Abstract
The carmine spider mite Tetranychus cinnabarinus is a major pest of crop and vegetable plants worldwide. Previous studies have shown that scopoletin is a promising acaricidal compound against Tetranychus cinnabarinus. However, the acaricidal mechanism of scopoletin remains unclear. In the present study, 12 full-length cDNAs of chitinase (CHIT) genes from Tetranychus cinnabarinus (designated TcCHITs) were cloned and characterized. Although TcCHITs were expressed throughout all life stages, their expression levels were significantly upregulated during the larval and nymphal stages. TcCHITs were downregulated 24 h after treatment with scopoletin and upregulated 24 h after treatment with diflubenzuron (DFB, a chitin synthesis inhibitor). Feeding double-stranded RNA effectively silenced TcCHIT transcription in Tetranychus cinnabarinus, thus increasing its susceptibility to scopoletin but reducing that to DFB. Meanwhile, TcCHIT silencing in larvae and adult resulted in an extremely low molting rate (7.3%) and high mortality rate (53.3%), respectively, compared with those in the control group. CHIT genes are closely related to arthropod survival, molting, and development in Tetranychus cinnabarinus, suggesting that acaricidal mechanisms of scopoletin and DFB may occur by inhibition and activation of CHIT gene expression, respectively. TcCHIT constitutes a possible target of scopoletin and DFB in Tetranychus cinnabarinus.
1. Introduction
Phytophagous mites of the genus Tetranychus and Panonychus are major pests on plants worldwide [1, 2]. The carmine spider mite Tetranychus cinnabarinus is of particular importance, because this extreme generalist species has been documented on more than 100 plant species, including food and economic crops, ornamental plants, and weeds [3–5]. Cinnabar spider mite is parthenogenic and exhibits strong fecundity and adaptability. This mite is also one of the pests that are most difficult to control because it easily develops resistance to pesticides [6].
Control of Tetranychus cinnabarinus in open-field crops primarily relies on synthetic chemical acaricides [2, 7–9]. Chemical acaricides have been extensively used to control mite pests because of their quick and efficient acaricidal effect [10]. However, spider mites rapidly develop resistance to almost all acaricidal agents, presenting a major factor that threatens efficient control of spider mites in agriculture [11, 12]. Furthermore, application of chemical acaricides has led to environmental and human health concerns [13]. Thus, controlling mite pests by traditional chemical acaricides has become challenging. Effective methods for controlling mite pests and environment-friendly acaricides should be developed. Phytogenous acaricides, which present low mammalian toxicity and can be rapidly degraded, are suitable for integrated mite management. Studies have also shown that these naturally occurring products may delay development of pesticide resistance in pests [14, 15].
Scopoletin, a coumarin compound, is an important secondary plant metabolite and phytogenous acaricidal compound with excellent contact-killing, systemic, repellent, and oviposition inhibition activities against Tetranychus cinnabarinus [16]. Studies have confirmed that scopoletin manifests growth-regulating, insecticidal, and antibacterial activities [17, 18]. The biological functions influenced by scopoletin are attributed to its various molecular targets, including transcription factors, growth factors, and their receptors, cytokines, enzymes, and genes that regulate cell proliferation and apoptosis [19]. Thus, understanding the mode of action of acaricides is crucial to identifying their molecular targets [20]. Although the acaricidal activity of scopoletin and its possible biochemical mechanism have been investigated, its molecular mechanism or molecular target(s) against Tetranychus cinnabarinus remains unknown.
Chitin is a polymer of β(1,4)-linked N-acetylglucosamine, which is the second most abundant natural polymer after cellulose. Chitin is extensively distributed as a structural component in arthropods, parasites, and microbes [21, 22]. Chitin is also the principal structural component of arthropod exoskeletons and the peritrophic membrane (PM) that lines the epithelium and envelopes gut contents [23]. New chitin is deposited and synthesized during insect growth and development. However, a part of the old cuticle is degraded. When insects or mites are treated with chitin synthesis inhibitors, such as diflubenzuron (DFB) and allosamidin, symptoms of death include development retardation, dysecdysis, and shrinkage [24, 25]. Findings showed that mites treated with scopoletin also exhibit similar death symptoms (Figure 6(c)) [26].
Figure 6.
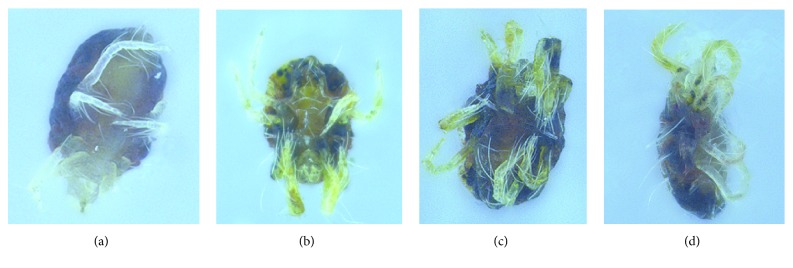
The phenotypes of T. cinnabarinus after DEPC-water exposure (a), dsRNA-TcCHITs feeding (b), scopoletin (c), and DFB (d) exposure for 48 h at the adult stage. Adult mites were treated with the LC50 of scopoletin and DFB. Note that dsRNA-TcCHITs feeding (b), scopoletin (c), and DFB (d) exposure exhibited similar death symptoms: the adult individuals integument shrinkage or damage.
In arthropods, the crucial step of chitin biodegradation pathway is associated with chitinases (CHITs). Insect CHITs, which belong to family 18 of glycosyl hydrolases, mediate digestion of chitin to chitooligosaccharides by hydrolyzing chitin via endo-type cleavage. CHITs are crucial in growth and development of insects and mites and act by hydrolyzing chitin of insect integument and midgut. A total of 16, 22, 20, and 12 CHIT and CHIT-like protein genes have been identified in Drosophila melanogaster, Anopheles gambiae, Tribolium castaneum, and Tetranychus urticae, respectively [27]. These genes have been classified into five or more groups on the basis of amino acid sequence similarity and phylogenetic analyses. CHIT and related proteins influence molting, digestion, cell proliferation, and tissue remodeling of mites and insects. Research showed that chitin is a critical component of insect cuticle and PM and that each period of growth and development of insects and mites requires a certain amount of chitin [23]. CHIT is a safe target of novel biological pesticides because chitin is absent in animals and plants [28]. CHIT inhibitors, such as allosamidin, argifin, and argadin, exert insecticidal effects by inhibiting CHIT activity, which interferes with normal growth and development of insects and mites [25, 29, 30]. Scopoletin has been reported for its insecticidal and growth inhibitory effects against Plutella xylostella, Spilarctia obliqua, and Diabrotica beetles [5, 17, 31]. In recent years, experimental evidence has shown that scopoletin inhibits the development of Tetranychus cinnabarinus, indicating that this compound may affect degradation of chitin by regulating expressions of CHIT genes [32].
