Abstract
Background
Nonalcoholic fatty liver disease (NAFLD) has become a leading cause of liver transplantation. Lingguizhugan decoction (LGZG), a classical Chinese herbal formula, has beneficial effects on NAFLD animal models. Our study examined the impact of LGZG on hepatic global transcriptome of high-fat-diet-induced NAFLD rats.
Methods
Three groups of Wistar rats were included: normal, NAFLD model, and LGZG-treated NAFLD groups. Four weeks for the treatment, liver tissues were harvested for RNA sequencing. Differentially expressed genes (DEGs) and enriched pathways were detected on hepatic global transcriptome profile. Real-time PCR validated the regulatory patterns of LGZG on NAFLD rats.
Results
DEGs between the NAFLD model and normal groups indicated the elevated peroxisome proliferator-activated receptor (PPAR) and hedgehog signaling pathways in NAFLD rats. In bile secretion pathway, genes involved in cholesterol secretion were activated by LGZG treatment. Increased expression of antioxidant OSIGN1 and decreased expression of genes (AHR, IRF2BP2, and RASGEF1B) that induce oxidative stress and inflammation were observed in NAFLD rats treated with LGZG. The regulatory patterns of LGZG treatment on these oxidative stress-related genes were confirmed by real-time PCR.
Conclusion
Our study revealed a “two-hits-targeting” mechanism of LGZG in the treatment for NAFLD: alleviating oxidative stress and activating cholesterol secretion.
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) encompasses a spectrum of pathological conditions, including simple steatosis, nonalcoholic steatohepatitis (NASH), fibrosis, and cirrhosis. NAFLD has been increasingly prevalent worldwide and has become a leading cause of liver transplantation, along with increasing obesity rate and metabolic syndrome [1]. According to the “two-hits” hypothesis of NAFLD pathogenesis, the first hit is represented by lipid accumulation in the hepatocytes, after which oxidative stress leads to severe NASH [2]. Additionally, recent studies demonstrated that other risk factors also contribute to the development of severe NASH, including altered gut microbiota [3, 4], endogenous alcohol metabolism [5–7], and endoplasmic reticulum stress [8]. Currently, many potential targets for the treatment of NAFLD are identified, including lipid metabolism, oxidative stress, inflammation, fibrosis, and altered gut microbiota [9]. However, clinical trials exclusively aimed at only one of these targets at a time and achieved limited effects. For example, vitamin E therapy targets the “second hit”—oxidative stress, and pioglitazone therapy targets insulin resistance. Both of these two therapies decrease serum AST/ALT and reduce lobular inflammation. They, however, have no impact on insulin resistance, portal inflammation, or liver fibrosis and did not obtain satisfactory sustained results [10].
Given NAFLD is a consequence of multiple risk factors, especially the major “two hits”—lipid accumulation and oxidative stress. A therapeutic strategy that targets both of “two hits” or several hits simultaneously could be more effective than one single therapeutic target. Traditional Chinese medicine (TCM) has been increasingly applied as the potential anti-NAFLD drugs and exhibited multipronged preventive and therapeutic effects [9, 11, 12]. LGZG is a classic TCM formula, which is a mixture of four herbs: Poria, Ramulus Cinnamomi, Atractylodes macrocephala Koidz, and Radix Glycyrrhizae. LGZG has a long time of clinical practice in the treatment of chronic congestive heart failure. In recent years, LGZG has been effectively used to treat obesity and hyperlipidemia [13] and our previous study showed that LGZG has a benefit in treating NAFLD [14].
TCM is an ancient medical practice system which emphasizes the integrity of the entire human body, and it usually exerts therapeutic effects via multiple targets or pathways. Recently, high-throughput omics technologies, especially transcriptomics RNA sequencing, have been increasingly applied in TCM research and revealed multipronged therapeutic mechanisms of TCM [11, 15]. Herein, to better understand the therapeutic mechanisms of LGZG, we examined the effects of LGZG treatment on the hepatic global transcriptome of HFD-induced NAFLD rats. Combined with real-time PCR validation, we identified a “two-hits-targeting” mechanism for LGZG in the treatment of NAFLD.
2. Materials and Methods
2.1. Experimental Animals and Treatment
This study was reviewed and approved by the Animal Experiment Ethics Committee of Shanghai University of TCM and carried out in accordance with their recommendations. A total of 24 male Wistar rats with weights of 130 g ± 10 g in specific pathogen free (SPF) grade were purchased from Shanghai Si-Lai-Ke Experimental Animal Ltd. (Shanghai, China). Animals were randomly divided into three groups (eight rats per group): normal, NAFLD model, and LGZG-treated NAFLD groups. Rats in the normal group were fed with a standard diet. Rats in the NAFLD model and LGZG-treated NAFLD groups were fed with HFD, which consists of 10% lard oil, 2% cholesterol, and 88% standard chow. Rats in the LGZG-treated NAFLD group received a dosage of 10 mL/kg/d (pure solution) via drinking freely. The dosage was 6 g crude medical material per kilogram body weight, approximately seven times of the standard dosage in practice, on the basis of the dose-equivalence equation between rats and humans [16]. Treatment lasted for four weeks. After a 12-hour fast, the animals were sacrificed under pentobarbital sodium (2%, 5.5 mL/kg) anesthesia. Liver tissues were harvested for subsequent analysis.
2.2. Drug Preparation
LGZG decoction is comprised of four Chinese herbs: Poria (20 g), Ramulus Cinnamomi (15 g), Rhizoma Atractylodis Macrocephalae (15 g), and Radix Glycyrrhizae (10 g). The dosage is determined according to the test book of “The Hndouts of JinguiYaoyue.” All herbs were purchased from Longhua Hospital affiliated to Shanghai University of TCM. LGZG decoction was made according to conventional TCM decocting methods [14]. Briefly, all herbs were boiled with 500 mL water after 30 min of soaking. After 20 min, the liquid was transferred by filtration as a first dose of medicine. The remaining of filtration was boiled after adding 400 mL water, and then liquid was transferred by filtration to make a second dose. Two doses were mixed to form 100 mL (pure solution) final decoction. The quality of LGZG was controlled with high-performance liquid chromatography (HPLC). HPLC-grade reagents were purchased from Burdick & Jackson. An Agilent 1100 HPLC system consisting of a G1354A pump, a G1313A autosampler, and a UV/VIS Photodiode Array G1315B detector was used for all analyses (Figure S1).
