–90. [DOI] [PubMed] [Google Scholar]
- Coscoy L, Ganem D, 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol 13:7–12. [DOI] [PubMed] [Google Scholar]
- Davison A, 2006. The ovotestis: an underdeveloped organ of evolution. BioEssays 28:642–650. [DOI] [PubMed] [Google Scholar]
- DeLuca SZ, O’Farrell PH, 2012. Barriers to male transmission of mitochondrial DNA in sperm development. Dev Cell 22:660–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguay BA, Smiley JR, 2013. Mitochondrial nucleases ENDOG and EXOG participate in mitochondrial DNA depletion initiated by Herpes Simplex Virus 1 UL12.5. J Virol 87:11787–11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn CW, Luo X, Wu Z, 2013. Phylogenetic analysis of gene expression. Integr Comp Biol 53:847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR, 1998. Profile hidden Markov models. Bioinformatics 14:755–763. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR, 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer D, Eisenberg D, 1999. Finding families for genomic ORFans. Bioinformatics 15:759–762. [DOI] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Passamonti M, 2011. Strict sex-specific mtDNA segregation in the germline of the DUI species Venerupis philippinarum (Bivalvia Veneridae). Mol Biol Evol 28:949–961. [DOI] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, et al. , 2012. De Novoassembly of the Manila clam Ruditapes philippinarumtranscriptome provides new insights into expression bias, mitochondrial doubly uniparental inheritance and sex determination. Mol Biol Evol 29:771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, et al. , 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol Evol 5:1535–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissi C, Iannelli F, Pesole G, 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101:301–320. [DOI] [PubMed] [Google Scholar]
- Howard MB, Ekborg NA, Taylor LE, Hutcheson SW, Weiner RM, 2004. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci 13:1422–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, 1992. Intragenomic conflict as an evolutionary force. Proc R Soc B 248:135–140. [Google Scholar]
- Jones P, Binns D, Chang HY, Fraser M, Li W, et al. , 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Pereira V, Tristem M, 2007. Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol 81:10712–10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenchington E, MacDonald B, Cao L, Tsagkarakis D, Zouros E, 2002. Genetics of mother-dependent sex ratio in blue mussels (Mytilusspp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics 161:1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz T, Pintard L, Willis JH, Hamill DR, Gönczy P, et al. , 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 15:1294–1298. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. , 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machordom A, Araujo R, Toledo C, Zouros E, Ladoukakis ED, 2015. Female-dependent transmission of paternal mtDNA is a shared feature of bivalve species with doubly uniparental inheritance (DUI) of mitochondrial DNA. J Zool Syst Evol Res 53:200–204. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, et al. , 2010. The Genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, 2015. Mitochondrial activity in gametes and transmission of viable mtDNA. Biol Direct 10:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Passamonti M, 2011. Doubly uniparental inheritance of mitochondria as a model system for studying germ line formation. PLoS ONE 6:e28194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Passamonti M, 2012. Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the Doubly Uniparental Inheritance (DUI) of mitochondria. J Exp Zool Part B 318:182–189. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M, 2013a. A comparative analysis of mitochondrial orfans: new clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol Evol 5:1408–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Nuzhdin SV, Passamonti M, 2013b. Nuclear genes with sex bias in Ruditapes philippinarum (Bivalvia, Veneridae): mitochondrial inheritance and sex determination in DUI Species. J Exp Zool Part B 320:442–454. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M, 2014a. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol Evol 6:391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Iannello M, Passamonti M, 2014b. Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr Genet 60:163–173. [DOI] [PubMed] [Google Scholar]
- Milani L, Ghiselli F, Pecci A, Maurizii MG, Passamonti M, 2015. The expression of a novel mitochondrially-encoded gene in gonadic precursors may drive paternal inheritance of mitochondria. PLoS ONE 10:e0137468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Chan DC, 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 15:634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K, 2013. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. Bioessays 35:955–964. [DOI] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, et al. , 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76:444–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamonti M, Ghiselli F, 2009. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol 28:79–89. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. , 2004. UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. [DOI] [PubMed] [Google Scholar]
- Ronkina N, Kotlyarov A, Dittrich-Breiholz O, Kracht M, Hitti E, et al. , 2007. The Mitogen-Activated Protein Kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol Cell Biol 27:170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronkina N, Kotlyarov A, Gaestel M, 2008. MK2 and MK3 – a pair of isoenzymes? Front Biosci 13:5511–5521. [DOI] [PubMed] [Google Scholar]
- Roux J, Rosikiewicz M, Robinson-Rechavi M, 2015. What to compare and how: comparative transcriptomics for Evo-Devo. J Exp Zool Part B 324:372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. , 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM, 1994a. Mitochondrial DNA inheritance. Nature 368:817–818. [DOI] [PubMed] [Google Scholar]
- Skibinski DO, Gallagher C, Beynon CM, 1994b. Sex-limited mitochondrial DNA transmission in the marine mussel Mytilus edulis. Genetics 138:801–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. , 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Beukeboom LW, 1998. Sex determination, sex ratios and genetic conflict. Annu Rev Ecol Syst 29:233–261. [Google Scholar]
- Wolf A, Mantri M, Heim A, Müller U, Fichter E, et al. , 2013. The polyserine domain of the lysyl-5 hydroxylase Jmjd6 mediates subnuclear localization. Biochem J 453:357–370. [DOI] [PubMed] [Google Scholar]
- Wright G, 1998. Sex change in the Mollusca. Trends Ecol Evol 3:137–140. [DOI] [PubMed] [Google Scholar]
- Yamano K, Youle RJ, 2011. Coupling mitochondrial and cell division. Nat Cell Biol. 13:1026–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa Y, Breton S, Hoeh WR, 2013. Population genetics of sex determination in Mytilus mussels: reanalyses and a model. J Hered 104:380–385. [DOI] [PubMed] [Google Scholar]
- Zhang Y, 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinform 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR, 1994a. Mitochondrial DNA inheritance. Nature 368:818. [DOI] [PubMed] [Google Scholar]
- Zouros E, Oberhauser Ball A, Saavedra C, Freeman KR, 1994b. An unusual type of mitochondrial DNA inheritance in the blue mussel Mytilus. Proc Natl Acad Sci USA 91:7463–7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouros E, 2000. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet Syst 75:313–318. [DOI] [PubMed] [Google Scholar]
- Zouros E, 2013. Biparental inheritance through uniparental transmission: the doubly uniparental Inheritance (DUI) of mitochondrial DNA. Evol Biol 40:1–31. [Google Scholar]