Our laboratory investigated the transcriptomics of Tetranychus cinnabarinus after treatment with scopoletin or a control solvent (Table S1 and S2, resp.). Interestingly, we observed that the CHIT gene family, which may be involved in the acaricidal mechanism of scopoletin against Tetranychus cinnabarinus, was differentially expressed. Thus, this study aimed to assess the role of differential expression of CHIT mRNA transcripts in acaricidal activity of scopoletin against Tetranychus cinnabarinus using RNA interference (RNAi). RNAi is a common mechanism of gene silencing in eukaryotic organisms. In recent years, this technique has shown considerable potential in controlling insect pests by silencing vital genes. Functional roles of CHIT genes in Tribolium castaneum and Panonychus citri were also evaluated using RNAi, and several CHIT genes were found to be essential for insect survival, molting, and development [33, 34], thereby revealing functional specialization among CHIT genes during molting. Thus, developing synthetic inhibitors that target CHITs is important for insect pest management.
In the present study, 12 full-length cDNAs of CHIT genes from Tetranychus cinnabarinus were cloned and characterized. Patterns of gene expressions in four developmental stages upon acaricidal treatment were analyzed. We adopted double-stranded RNA (dsRNA) feeding as gene knockdown strategy to investigate the role of CHITs in the acaricidal action of scopoletin and DFB, a chitin synthesis inhibitor, against Tetranychus cinnabarinus. This study demonstrated that suppressing TcCHIT transcription increases susceptibility of T. cinnabarinus to scopoletin but reduces that to DFB. This study also clarified the role of CHITs in the acaricidal mechanism of scopoletin against Tetranychus cinnabarinus.
2. Materials and Methods
2.1. Mite Rearing
The Tetranychus cinnabarinus colony used in this study was collected from cowpeas in Beibei, Chongqing, China, and has been maintained for more than 16 years without exposure to any pesticides [35]. The mites were reared on potted cowpea seedlings (Vigna unguiculata) in the insectary at 25 ± 1°C, 50% ± 5% RH, and 14 : 10 h (L : D) photoperiod.
2.2. Bioassays and RNA-seq Data
Food and Agriculture Organization-recommended slip-dip method was used to measure scopoletin (purity, 95%; Southwest University, Beibei, Chongqing, China) and DFB (purity, 98.5%; Taitan, Shanghai, China) (Figure S1) toxicity against adult female Tetranychus cinnabarinus [36]. We adopted the bioassay procedure described by Ding et al. [35]. A total of 30 adult female mites (3–5 days old) were briefly placed on their backs on double-sided tape on glass. The mites were then dipped into each test solution for 5 s. Each dose was performed in triplicate. The use of sterile distilled water with 0.1% (v/v) Tween 80 and 3% (v/v) acetone was designated as control treatment. The mites were observed under an anatomical microscope after 48 h of rearing under controlled growth conditions as described in mite rearing. Mites that exhibited immobility or irregular trembling of legs were considered dead. Lethal and sublethal concentrations for subsequent experiments were determined on the basis of log-probit analysis of concentration–mortality data. RNA-seq was employed to analyze transcriptome changes in Tetranychus cinnabarinus treated with median lethal concentration (LC50) of scopoletin and the solvent against Tetranychus cinnabarinus for 24 and 48 h, respectively. For scopoletin treatment, more than 300 female adults were transferred onto three freshly potted cowpea leaves, which were placed in a small Petri dish containing water. The leaves were sprayed with scopoletin solution with the abovementioned concentration. Sterile distilled water with 0.1% Tween 80 and 3% acetone was used as solvent for the control group. Three Petri dishes from one independent experiment comprised a replicate, and two biological replicates were used for RNA purification and sequencing. All sequencing data were submitted to the GEO website (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE92959 (unpublished data).
2.3. Total RNA Extraction, cDNA Synthesis, and TcCHIT Cloning
Total RNA was extracted from 300 adult (3–5 days old) Tetranychus cinnabarinus females. Extraction was performed by using RNeasy® Plus Micro Kit (Tiangen, Beijing, China). To determine RNA quantity, the absorbance at 260 nm and absorbance ratio of OD260/280 were measured by using a Nanovue UV-Vis spectrophotometer (GE Healthcare, Fairfield, CT). RNA integrity was further confirmed by 1% agarose gel electrophoresis. Reverse transcription was performed by using a PrimeScript® 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). Synthesized cDNA was stored at −20°C. To obtain full-length TcCHITs, specific primers were designed and synthesized (Table S3) on the basis of complete genomic sequences from sister species Tetranychus urticae (http://bioinformatics.psb.ugent.be/orcae/overview/Teur). Specific polymerase chain reactions (PCRs) were performed in a C1000™ Thermal Cycler (BIO-RAD, Hercules, CA, USA). PCRs were performed with a 25 μL reaction volume with 2.5 μL 10x PCR buffer (Mg2+-free), 2.0 μL dNTPs (2.5 mM), 2.5 μL MgCl2 (25 mM), 1 μL cDNA templates, 1 μL each primer (10 mM), 0.2 μL rTaq™ polymerase (TaKaRa), and 14.8 μL ddH2O. PCR program was 94°C for 3 min, followed by 35 cycles of 94°C for 30 s, 48°C to 60°C (based on primer annealing temperatures) for 30 s, 72°C extension for 1 min to 2 min (based on the predicted length of amplified products), and a final extension of 10 min at 72°C. Amplified PCR fragments were gel-purified by using a gel extraction mini kit (Tiangen, Beijing, China), ligated into pMD™ 19-T vector (Takara, Dalian, China) and then transformed into Trans5α competent cells of Escherichia coli (Tiangen, Beijing, China). Recombinant plasmids were sequenced at the Beijing Genomics Institute (Beijing, China).
2.4. Gene Characterization and Phylogenetic Analysis
Nucleotide sequences of TcCHITs were edited by DNAMAN 5.2.2. The deduced amino acid sequences of 12 CHIT proteins were aligned with ClustalW program [31, 37]. Molecular weight and isoelectric point of the proteins were calculated by using ExPASy Proteomics Server (http://cn.expasy.org/tools/pi_tool.html) [38]. The signal peptide was predicted by using SignalP 4.1 (http://www.cbs.dtu.dk/service/SignalP/) [39], and the transmembrane region was analyzed by using TMHMM Server (v.2.0) (http://www.cbs.dtu.dk/services/TMHMM/) [40]. N-glycosylation sites were predicted by using NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/) [41]. The phylogenetic tree was constructed by using MEGA 5.0 via neighbor-joining method with 1000 bootstrap replicates [42].