2.3. RNA Sequencing Analysis
Liver tissue from the right lobe was collected after four weeks of LGZG treatment as described above. Total RNA was isolated with NanoPhotometer spectrophotometer (IMPLEN, CA, USA) and qualified on the Qubit RNA Assay (Qubit 2.0 Fluorometer, Life Technologies, CA, USA). The RNA libraries were sequenced on IlluminaHiseq 4000 platform with paired-end 150 base pair long reads. Clean data were obtained from raw data by removing reads containing adapter, N base, and low-quality reads with NGS QC Toolkit (version: 2.3.3). Clean data were mapped to the reference genome of R.norvegicus6.0 and estimated for gene expression level using TopHat2 and cufflinks (version: 2.2.1).
2.4. Real-Time PCR
OSGIN1, IRF2BP2, AHR, and RASGEF1B mRNA levels were determined by real-time PCR. Primers were designed with the primer premier 5.0 software (Table S1). Total RNA of liver tissues was extracted with TRIzol reagent (Invitrogen, USA). The concentration of RNA was measured with NanoDrop 2000 (Thermo Scientific, USA). Quantitative measurement was performed with the Premix Ex Taq kit (TakaRa) according to the manufacturer's instructions on Applied Biosystems StepOne Plus Sequence Detection System. The real-time cycler conditions were as follows: first denatured at 95°C for 30 s and then amplified with 40 cycles (each cycle was denaturated at 90°C for 5 s and annealing/extension at 60°C for 30 min). Product purity was determined by dissociation curve analysis. Gene expression was quantified relative to the values of the control group after adjusting for β-actin by the 2−ΔΔCT method [17].
2.5. Statistical Analysis
Differentially expressed genes (DEGs) were identified by cuffdiff (version: 2.2.1), with a Q value < 0.05 and an absolute value of log2 fold change > 0.58. Pathway enrichment analysis was performed with clusterProfiler R package (version: 2.4.3) with P value < 0.05. Data were denoted as mean ± standard deviation (SD). In biochemical analysis, statistical analysis was performed with one-way analysis of variance (ANOVA) and Dunnett's test. In RT-PCR validation, statistical analysis was performed with Mann–Whitney test with a two-tailed distribution. P values < 0.05 were considered statistically significant. Statistical analysis was performed in R 3.2.3 software.
3. Results
3.1. Genes and Pathways Associated with NAFLD Pathology
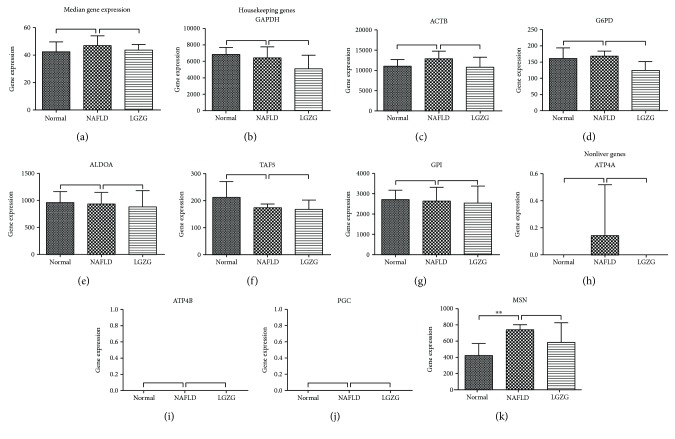
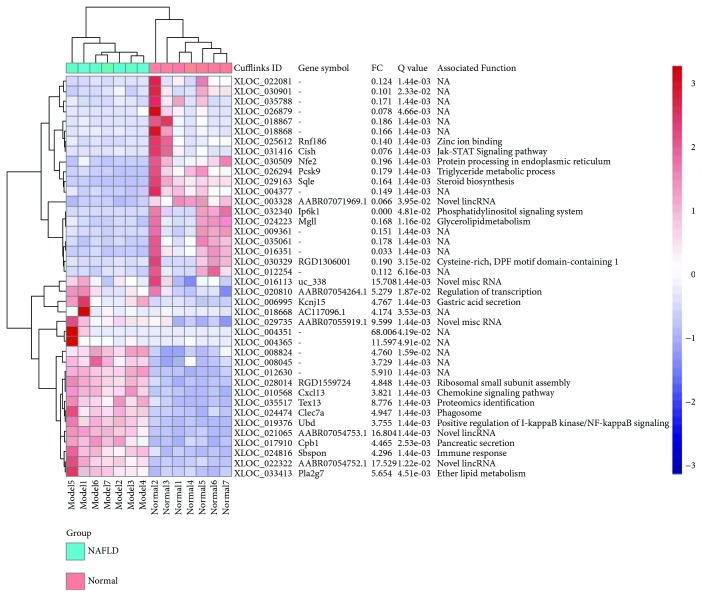
Expression levels of about 22,077 genes of rat livers were quantified on a global RNA sequencing. Similar median gene expression and expression levels for housekeeping genes and nonliver genes are indicative of the qualified RNA-seq data in the present study (Figure 1). A total of 931 genes were differentially expressed between the NAFLD model and normal groups, including 494 upregulated and 437 downregulated genes. These 931 DEGs were regarded as NAFLD-regulated genes. The top 40 regulated genes (20 most upregulated and 20 most downregulated) were depicted in Figure 2. These genes are known to be associated with lipid metabolism, including triglyceride metabolic process (PCSK9), steroid biosynthesis (SQLE), glycerolipid metabolism (MGLL), ether lipid metabolism (PLA2G7), and phosphatidylinositol signaling system (IP6K1). Additionally, the chemokine CXCL13, ubiquitin UBD, and somatomedin SBSPON play roles in inflammatory responses, and their expressions were elevated in NAFLD rats (Figure 2).
Figure 1.
Median gene expression (a) was similar among all study groups. Housekeeping genes GAPDH (b), ACTB (c), G6PD (d), ALDOA (e), TAF (f), and GPI (g) and nonliver genes ATP4A (h), ATP4B (i), PGC (j), and MSN (k) exhibited similar expression among all study groups. ∗∗Q value < 0.01 calculated from cuffdiff.
Figure 2.
Differential expression of top 40 NAFLD-regulated genes. The samples are broadly divided into two groups, normal and NAFLD model groups. The color scale shown at the top right illustrates the relative expression level of the genes across all samples. FC, fold change between NAFLD model and normal groups.