2.5. dsRNA Synthesis, dsRNA Feeding, and Knockdown of TcCHIT Expression by RNAi
A set of T7 RNA polymerase promoter primers (Table S3) were designed to amplify 160–600 bp lengths of target genes to generate PCR products for in vitro transcription and dsRNA production (Table S3). TcCHITs and green fluorescent protein (GFP) (ACY56286) genes were amplified by PCR. The PCR program was as described in Section 2.3. Recombinant plasmids were used as templates. The GFP gene was used as negative control. Amplified segments were gel-purified and used in application of TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, Lithuania, EU). dsRNAs were further purified by using GeneJET RNA Purification Kit (Thermo Scientific, Lithuania, EU). Size of dsRNA products was determined by 1% agarose gel electrophoresis. Concentration of dsRNAs was determined by using a spectrophotometer. dsRNAs were stored at −70°C. Systemic delivery of TcCHIT dsRNAs via leaf-disc feeding was used to knock down TcCHIT expression. In this study, to investigate whether knock down of expression of target genes affects transcript levels of nontarget genes via leaf-disc feeding method, the chitin metabolic pathways related to two chitin synthetase genes (tetur03g08510 and tetur08g00170, designated CHS) were detected when a mixture of 12 TcCHIT dsRNAs was applied. The mites were fed with a mixture of 12 different TcCHIT dsRNAs for 48 h. Figure S2 shows the schematic diagram of artificial feeding of dsRNA. Briefly, cowpea leaves were cut to a feeding arena (2 cm in diameter) and dehydrated via incubation at 60°C for 3–5 min. The leaves were then treated with diethylpyrocarbonate- (DEPC-) water, dsRNA-GFP, or TcCHIT dsRNAs (1000 ng/μl) for 3-4 h at room temperature. After complete absorption of liquids, the leaves were placed on wet filter paper. The leaf discs were then placed on water-saturated sponges. Thirty female adults (3–5 days old and starved for 24 h) were placed on each pretreated leaf-disc. The leaf discs were then placed upside down on Petri dishes (7 cm in diameter) to prevent mites from escaping. The dsRNA-treated leaf discs, which were infested with Tetranychus cinnabarinus, were placed under controlled growth conditions as described in Section 2.1. The mites were finally collected for subsequent experiments 48 h after feeding.
2.6. Quantitative Real-Time PCR (qPCR)
To detect TcCHIT expression throughout the different life stages of mites, approximately 2000 eggs, 1500 larvae, 800 nymphs, and 200 adults were collected per sample in triplicate. To quantify TcCHIT expression in response to scopoletin and DFB exposure, we collected 200 female adults per sample in triplicate. To examine the effects of scopoletin and DFB exposure on TcCHIT expression, female adults were treated with scopoletin or DFB, with 0.1% (v/v) Tween 80 and 3% (v/v) acetone as surfactant. As in the slip-dip assay, LC10, LC30, and LC50 of scopoletin and LC50 of DFB corresponded to 0.099, 0.374, 0.938, and 0.477 mg/mL, respectively. For scopoletin and DFB exposure experiment, we adopted a slightly modified version of the leaf-disc dipping method described by Michel et al. [43]. More than 200 female adults (3–5 days old) were briefly transferred to three freshly potted cowpea leaves in a small Petri dish with water. Each detached cowpea leaf was dipped for 5 s in the test solution with the abovementioned concentration. When the liquid had dried around the mites, they were subjected again under the abovementioned conditions. Sterile distilled water with 0.1% Tween 80 and 3% acetone was then used as the control treatment (CK). After a 24 h interval, only surviving female adult mites from the treated and control groups were collected and frozen at −80°C for RNA extraction. Each experiment was performed at least in triplicate and utilized independent biological samples. To examine the effectiveness of RNAi, approximately 200 female adult mites were collected per sample 48 h after dsRNA feeding. Samples were prepared in triplicate. The specific primers used for qPCR of TcCHITs were designed by using Primer 3.0 (http://frodo.wi.mit.edu/) (Table S3) [44]. RPS18 (FJ608659) was used as the stable reference gene for all qPCR assays (Table S3) [45]. qPCR was performed by using a Mx3000P thermal cycler (Agilent Technologies, Inc., Wilmington, NC, USA) on 20 μL reaction mixtures containing 1 μL cDNA template (200 ng/μL), 10 μL iQ™ SYBR® Green Supermix (BIO-RAD, Hercules, CA, USA), 1 μL of each gene-specific primer (0.2 mM), and 7 μL ddH2O. The optimized qPCR protocol used for amplification was 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 15 s, 60°C for 30 s, and elongation at 72°C for 30 s. Melt curve analyses (from 60°C to 95°C) were performed to ensure consistency of amplified products. Quantification of expression level was analyzed using 2−ΔΔCt method [46].
2.7. Susceptibility Test of Tetranychus cinnabarinus to Acaricides after RNAi of TcCHITs
Sublethal doses of scopoletin and DFB (LC30 and LC50 of scopoletin and DFB, resp.) were applied in bioassays. We also adopted the slip-dip method described above and the detailed bioassay procedure described by Ding et al. [35]. LC30 and LC50 values of two acaricides were used as diagnostic doses to compare changes in susceptibility to acaricides in Tetranychus cinnabarinus 48 h after feeding of TcCHIT dsRNAs.
2.8. Statistical Analysis
All experiments included at least three biological replicates. Differences in expression levels of TcCHITs during the four developmental stages and mortality rates were analyzed by one-way analysis of variance, followed by Duncan's multiple tests in SPSS (v.16.0, SPSS Inc., Chicago, IL, USA), at alpha = 0.05.
3. Results
3.1. Analysis of Acaricidal Toxicity
Table 1 presents the LC50 values calculated for the two acaricides against adult Tetranychus cinnabarinus. Estimated LC50 values of scopoletin and DFB reached 0.938 and 0.477 mg/mL, respectively. These results showed that DFB exhibited more significant acaricidal efficiency compared with scopoletin. However, LC50 of scopoletin indicated its excellent toxic effects as a botanical acaricide.
Table 1.
Toxicity of acaricides against adult T. cinnabarinus after 48 h of exposure time.
| Acaricide | N | LC50 (mg·mL−1)a 95% CIb | Slope (±SE) | χ 2 c | P |
|---|---|---|---|---|---|
| Scopoletin | 540 | 0.938 (0.576~2.292) | 1.314 (±0.15) | 6.321 | 0.097 |
| DFB | 540 | 0.477 (0.118~0.902) | 2.254 (±0.24) | 5.939 | 0.051 |
aLC50: median lethal concentration. bCI: 95% confidence interval. cChi-square testing linearity, alpha = 0.05.