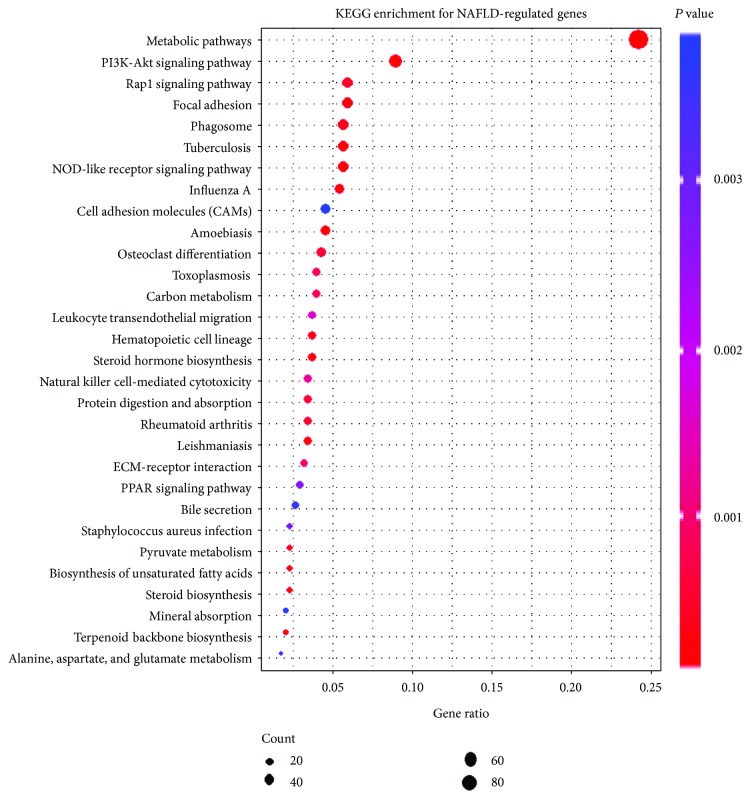
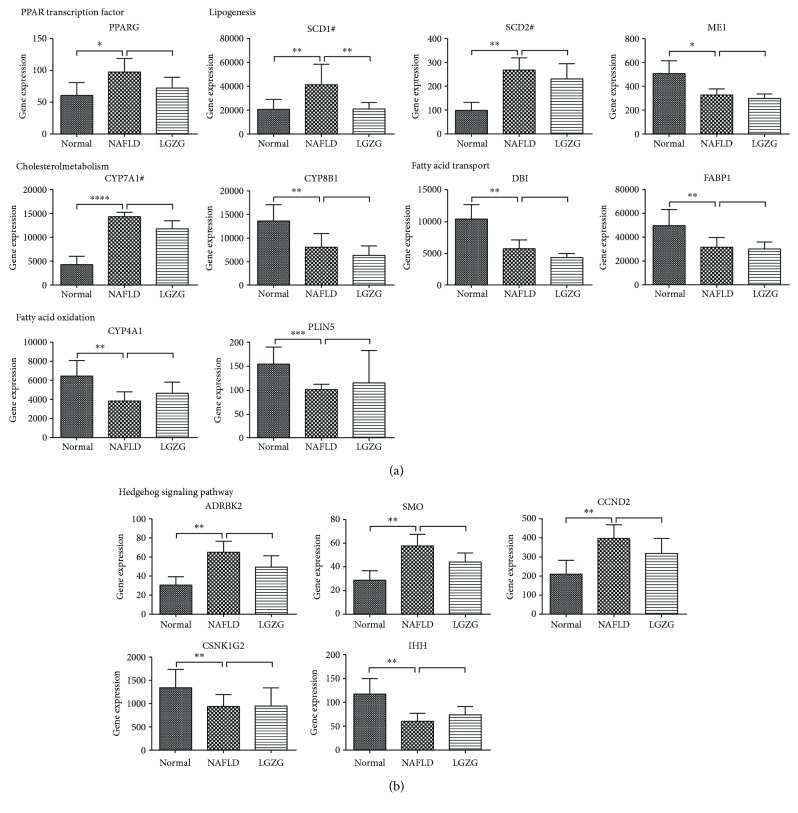
Further, 76 KEGG pathways were enriched with NAFLD-regulated genes (Table S2). The top 30 enriched KEGG pathways were shown in Figure 3. In addition to several well-known pathways that are related to NAFLD pathology (including fatty acid elongation, AMPK signaling pathway, and NF-kappa B signaling pathway), peroxisome proliferator-activated receptor (PPAR) and hedgehog (Hh) signaling pathways (Table S2) are of special interest because of their essential roles in hepatic fibrosis of NAFLD progression. As to individual genes of PPAR signaling pathway (Figure 4(a)), NAFLD rats exhibited increased expression of one of the nuclear receptors—PPARG, which regulates lipogenesis and cholesterol metabolism and represents a good candidate gene for NAFLD [18]. Consistently, increased expression of stearoyl-CoA desaturase (SCD1 and SCD2) was observed in NAFLD rat livers, which are regulated by PPARG and the rate-limiting enzymes in lipid biosynthesis [19, 20]. CYP7A1, a rate-limiting enzyme in cholesterol metabolism, was upregulated in NAFLD rats. However, some target genes of PPARG were downregulated in NAFLD rats. And these decreased expressions of the target genes resulted in impaired fatty acid transport (DBI and FABP1) and oxidation (CYP4A1), which contributed to fatty acid accumulation in the NAFLD liver.
Figure 3.
Scatter plot of the top 30 enriched KEGG pathways for NAFLD-regulated genes. The x-axis represents the ratio of NAFLD-regulated gene and all gene numbers annotated in this KEGG pathway. The y-axis is enriched KEGG pathways. P value was calculated from hypergeometric test. A smaller P value indicates higher significance (P < 0.05).
Figure 4.
Differential gene expression of (a) PPAR and (b) hedgehog signaling pathways in a NAFLD rat model. ∗Q value < 0.05; ∗∗Q value < 0.01; ∗∗∗Q value < 0.001; ∗∗∗∗Q value < 0.0001 calculated from cuffdiff. #Rate-limiting enzyme.
G protein-coupled transmembrane receptor Smoothened (SMO) is one of important components of canonical Hh signaling pathway. Our results showed upregulation of SMO in NAFLD rats, compared with normal rats. A trend of decreased expression of SMO was shown in LGZG-treated rats. In addition, ADRBK2 (upstream regulator of SMO) and CCND2 (Hh downstream target gene) were also significantly elevated in NAFLD rats (Figure 4(b)).