3.2. cDNA Cloning and Characterization of TcCHITs
The deduced amino acid sequences and full-length cDNAs of 12 TcCHITs, which contained open reading frames (ORFs), were deposited in GenBank under the accession numbers indicated in Table 2. Table 2 summarizes the lengths of deduced amino acid sequences, predicted protein molecular weights, and theoretical isoelectric points. A signal peptide was detected at the N-terminal end of TcCHIT1, TcCHIT2, TcCHIT3, TcCHIT4, TcCHIT7, TcCHIT8, TcCHIT9, TcCHIT10, TcCHIT11, and TcCHIT12 (Figure 1). Meanwhile, TcCHIT1, TcCHIT2, TcCHIT3, TcCHIT6, TcCHIT7, TcCHIT8, TcCHIT9, TcCHIT10, TcCHIT11, and TcCHIT12 were predicted to contain a chitin-binding domain (Figure 1). TcCHIT1, TcCHIT2, TcCHIT4, TcCHIT5, TcCHIT6, TcCHIT7, TcCHIT8, and TcCHIT9 were predicted to include one catalytic domain; TcCHIIT10 comprised three catalytic domains; and TcCHIT3, TcCHIT11, and TcCHIT12 comprised two catalytic domains. Furthermore, TcCHT3, TcCHIT5, and TcCHIT6 were predicted to contain one transmembrane span domain. These genes, except for TcCHIT7, were also observed to possess potential N-glycosylation sites (Figure 1).
Table 2.
Complete sequence information of the 12 CHIT genes of T. cinnabarinus.
| Gene | Accession numbers | Coding sequence (bp) | Deduced full-length of amino acid | Calculated full-length of molecular (kDa) | Isoelectric point |
|---|---|---|---|---|---|
| TcCHIT1 | KT956964 | 1632 | 543 | 60.8 | 5.44 |
| TcCHIT2 | KT956965 | 1887 | 628 | 69.03 | 8.65 |
| TcCHIT3 | KT956966 | 2793 | 930 | 104.32 | 6.23 |
| TcCHT4 | KT956967 | 1593 | 530 | 59.75 | 6.07 |
| TcCHT5 | KT956968 | 1194 | 397 | 45.63 | 6.12 |
| TcCHT6 | KT956969 | 1272 | 423 | 49.05 | 8.27 |
| TcCHIT7 | KY084261 | 1233 | 410 | 47.34 | 9.19 |
| TcCHIT8 | KY084262 | 1260 | 419 | 48.51 | 9.64 |
| TcCHIT9 | KY084263 | 834 | 277 | 32.02 | 8.92 |
| TcCHT10 | KY084264 | 1881 | 627 | 69.55 | 6.65 |
| TcCHIT11 | KY084265 | 2748 | 915 | 99.42 | 6.46 |
| TcCHT12 | KY084266 | 1293 | 430 | 48.75 | 6.98 |
Figure 1.
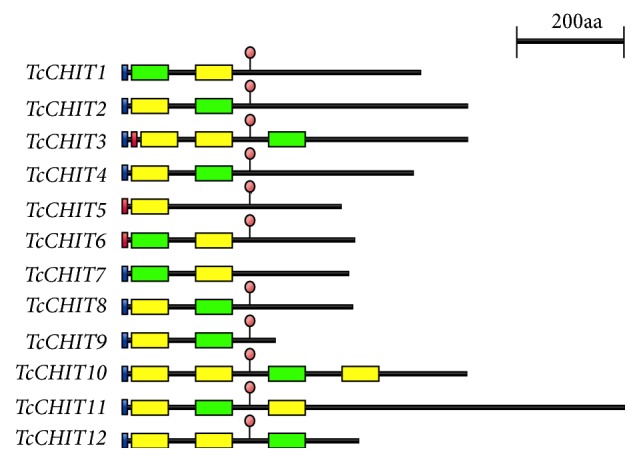
Domain architecture of putative chitinases of T. cinnabarinus. Blue boxes: signal peptide; yellow boxes: catalytic domain; green boxes: chitin-binding domain; red boxes: transmembrane span; red circles: the N-glycosylation sites; lines: linker regions.
3.3. Phylogenetic Analysis of TcCHITs
Phylogenetic analysis was performed by using MEGA 5.0 with the maximum likelihood method on the basis of deduced amino acid sequences of TcCHITs and other known CHIT proteins, including orthologs from the family of Tetranychus urticae, Anopheles gambiae, and Drosophila melanogaster. All CHIT sequences, which possess complete ORFs, were obtained from the Tetranychus urticae genome and the National Center for Biotechnology Information (Bethesda, MD) (https://www.ncbi.nlm.nih.gov/) (Table S4). Results of phylogenetic analysis revealed that CHIT genes for Tetranychus cinnabarinus can be divided into four groups (Figure 2): TcCHIT4, TcCHIT5, TcCHIT6, TcCHIT7, TcCHIT8, and TcCHIT9 under Group I CHITs; TcCHT1, TcCHIT11 in Group II CHITs; TcCHIT3, TcCHIT12 under Group III CHITs; TcCHIT2, TcCHIT10 under Group IV CHITs. CHIT genes from Tetranychus cinnabarinus and Tetranychus urticae clustered into the CHIT family and shared a single clade (Figure 2). This result suggests that TcCHITs and TuCHITs are evolutionarily related and possibly share similar physiological functions.
Figure 2.