3.2. Genes and Pathways Mediating the Therapeutic Effects of LGZG
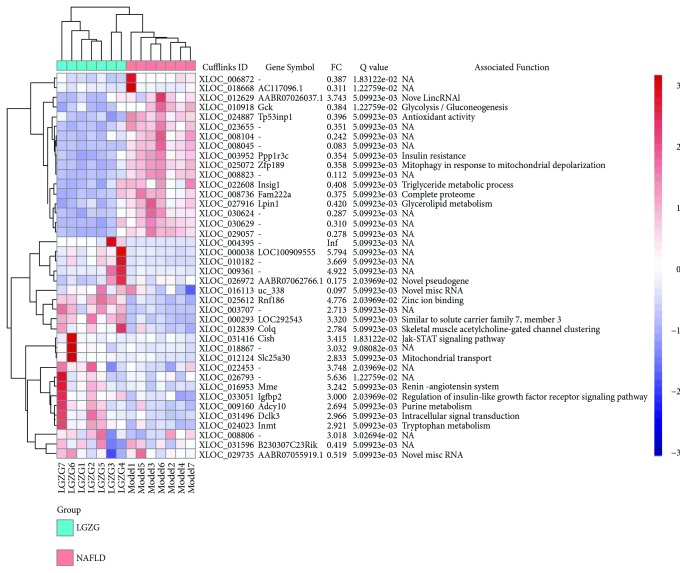
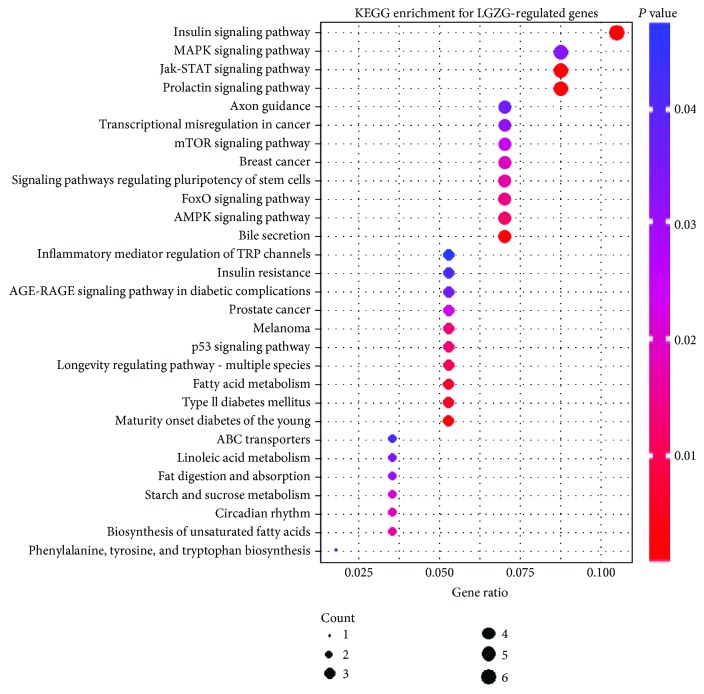
Compared with the NAFLD model group, elevated expression of 110 genes and decreased expression of 89 genes were observed in the livers of the LGZG-treated NAFLD rats. These total 199 genes were considered as LGZG-regulated genes. Figure 5 showed the top 40 LGZG-regulated genes (20 most upregulated and 20 most downregulated). The LGZG-treated NAFLD rats exhibited reduced expression of INSIG1 and LPIN1, which indicated the decreased cholesterol biosynthesis and triglyceride accumulation in the liver. Pathway enrichment analysis was performed with LGZG-regulated genes and unearthed 29 enriched pathways (Figure 6, Table S3). In bile secretion pathway, genes required for cholesterol secretion were elevated, including ABCG8, ABCG5, and NCEH1. Some metabolism and signaling pathways were also enriched, including fatty acid metabolism, Jak-STAT, and FoxO signaling pathways.
Figure 5.
Differential expression of top 40 LGZG-regulated genes. The samples of the NAFLD model group formed a clade and were separated from the samples of the LGZG-treated NAFLD groups. The color scale shown at the top right illustrates the relative expression level of the genes across all samples. FC, fold change between LGZG-treated NAFLD and NAFLD model groups.
Figure 6.
Scatter plot of the enriched KEGG pathways for LGZG-regulated genes. The x-axis represents the ratio of LGZG-regulated gene numbers annotated in this KEGG pathway to all gene numbers annotated in this KEGG pathway. The y-axis is enriched KEGG pathways. P value was calculated from hypergeometric test. A smaller P value indicates higher significance (P < 0.05).
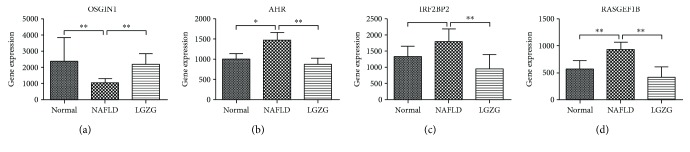
Furthermore, four (OSGIN1, AHR, IRF2BP2, and RASGEF1B) were shown to be strongly regulated upon LGZG therapy, which may play potential roles in NAFLD physiopathology (Figure 7, Table 1). These four genes also showed differential expression changes in NAFLD rats, compared with normal rats. Also, the abnormal expression change in NAFLD rats was significantly reversed by the LGZG therapy. Thus, they were considered as the important target genes of LGZG.
Figure 7.
Differential gene expression of LGZG-regulated genes. ∗Q value < 0.05; ∗∗Q value < 0.01 calculated from cuffdiff.
Table 1.
Important LGZG-regulated genes.
| Gene | Q value∗ | FC# | Q value$ | FC+ | Associated function |
|---|---|---|---|---|---|
| OSGIN1 | 1.44e-03 | 0.43 | 5.10e-03 | 2.31 | Oxidoreductase activity |
| IRF2BP2 | 6.62e-02 | 1.43 | 5.10e-03 | 0.52 | Interferon regulatory factor 2-binding protein 1 & 2 |
| AHR | 2.20e-02 | 1.37 | 5.10e-03 | 0.67 | Blood vessel development |
| RASGEF1B | 1.44e-03 | 1.63 | 5.10e-03 | 0.46 | Ras guanyl-nucleotide exchange factor activity |
∗ Q value of comparison between the NAFLD model and normal groups, calculated from cuffdiff. #Fold change of comparison between the NAFLD model and normal groups, calculated from cuffdiff. $Q value of comparison between the LGZG-treated NAFLD and NAFLD model groups, calculated from cuffdiff. +Fold change of comparison between the LGZG-treated NAFLD and NAFLD model groups, calculated from cuffdiff.
3.3. Validation of LGZG-Regulated Genes with Real-Time PCR
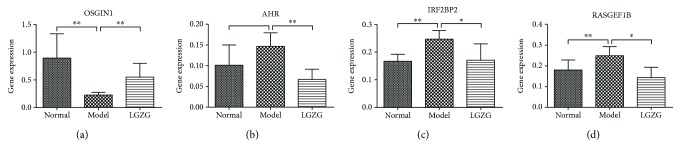
Expression levels of four LGZG-regulated genes were validated with real-time PCR, including OSGIN1, AHR, IRF2BP2, and RASGEF1B (Figure 8). The real-time PCR results confirmed that the expression of OSGIN1 was reversely elevated, and the expression levels of IRF2BP2, AHR, and RASGEF1B were inversely suppressed under the LGZG treatment.
Figure 8.
Differential gene expression of LGZG-regulated genes was examined by real-time PCR. Plotted values are the mean ± SD of mRNA expression levels in the livers of normal rats (normal), NAFLD model rats, and NAFLD rats treated with LGZG decoction (LGZG). N = 6 per group. ∗P < 0.05; ∗∗P < 0.01 Mann–Whitney test.
4. Discussion
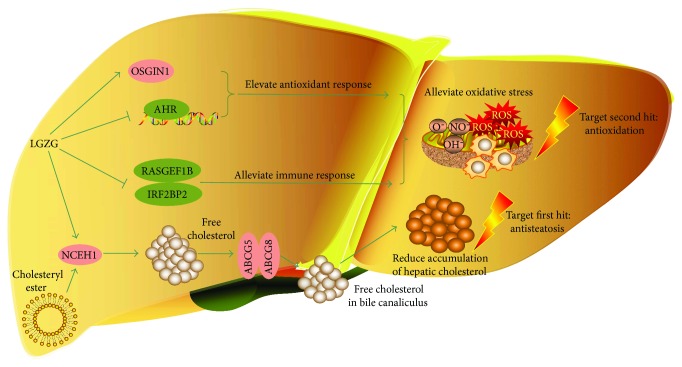
LGZG is a classic TCM formula that has been effectively used to treat obesity and hyperlipidemia. Recently, it has exhibited potent effects on HFD-induced NAFLD [14]. In this study, we examined the comprehensive effects of LGZG on the hepatic global gene expression profile in NAFLD progression. Many of the changes in expression profile reflected decreased hepatic cholesterol, oxidative stress, and inflammation. Subsequently, the elevated gene expression in PPAR and Hh signaling pathways of NAFLD rats provided an assurance of RNA-seq dataset quality. Two potential mechanisms under the efficacy of LGZG for NAFLD were identified: (i) alleviated oxidative stress and (ii) promoted cholesterol secretion to reduce hepatic cholesterol accumulation (Figure 9). These observations indicated a “two-hits-targeting” [2, 21] mechanism for LGZG in the treatment of NAFLD.
Figure 9.
Summary of the “two-hits-targeting” therapeutic mechanisms of LGZG for NAFLD. Data presented in this study supports two independent mechanisms: (i) LGZG activates antioxidant (OSGIN1) and suppresses prooxidant and proinflammatory genes (AHR, IRF2BP2, and RASGEF1B), causing alleviated oxidative stress, and (ii) LGZG induces cholesterol secretion genes in the liver, leading to reduced accumulation of liver lipid. These mechanisms reflect that LGZG treatment prominately impeded classic “two hits” of NASH physiopathology. Red genes indicate increased expression; green genes indicate decreased expression. Normal arrows indicate activation. Arrows with a vertical line at the end indicate suppression.
4.1. Characteristics of HFD-Induced NAFLD
NAFLD physiopathology-associated pathways were significantly enriched with NAFLD-regulated genes. Among these pathways, activation of PPAR and Hh signaling pathways is of special interest because their deregulation contributes to liver damage and metabolic syndrome [22–27]. Hh signaling is significantly upregulated in NASH, compared with the normal healthy liver [28]. Recent study showed that activation of SMO could induce Hh-responsive hepatocytes in NAFLD [28], which lends strong support to our results (Figure 4(b)). Notably, multiple studies showed that in different rodent models of diet-induced NASH, pharmacological inhibition of SMO (vismodebig or LDE225) can deactivate Hh signaling pathway and consistently improve liver inflammation and fibrosis [25, 28–30]. Despite no significant difference was observed between NAFLD and LGZG treatment groups for SMO, a trend of decreased expression was shown in LGZG-treated NAFLD rats. In addition, lipid metabolism-related genes were markedly altered in NAFLD rats, which contributed to hepatic TG accumulation. Genes and pathways associated with NAFLD pathology were the manifestation of the RNA-seq dataset quality.
4.2. Alleviation of Oxidative Stress
Significant expression changes of four LGZG-regulated genes (OSGIN1, AHR, IRF2BP2, and RASGEF1B) exhibited alleviation of oxidative stress upon LGZG treatment, compared with the NAFLD group. As an antioxidant, OSGIN1 is a cell growth inhibitor to resist oxidative stress [31]. Liu et al. reported that the mRNA level of OSGIN1 was reduced in HCC specimens, and in HCC pateints, the inhibition of OSGIN1 was related to shorter overall and disease-free survival times [32], suggesting that the significant upregulation of OSGIN1 is critical for antioxidant response. AHR is identified to induce cellular oxidative stress and increase lipid peroxidation in NAFLD [33–36], and activation of AHR has pleotropic effects on steatosis of NAFLD [37, 38].
RASGEF1B and IRF2BP2 were involved in inflammatory responses and were dramatically inhibited by the LGZG treatment. RASGEF1B was identified as a Ras-associated guanine nucleotide exchange factor and upregulated in macrophages stimulated with bacterial lipopolysaccharides (LPS) [39]. LPS is currently considered one of the major “hits” in NAFLD pathogenesis and progression [40]. Our current study showed significant upregulation of RASGEF1B in NAFLD model rats and reverse inhibition of RASGEF1B by LGZG treatment. Increased expression of RASGEF1B may be a strong defense response to LPS in NAFLD progression, and its restoration indicated the alleviation of inflammatory responses. IRF2BP2 acts as a negative regulator of the nuclear factor of activated T cell (NFAT) transcription factor [41]. Restoration of IRF2BP2 by LGZG treatment also suggested the alleviation of inflammatory responses in NAFLD. Continuous oxidative stress may lead to chronic inflammation. Thus, the alleviation of inflammatory responses also indicates the reduced oxidative stress upon LGZG treatment.
4.3. Activation of Cholesterol Secretion
Liver lipid accumulation is the first “hit” in the pathogenesis of NAFLD, and its removal is a desired intervention for NAFLD. Cholesterol is one type of important lipid in the liver, which has been shown as an emerging factor involved in the development of many metabolic diseases [8]. Free cholesterol stores in the liver in the formation of cholesterol esters. In the cholesterol secretion pathway, the upregulation of neutral cholesteryl ester hydrolase 1 (NCEH1) can accelerate the transformation of cholesterol esters into free cholesterol. ABCG5 and ABCG8 are two half-transporters that dimerize to create a cholesterol transporter, and their activation promotes the excretion of hepatic cholesterol [42]. In addition, NCEH1, ABCG5, and ABCG8 are reported to be drug targets of pioglitazone [43] and ezetimibe [42] in the therapy of human gallbladder cholesterolosis and HFD-induced fatty liver.