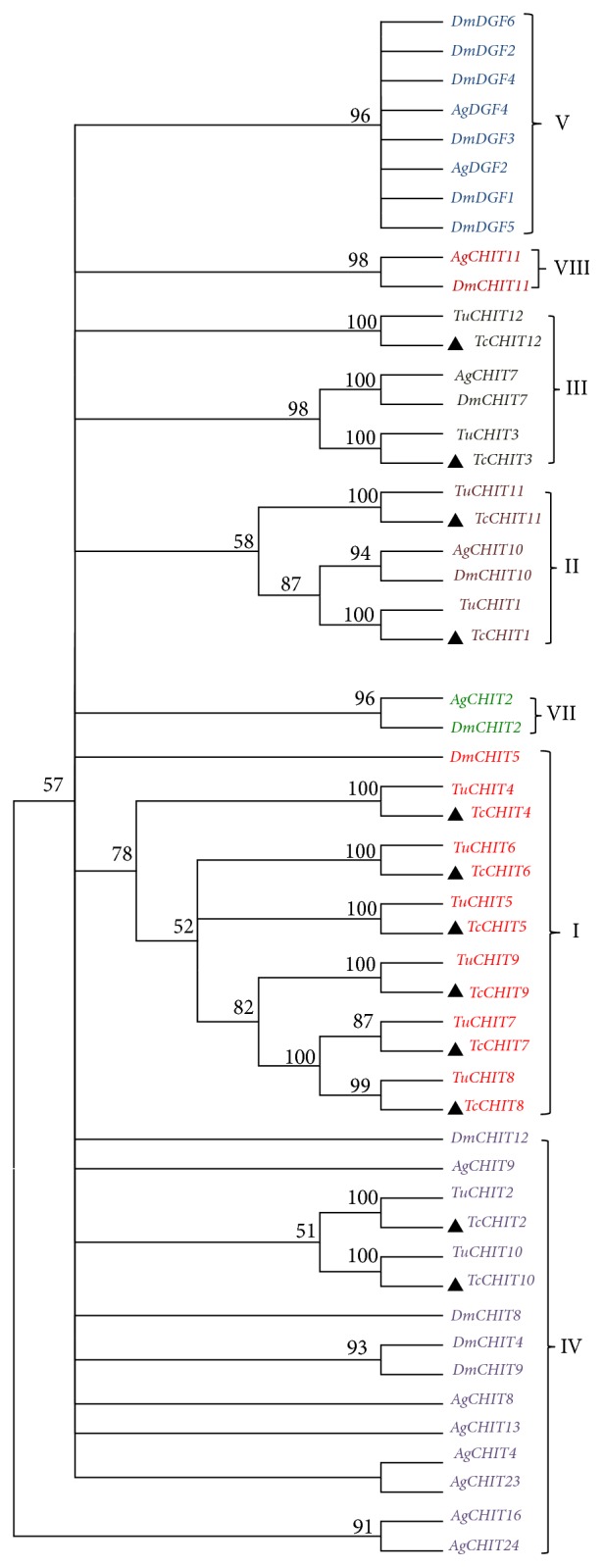
Phylogenic analysis of TcCHITs. Maximum likelihood tree constructed by MEGA 5.0. Phylogeny testing was conducted via the bootstrap method with 1000 replications. Sequences used for constructing the tree are listed in Supplementary Table S4.
3.4. Expression Patterns of TcCHITs in Different Developmental Stages and upon Acaricide Treatment
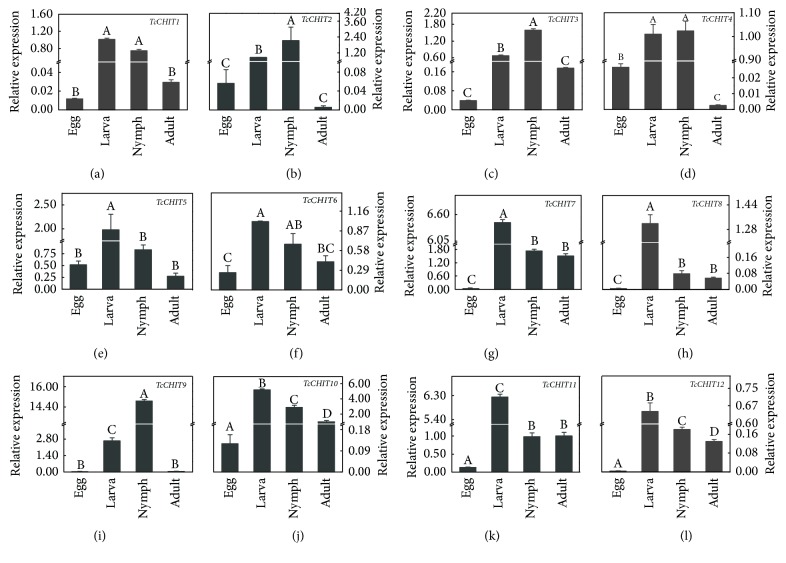
qPCR was performed to evaluate TcCHIT gene expression levels during different developmental stages (egg, larva, nymph, and adult) and upon acaricidal treatment. Results showed that the 12 CHIT genes (TcCHIT1 to -12) were expressed throughout all life stages, suggesting the involvement of TcCHITs in biological processes in all developmental and growth stages. Specifically, TcCHITs were highly expressed during the larval and nymphal stages compared to in other developmental stages; TcCHIT expression levels were the lowest during the egg stage (Figure 3). Statistical analysis suggests that relative expression levels of TcCHITs totaled 0.012, 0.056, 0.038, 0.027, 0.514, 0.254, 0.029, 0.004, 0.019, 0.120, 0.126, and 0.004 during the egg stage; 1.009, 0.845, 0.677, 1.009, 1.982, 1.006, 6.434, 1.318, 2.648, 5.192, 6.243, and 0.650 during the larval stage; 0.745, 2.130, 1.583, 1.024, 0.834, 0.672, 1.730, 0.078, 14.873, 2.902, 0.981, and 0.181 during the nymphal stage; and 0.029, 0.006, 0.175, 0.002, 0.275, 0.416, 1.504, 0.057, 0.051, 1.000, 1.000, and 0.129 during the adult stage (Figure 3).
Figure 3.
RT-qPCR evaluation of the developmentally specific expression patterns of the 12 CHIT genes in T. cinnabarinus. The following life stages were analyzed: egg, larvae, nymph, and adult. Error bars represent the standard error of the calculated mean based on three biological replicates. Different letters on the error bars indicate significant difference according to Duncan's multiple tests (alpha = 0.05), that is, no statistical difference between “A” and “A”; significant difference among “A,” “B,” “C,” and “D.” RPS18 was used as the reference gene. (a) to (l) were TcCHIT1~12, respectively.
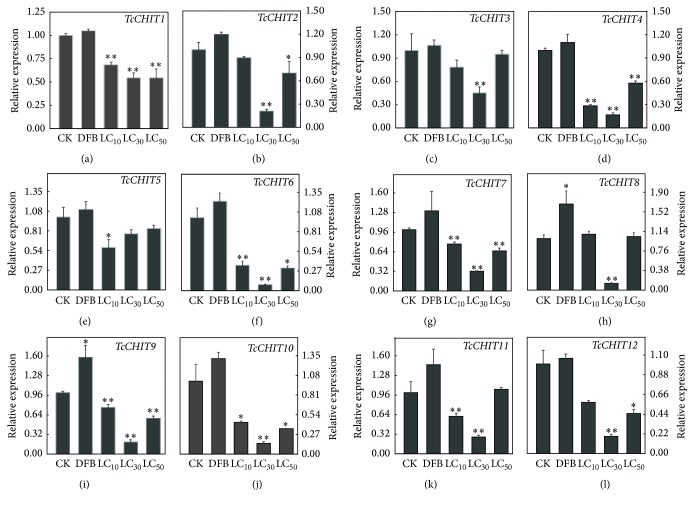
Results of scopoletin treatment experiment showed that, compared with the genes in the control group, all 12 CHIT genes (TcCHIT1 to -12) were downregulated after 24 h of exposure to scopoletin (Figure 4). Statistical analysis suggests that compared with expression levels of the control (CK), relative expression levels of TcCHITs were 1.5-, 1.1-, 1.3-, 3.5-, 1.7-, 2.9-, 1.3-, 0.9-, 1.3-, 2.3-, 1.6-, and 1.7-fold lower at LC10 doses of scopoletin; 1.8-, 4.8-, 2.3-, 6.0-, 1.3-, 12.4-, 2.3-, 7.7-, 5.2-, 6.7-, 3.7-, and 5.2-fold lower at LC30 doses of scopoletin; and 1.8-, 1.4-, 1.1-, 1.7-, 1.2-, 3.2-, 1.1-, 1.0-, 1.7-, 2.9-, 0.9-, and 2.2-fold lower at LC50 doses of scopoletin. However, relative expression levels of all the 12 CHIT genes were upregulated and were 1.1-, 1.2-, 1.1-, 1.3-, 1.1-, 1.2-, 1.3-, 1.7-, 1.6- 1.3-, 1.5, and 1.1-fold higher than those in the control (CK) after treatment with DFB at LC50.
Figure 4.
Expression profiles of TcCHITs transcripts after scopoletin treatment for 24 h at three different concentrations. Error bars represent the standard error of the calculated mean based on three biological replicates. Water containing 0.1% Tween-80 was used as the control treatment (CK). LC50 of DFB was used as the positive control. Asterisk (∗) on the error bar indicates a significant difference between the treatment and group (CK) according to t-tests, (∗p < 0.05) or (∗∗p < 0.01). RPS18 was used as the reference gene. (a) to (l) were TcCHIT1~12, respectively.
3.5. RNAi via dsRNA Knockdown
To validate existence of offsite effects, expressions of all 12 CHIT genes and 2 CHS genes were detected when a mixture of 12 TcCHIT dsRNAs was applied (Figure 5). mRNA expressions of TcCHITs significantly decreased but not those of TcCHSs when TcCHIT dsRNAs were applied at the adult stage in mites (Figure 5). Results showed that transcript levels of TcCHITs significantly decreased to 57.30%, 53.87%, 32.23%, 60.37%, 72.32%, 61.78%, 55.49%, 70.14%, 81.16%, 51.02%, 77.57%, and 35.50% compared with transcript levels of TcCHITs after DEPC-water treatment (Figure 5). No significant difference in transcript efficiency existed between the two controls (water and dsGFP) (Figure 5). These results indicate the absence of offsite effect of RNAi experiments in this study. At 48 h after feeding of TcCHIT dsRNAs, 53.3% of mites died because of integument shrinkage or damage (Figures 6(b) and 7). By contrast, the mites fed with dsGFP showed 4.7% mortality. These results reveal successful knockdown of TcCHIT transcripts by RNAi in Tetranychus cinnabarinus.
Figure 5.
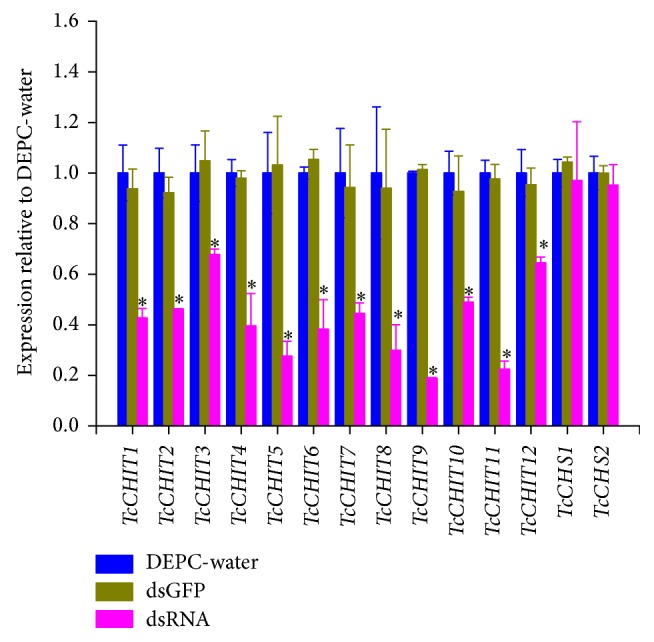
Quantitative PCR detection of target gene expression after dsRNA-TcCHITs feeding relative to expression levels after DEPC-water treatment. dsGFP was adopted as negative control. RPS18 was used as the reference gene. Asterisk (∗) on the error bar indicates a significant difference between the treatment and group (DEPC-water) according to t-tests, p < 0.05.
Figure 7.
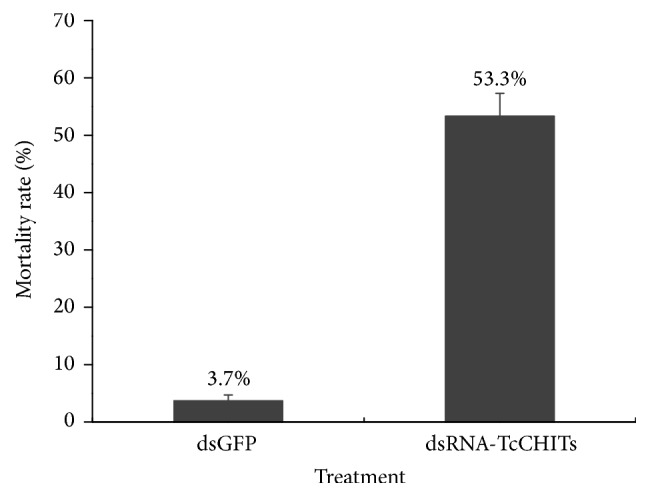
Mortality rate of T. cinnabarinus after being artificially fed with dsGFP or dsRNA-TcCHITs at the adult stage after 48 h. Means were compared by t-tests, p < 0.05 (n = 3, including 180 mites).
To explore the biological functions of TcCHITs, RNAi method was applied to knock down TcCHITs expression at the larval stage. Consequently, at 48 h after dsRNA feeding, 92.7% of mites died because of failure to molt or to undergo dysecdysis (Figures 8 and 9, resp.). By contrast, the mites fed with dsGFP exhibited only 1.4% mortality. At 72 h after dsRNA feeding, the remaining mites still failed to molt after treatment with TcCHIT dsRNAs, whereas all the mites in the control group successfully turned into nymphs. At 72 h after dsRNA feeding, molting rate reached 7.3% during treatment of TcCHITs dsRNAs and totaled 98.6% in dsGFP treatment (Figure 9).