5. Conclusion
Based on transcriptome analysis and experimental validation, our study examined the comprehensive effects of LGZG on hepatic global gene expression profile in HFD-induced NAFLD rats. Compared with normal rats, our data revealed significant upregulation of PPAR and Hh signaling pathways in NAFLD rats, which are known to be involved in NAFLD pathology and thus provide an assurance of the quality of our RNA-seq dataset. Of particular interest, NAFLD rats with LGZG treatment exhibited elevated expression of antioxidant and suppressed expression of prooxidant and proinflammatory genes in oxidative stress. Additionally, genes required for cholesterol secretion were increased by the LGZG treatment. These findings supported a “two-hits-targeting” mechanism for LGZG in the treatment of NAFLD: alleviating oxidative stress and activating cholesterol secretion.
Acknowledgments
This work was supported by the National Natural Science Foundation of China 81620108030 and Natural Science Foundation of Shanghai 15ZR1441900.
Abbreviations
- NAFLD:
Nonalcoholic fatty liver disease
- NASH:
Nonalcoholic steatohepatitis
- LGZG:
Lingguizhugan
- EBP/BW:
Epididymal fat pad/body weight ratio
- SPF:
Specific pathogen free
- HFD:
High-fat diet
- HPLC:
High-performance liquid chromatography
- H&E:
Hematoxylin and eosin
- ALT:
Alanine aminotransferase
- AST:
Aspartate aminotransferase
- TG:
Triglyceride
- TC:
Total cholesterol
- VLDL:
Very-low-density lipoprotein
- RT-PCR:
Real-time PCR
- DEG:
Differentially expressed genes
- SD:
Standard deviation
- ANOVA:
One-way analysis of variance
- CV:
Central vein
- PCSK9:
Proprotein convertase subtilisin/kexin type 9
- SQLE:
Squalene monooxygenase
- MGLL:
Acylglycerol lipase
- PLA2G7:
Platelet-activating factor acetylhydrolase
- IP6K1:
Inositol hexakisphosphate 5-kinase
- CXCL13:
C-X-C motif chemokine 13
- UBD:
Ubiquitin D
- SBSPON:
Somatomedin B and thrombospondin type 1 domain containing
- PPAR:
Peroxisome proliferator-activated receptor
- PPARG:
Peroxisome proliferator-activated receptor gamma
- SCD:
Stearoyl-CoA desaturase
- SCD2:
Stearoyl-Coenzyme A desaturase 2
- CYP7A1:
Cholesterol 7alpha-monooxygenase
- DBI:
Diazepam-binding inhibitor (GABA receptor modulator, acyl-CoA-binding protein)
- FABP1:
Fatty acid binding protein 1, liver
- CYP4A1:
Alkane 1-monooxygenase
- SMO:
Smoothened
- Hh:
Hedgehog
- ADRBK2:
Beta-adrenergic receptor kinase
- CCND2:
Cyclin D2
- INSIG1:
Insulin-induced gene 1
- LPIN1:
Phosphatidate phosphatase LPIN
- ABCG8:
ATP-binding cassette, subfamily G (WHITE), member 8 (sterolin 2)
- ABCG5:
ATP-binding cassette, subfamily G (WHITE), member 5 (sterolin 1)
- NCEH1:
Neutral cholesterol ester hydrolase 1
- OSGIN1:
Oxidative stress-induced growth inhibitor 1
- AHR:
Aryl hydrocarbon receptor
- IRF2BP2:
Interferon regulatory factor 2 binding protein 2
- RASGEF1B:
RasGEF domain family, member 1B
- HCC:
Hepatocellular carcinoma.
Contributor Information
Tao Liu, Email: lh2488@126.com.
Peiyong Zheng, Email: zpychina@sina.com.
Disclosure
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Authors' Contributions
Each author has contributed significantly to the submitted work. Tao Liu and Peiyong Zheng conceived and designed the experiments. Lili Yang and Weili Lin performed the experiments. Lili Yang, Weili Lin, Colleen A. Nugent, Shijun Hao, and Haiyan Song analyzed the data. Lili Yang, Weili Lin, Colleen A. Nugent, Shijun Hao, Haiyan Song, and Tao Liu wrote the paper. All authors read and approved the final manuscript. Lili Yang and Weili Lin contributed equally to this work.
Supplementary Materials
Figure S1. Quality control of Lingguizhugan Decoction under High Performance Liquid Chromatography (HPLC). A. Reference substance; B. Lingguizhugan Decoction 1. glycyrrhizic acid. Standardization of extract by HPLC glycyrrhizic acid was purchased from Shanghai R&D Centre for standardization of Chinese Medicines (Shanghai, China), and the purity was higher than 98.0%;. HPLC-grade reagents were purchased from Burdick & Jackson. An Agilent 1100 HPLC system consisting of a G1354A pump, a G1313A auto-sampler, and a UV/VIS Photodiode Array G1315B Detector was used for all analyses. Chromatographic separations were carried out on an Merck C18 hibar column (4.6 mm × 250mm, 5 μm) with methanol:acetonitrile : water :acetid acid(as solvent ) (15:35:45:0.9,v/v) in the mobile phase at a flow rate of 0.8 mL/min at 25°C for 40 min. Twenty μL (after a high speed centrifugation) of the sample was injected, and the signals were detected at 254 nm with UV detection. The HPLC fingerprint of Lingguizhugan decoction revealed the major peaks (glycyrrhizic acid) at 254 nm. The content of glycyrrhizic acid were 0.9548%. Figure S2. Hierarchical clustering of replicate samples based on correlation matrix in three groups: (A) Normal group; (B) NAFLD model group; (C) LGZG-treated group. Each column and row represents samples in each group. Each cell of the heatmap reflects the distance of two samples. Distance was measured with Euclidean metric. The legend of distance value is shown on the upper right. Figure S2A and figure S2B revealed that s12 and s24 were the outlier samples of normal group and NAFLD model group, respectively. Figure S2C showed no outlier samples, but the data in s37 was missed during assessment of serum ALT and AST. Thus, s37 in LGZG-treated group was also removed in the RNA-seq analysis. Table S1. Primer pairs for real-time PCR analysis. Table S2. Enriched KEGG pathways with NAFLD regulated genes. Table S3. Enriched KEGG pathways with LGZG regulated genes.