Figure 8.
The phenotypes of T. cinnabarinus after being artificially fed with dsGFP or dsRNA-TcCHITs at the larval stage after 72 h. The artificial feeding of dsTcCHITs resulted in a lethal phenotype: the larva individuals failed molting or undergoing dysecdysis, whereas mites fed with dsGFP developed normally.
Figure 9.
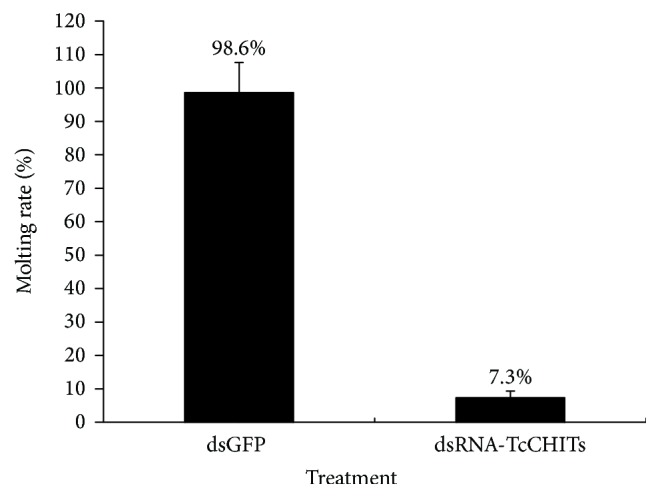
Molting rate of T. cinnabarinus after being artificially fed with dsGFP or dsRNA-TcCHITs at the larval stage after 72 h. Means were compared by t-tests, p < 0.05.
3.6. Susceptibility Test of Tetranychus cinnabarinus to Acaricides after RNAi of TcCHITs
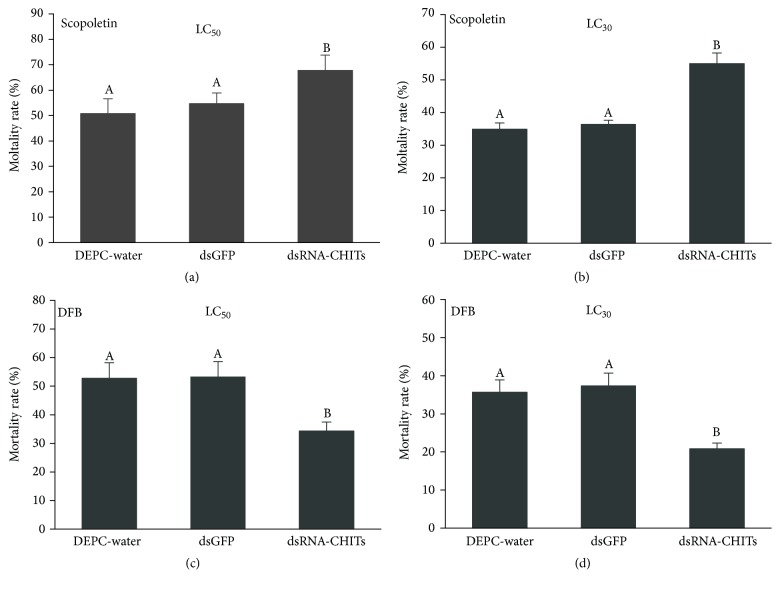
Susceptibilities to the two acaricides at 48 h after TcCHITs dsRNA feeding at the adult stage were detected by the slip-dip method. TcCHITs transcripts in the LC50 and LC30 assays of scopoletin were knocked down by RNAi in Tetranychus cinnabarinus. Mortality significantly increased to 16.92% and 20.12% in the mites fed with TcCHITs dsRNAs compared with mites treated with DEPC-water (Figure 10). Contradictory results were generated when TcCHITs transcripts in the LC50 and LC30 assays of DFB were knocked down by RNAi in Tetranychus cinnabarinus. Mortality significantly decreased to 18.43% and 14.88% in the mites fed with TcCHITs dsRNAs compared with those treated with DEPC-water (Figure 10). No significant difference in mortality existed between DEPC-water and dsRNA-GFP treatments (Figure 10). These results demonstrate that RNAi of CHIT genes increased susceptibility of Tetranychus cinnabarinus to scopoletin but reduced that to DFB. These results indicate that CHIT genes may play a crucial role in the acaricidal effects of scopoletin and DFB.
Figure 10.
Knockdown of TcCHITs expressions increased susceptibility to scopoletin and reduced susceptibility to DFB in mites. (a, b) Mortality of TcCHITs-silenced T. cinnabarinus to scopoletin at LC50 and LC30, respectively. (c, d) Mortality of TcCHITs-silenced T. cinnabarinus to DFB at LC50 and LC30, respectively. dsGFP was adopted as negative control. Error bars represent the standard error of the calculated mean based on three biological replicates. Different letters on the error bars indicate significant difference according to Duncan's multiple tests (alpha = 0.05) that is, no statistical difference between “A” and “A”; significant difference between “A” and “B”.
4. Discussion
As an important phenolic phytoalexin in plants, scopoletin features numerous pharmacological activities, such as antitumor activity. Scopoletin can affect and disrupt growth, proliferation, metastasis, and metabolism of tumor cells and induce apoptosis [47, 48]. However, the potential acaricidal mechanism of scopoletin as a plant-derived acaricide against Tetranychus cinnabarinus remains unknown.
Identification and characterization of CHIT genes from mites will aid in determining the involvement of CHITs in responses of mites to specific acaricides. Findings of the present study will also help us to better understand the biological functions of CHITs. We cloned and characterized 12 full-length CHIT cDNAs in Tetranychus cinnabarinus (TcCHIT1 to -6; Wang et al. [26]). In 1993, full-length cDNA of the first insect CHIT gene was cloned and identified from the tobacco hornworm Manduca sexta [49]. Since then, CHIT genes of various insects have been cloned and identified. The encoded CHITs in Tribolium castaneum have been divided into eight subgroups on the basis of sequence similarity and domain architecture [50]. Structural analysis of TcCHITs demonstrated that these genes possess a multidomain structural organization, which includes 1–3 catalytic domains (GH-18 domain), 0-1 cysteine-rich chitin-binding domains (peritrophin-A domain/CBM-14 domain), and serine/threonine-rich regions that can be heavily glycosylated (Figure 1). TcCHITs are predicted to feature a signal peptide (TcCHT1, TcCHT2, TcCHT3, TcCHT4, TcCHT7, TcCHT8, TcCHT9, TcCHT10, TcCHT11, and TcCHT12) or a transmembrane (TcCHT3, TcCHT5, and TcCHT6) span domain as they are targeted either to the extracellular space or sorted into the plasma membrane, in both cases facing carbohydrates of the extracellular matrix (Figure 1). Differences in their domain architecture indicate distinctive biological functions for specific CHITs. For example, Xia et al. [34] reported that CHIT5, TcCHT7, and TcCHT10, which are also expressed at all developmental stages, play critical roles in digesting the old pupal cuticle, wing/elytra extension, and molting in Tribolium castaneum, respectively.