References
- 1.Wree A., Broderick L., Canbay A., Hoffman H. M., Feldstein A. E. From NAFLD to NASH to cirrhosis–new insights into disease mechanisms. Nature Reviews Gastroenterology & Hepatology. 2013;10(11):627–636. doi: 10.1038/nrgastro.2013.149. [DOI] [PubMed] [Google Scholar]
- 2.Day C. P., James O. F. Steatohepatitis: a tale of two “hits”? Gastroenterology. 1998;114(4):842–845. doi: 10.1016/s0016-5085(98)70599-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L., Baker S. S., Gill C., et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 4.Zhu L., Baker R. D., Zhu R., Baker S. S. Gut microbiota produce alcohol and contribute to NAFLD. Gut. 2016;65(7, article 1232) doi: 10.1136/gutjnl-2016-311571. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y., Baker S. S., Baker R. D., Zhu R., Zhu L. Systematic analysis of the gene expression in the livers of nonalcoholic steatohepatitis: implications on potential biomarkers and molecular pathological mechanism. PLoS One. 2012;7(12, article e51131) doi: 10.1371/journal.pone.0051131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu R., Baker S. S., Moylan C. A., et al. Systematic transcriptome analysis reveals elevated expression of alcohol-metabolizing genes in NAFLD livers. The Journal of Pathology. 2016;238(4):531–542. doi: 10.1002/path.4650. [DOI] [PubMed] [Google Scholar]
- 7.Baker S. S., Baker R. D., Liu W., Nowak N. J., Zhu L. Role of alcohol metabolism in non-alcoholic steatohepatitis. PLoS One. 2010;5(3, article e9570) doi: 10.1371/journal.pone.0009570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sozen E., Ozer N. K. Impact of high cholesterol and endoplasmic reticulum stress on metabolic diseases: an updated mini-review. Redox Biology. 2017;12:456–461. doi: 10.1016/j.redox.2017.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L., Baker R. D., Baker S. S. Editorial: from multiple hits to multiple therapeutic targets of non-alcoholic fatty liver disease. Current Drug Targets. 2015;16(12):1272–1273. doi: 10.2174/1389450116999150928154553. [DOI] [PubMed] [Google Scholar]
- 10.Sanyal A. J., Chalasani N., Kowdley K. V., et al. Pioglitazone, vitamin E, or placebo for nonalcoholic steatohepatitis. New England Journal of Medicine. 2010;362(18):1675–1685. doi: 10.1056/NEJMoa0907929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Q., Liu W., Baker S. S., et al. Multi-targeting therapeutic mechanisms of the Chinese herbal medicine QHD in the treatment of non-alcoholic fatty liver disease. Oncotarget. 2017;8(17):27820–27838. doi: 10.18632/oncotarget.15482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J.-Y., Zhang L., Li Z.-P., Ji G. Natural products on nonalcoholic fatty liver disease. Current Drug Targets. 2015;16(12):1347–1355. doi: 10.2174/1389450116666150531155711. [DOI] [PubMed] [Google Scholar]
- 13.Chen D., Ke B., Huang Y., et al. Effects of the modified Linggui Zhugan Decoction () combined with short-term very low calorie diets on glycemic control in newly diagnosed type 2 diabetics. Journal of Traditional Chinese Medicine. 2011;31(3):185–188. doi: 10.1016/S0254-6272(11)60038-1. [DOI] [PubMed] [Google Scholar]
- 14.Liu T., Yang L.-L., Zou L., et al. Chinese medicine formula Lingguizhugan decoction improves beta-oxidation and metabolism of fatty acid in high-fat-diet-induced rat model of fatty liver disease. Evidence-Based Complementary and Alternative Medicine. 2013;2013:1–9. doi: 10.1155/2013/429738.429738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y., Lin W., Li C., et al. Multipronged therapeutic effects of Chinese herbal medicine Qishenyiqi in the treatment of acute myocardial infarction. Frontiers in Pharmacology. 2017;8:p. 98. doi: 10.3389/fphar.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C. X. Pharmacology of Traditional Chinese Medicine. Shanghai, China: Shanghai Scientific and Technical Publishers; 2006. [Google Scholar]
- 17.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Gawrieh S., Marion M. C., Komorowski R., et al. Genetic variation in the peroxisome proliferator activated receptor-gamma gene is associated with histologically advanced NAFLD. Digestive Diseases and Sciences. 2011;57(4):952–957. doi: 10.1007/s10620-011-1994-2. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y. C., Ntambi J. M. Regulation of stearoyl-CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochemical and Biophysical Research Communications. 1999;266(1):1–4. doi: 10.1006/bbrc.1999.1704. [DOI] [PubMed] [Google Scholar]
- 20.Ntambi J. M. Regulation of stearoyl-CoA desaturase by polyunsaturated fatty acids and cholesterol. Journal of Lipid Research. 1999;40(9):1549–1558. [PubMed] [Google Scholar]
- 21.Marzuillo P., Grandone A., Perrone L., del Giudice E. M. Understanding the pathophysiological mechanisms in the pediatric non-alcoholic fatty liver disease: the role of genetics. World Journal of Hepatology. 2015;7(11, article 1439):1443. doi: 10.4254/wjh.v7.i11.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamed A. M., El-Kharashi O. A., Boctor S. S., Abd-Elaziz L. F. Potential involvement of PPAR α activation in diminishing the hepatoprotective effect of fenofibrate in NAFLD: accuracy of non-invasive panel in determining the stage of liver fibrosis in rats. Biomedicine & Pharmacotherapy. 2017;85:68–78. doi: 10.1016/j.biopha.2016.11.114. [DOI] [PubMed] [Google Scholar]
- 23.Federico A., Dallio M., Masarone M., Persico M., Loguercio C. The epidemiology of non-alcoholic fatty liver disease and its connection with cardiovascular disease: role of endothelial dysfunction. European Review for Medical and Pharmacological Sciences. 2016;20(22):4731–4741. [PubMed] [Google Scholar]
- 24.Liss K. H., Finck B. N. PPARs and nonalcoholic fatty liver disease. Biochimie. 2017;136:65–74. doi: 10.1016/j.biochi.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verdelho Machado M., Diehl A. M. Role of hedgehog signaling pathway in NASH. International Journal of Molecular Sciences. 2016;17(6) doi: 10.3390/ijms17060857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machado M. V., Michelotti G. A., Xie G., et al. Mouse models of diet-induced nonalcoholic steatohepatitis reproduce the heterogeneity of the human disease. PLoS One. 2015;10(5, article e0127991) doi: 10.1371/journal.pone.0127991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guy C. D., Suzuki A., Zdanowicz M., et al. Hedgehog pathway activation parallels histologic severity of injury and fibrosis in human nonalcoholic fatty liver disease. Hepatology. 2012;55(6):1711–1721. doi: 10.1002/hep.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon H., Song K., Han C., et al. Inhibition of hedgehog signaling ameliorates hepatic inflammation in mice with nonalcoholic fatty liver disease. Hepatology. 2015;63(4):1155–1169. doi: 10.1002/hep.28289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirsova P., Ibrahim S. H., Bronk S. F., Yagita H., Gores G. J. Vismodegib suppresses TRAIL-mediated liver injury in a mouse model of nonalcoholic steatohepatitis. PLoS One. 2013;8(7, article e70599) doi: 10.1371/journal.pone.0070599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Syn W. K., Jung Y., Omenetti A., et al. Hedgehog-mediated epithelial-to-mesenchymal transition and fibrogenic repair in nonalcoholic fatty liver disease. Gastroenterology. 2009;137(4):1478–1488.e8. doi: 10.1053/j.gastro.2009.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh H., Ng C., Ong C., Lim K., Chan T. M. Cloning and characterization of a novel pregnancy-induced growth inhibitor in mammary gland. Endocrinology. 2001;142(8):3607–3615. doi: 10.1210/endo.142.8.8297. [DOI] [PubMed] [Google Scholar]
- 32.Liu M., Li Y., Chen L., et al. Allele-specific imbalance of oxidative stress-induced growth inhibitor 1 associates with progression of hepatocellular carcinoma. Gastroenterology. 2014;146(4):1084–1096.e5. doi: 10.1053/j.gastro.2013.12.041. [DOI] [PubMed] [Google Scholar]
- 33.He J., Hu B., Shi X., et al. Activation of the aryl hydrocarbon receptor sensitizes mice to nonalcoholic steatohepatitis by deactivating mitochondrial sirtuin deacetylase Sirt3. Molecular and cellular biology. 2013;33(10):2047–2055. doi: 10.1128/MCB.01658-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalton T. P., Puga A., Shertzer H. G. Induction of cellular oxidative stress by aryl hydrocarbon receptor activation. Chemico-Biological Interactions. 2002;141(1-2):77–95. doi: 10.1016/s0009-2797(02)00067-4. [DOI] [PubMed] [Google Scholar]
- 35.Lee J. H., Wada T., Febbraio M., et al. A novel role for the dioxin receptor in fatty acid metabolism and hepatic steatosis. Gastroenterology. 2010;139(2):653–663. doi: 10.1053/j.gastro.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu H., Cui W., Klaassen C. D. Nrf2 protects against 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin (TCDD)-induced oxidative injury and steatohepatitis. Toxicology and Applied Pharmacology. 2011;256(2):122–135. doi: 10.1016/j.taap.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He J., Lee J. H., Febbraio M., Xie W. The emerging roles of fatty acid translocase/CD36 and the aryl hydrocarbon receptor in fatty liver disease. Experimental Biology and Medicine. 2011;236(10):1116–1121. doi: 10.1258/ebm.2011.011128. [DOI] [PubMed] [Google Scholar]
- 38.Wahlang B., Song M., Beier J. I., et al. Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicology and Applied Pharmacology. 2014;279(3):380–390. doi: 10.1016/j.taap.2014.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferreira L. R., Abrantes E. F., Rodrigues C. V., et al. Identification and characterization of a novel mouse gene encoding a Ras-associated guanine nucleotide exchange factor: expression in macrophages and myocarditis elicited by Trypanosoma cruzi parasites. Journal of Leukocyte Biology. 2002;72(6):1215–1227. [PubMed] [Google Scholar]
- 40.Ceccarelli S., Panera N., Mina M., et al. LPS-induced TNF-α factor mediates pro-inflammatory and pro-fibrogenic pattern in non-alcoholic fatty liver disease. Oncotarget. 2015;6(39):41434–41452. doi: 10.18632/oncotarget.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller M. D., Pandey R., Li D., et al. Mutation in IRF2BP2 is responsible for a familial form of common variable immunodeficiency disorder. Journal of Allergy and Clinical Immunology. 2016;138(2):544–550.e4. doi: 10.1016/j.jaci.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bae J. S., Park J. M., Lee J., et al. Amelioration of non-alcoholic fatty liver disease with NPC1L1-targeted IgY or n-3 polyunsaturated fatty acids in mice. Metabolism. 2017;66:32–44. doi: 10.1016/j.metabol.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 43.Wang J. M., Wang D., Tan Y. Y., Zhao G., Ji Z. L. Pioglitazone reduces lipid droplets in cholesterolosis of the gallbladder by increasing ABCA1 and NCEH1 expression. Molecular and Cellular Biochemistry. 2015;399(1-2):7–15. doi: 10.1007/s11010-014-2225-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Quality control of Lingguizhugan Decoction under High Performance Liquid Chromatography (HPLC). A. Reference substance; B. Lingguizhugan Decoction 1. glycyrrhizic acid. Standardization of extract by HPLC glycyrrhizic acid was purchased from Shanghai R&D Centre for standardization of Chinese Medicines (Shanghai, China), and the purity was higher than 98.0%;. HPLC-grade reagents were purchased from Burdick & Jackson. An Agilent 1100 HPLC system consisting of a G1354A pump, a G1313A auto-sampler, and a UV/VIS Photodiode Array G1315B Detector was used for all analyses. Chromatographic separations were carried out on an Merck C18 hibar column (4.6 mm × 250mm, 5 μm) with methanol:acetonitrile : water :acetid acid(as solvent ) (15:35:45:0.9,v/v) in the mobile phase at a flow rate of 0.8 mL/min at 25°C for 40 min. Twenty μL (after a high speed centrifugation) of the sample was injected, and the signals were detected at 254 nm with UV detection. The HPLC fingerprint of Lingguizhugan decoction revealed the major peaks (glycyrrhizic acid) at 254 nm. The content of glycyrrhizic acid were 0.9548%. Figure S2. Hierarchical clustering of replicate samples based on correlation matrix in three groups: (A) Normal group; (B) NAFLD model group; (C) LGZG-treated group. Each column and row represents samples in each group. Each cell of the heatmap reflects the distance of two samples. Distance was measured with Euclidean metric. The legend of distance value is shown on the upper right. Figure S2A and figure S2B revealed that s12 and s24 were the outlier samples of normal group and NAFLD model group, respectively. Figure S2C showed no outlier samples, but the data in s37 was missed during assessment of serum ALT and AST. Thus, s37 in LGZG-treated group was also removed in the RNA-seq analysis. Table S1. Primer pairs for real-time PCR analysis. Table S2. Enriched KEGG pathways with NAFLD regulated genes. Table S3. Enriched KEGG pathways with LGZG regulated genes.