TcCHIT transcripts were detected in all four tested developmental stages of Tetranychus cinnabarinus, indicating that the CHIT gene family plays an important role during the entire life cycle of mites. However, TcCHIT expression levels during larval and nymphal stages were significantly higher than in other developmental stages of Tetranychus cinnabarinus, agreeing with the results of Yang et al. [51]. Our present results showed that the CHIT gene family exhibits different developmental patterns of expression in Tetranychus cinnabarinus. Expression level of TcCHIT9 during the larval and nymphal stages was approximately 250-fold higher than those during the egg and adult stages, whereas TcCHT3, TcCHT7, TcCHT10, and TcCHT11 were approximately 10-fold higher. Differences in expression levels may be related to the structure and function of CHIT genes. Different developmental patterns of expression indicate functional specialization in the CHIT gene family of Tetranychus cinnabarinus during molting. In addition, RNAi studies in Tribolium castaneum have provided strong experimental evidence for different developmental patterns of expression and tissue-specific expression of different CHIT genes [34].
Results of the present study further showed that 24 h after scopoletin exposure, TcCHIT expression levels were downregulated and that the challenge of inhibiting TcCHIT1, TcCHT6, TcCHT7, TcCHT9, and TcCHT10 expressions was more significant than that of inhibiting the remaining CHIT genes in Tetranychus cinnabarinus. Specifically, statistical analysis suggests that TcCHIT expression levels were downregulated more significantly with LC30 dose of scopoletin than with LC10 and LC50 doses of scopoletin than those of the control. This result suggests the possible benefits of using appropriate doses of scopoletin. This study also showed that TcCHITs expression levels were significantly downregulated during the adult stage of mites. The preceding results indicate that the acaricidal mechanism of scopoletin may degrade chitin biodegradation in Tetranychus cinnabarinus by decreasing CHIT gene expression. However, relative expression levels of TcCHIT8 and TcCHT9 were significantly upregulated after DFB treatment (positive control). Several studies have shown that the action mechanism of DFB may occur through direct inhibition of chitin synthase activity; inhibition of zymogen activation process; activation of CHITs; interference with hormonal balance; interference with nerve-secreting cells of the brain; and interference with nucleic acid, protein synthesis, and metabolism in several different insect species [52]. However, the acaricidal mechanism of DFB remains unknown. These results suggest that TcCHITs play an essential role in acaricidal mechanisms of scopoletin and DFB in Tetranychus cinnabarinus.
In this study, we employed RNAi to investigate the biological roles of mite CHIT genes in Tetranychus cinnabarinus. RNAi has become an increasingly common method to knock down expression of genes of interest in insects and mites. RNAi also features potential applications in screening and identification of pharmaceutical targets. Previous studies have demonstrated that expressions of specific CHIT genes in Tribolium castaneum can be knocked down by microinjection assays with gene-specific RNAi [34]. For example, Xia et al. [34] reported that specific knockdown of CHT10 transcripts of Tribolium castaneum, which contains multiple catalytic domains, prevented embryo hatching, larval molting, pupation, and adult metamorphosis. In this study, results of qPCR analyses showed that TcCHIT transcript levels significantly decreased to 30%–80% in mites 48 h after feeding with TcCHITs dsRNAs. In this study, TcCHITs silencing in larvae and adult samples resulted in high mortality rate. These results not only demonstrated successful dsRNA-mediated knockdown of TcCHITs transcripts but also showed suitability of dsRNA delivery via the leaf-disc method for RNAi in Tetranychus cinnabarinus.
Several studies have shown that activation or inhibition of CHITs kills insects or mites. Ding et al. [53] reported that mortality of M. sexta larvae was higher when treated with a sublethal dose of Bacillus thuringiensis (Bt) toxin on transgenic tobacco lines that express M. sexta CHIT (Group I). This result suggests that overexpression of CHIT increases susceptibility of M. Sexta to Bt. In addition, Sakuda et al. [25] showed that allosamidin strongly inhibits insect CHITs and exerts insecticidal effects by preventing molting of insect larvae and pupae. These results indicate that inhibition of CHITs can lead to insect death. Our results showed that TcCHITs play a similar role in acaricidal mechanisms of scopoletin and DFB. Expressions of TcCHITs were downregulated 24 h after scopoletin exposure. Specifically, scopoletin susceptibility increased when TcCHITs in LC50 and LC30 assays were suppressed via RNAi. These results suggest that the acaricidal mechanism of scopoletin may transpire by inhibition of expressions of CHIT genes. However, TcCHIT expressions were upregulated 24 h after DFB treatment. Meanwhile, DFB susceptibility decreased when TcCHIT transcripts were knocked down by RNAi in Tetranychus cinnabarinus. These results indicate that the acaricidal mechanism of DFB may transpire by activation of CHIT gene expression. The function of CHIT genes of Tetranychus cinnabarinus has not been reported at present. However, Zhang et al. [27] reported that CHITs are closely related to insect molting in Tribolium castaneum, suggesting that acaricidal mechanism of scopoletin and DFB possibly prevents mites from undergoing normal growth and development by destroying the integument of Tetranychus cinnabarinus. This paper is the first to report that knockdown of CHIT gene expression in Tetranychus cinnabarinus increases susceptibility to scopoletin but reduces that to DFB.
Acknowledgments
This research was partially supported by a combination of funding from the National Science Foundation of China (31272058, 31572041, and 31601674).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Hong Zhou and Yong-qiang Zhang contributed equally to this work. Hong Zhou, Yong-qiang Zhang, Dan Wang, and Wei Ding conceived and designed the experiments; Hong Zhou, Ting Lai, Fu-you Guo, and Jin-lin Liu performed the experiments and analyzed the data; Hong Zhou, Yong-qiang Zhang, and Wei Ding wrote the paper; Hong Zhou, Wei Ding, and Yong-qiang Zhang revised the paper.
Supplementary Materials
Table S1. The differentially expressed genes between scopoletin- and solvent-treated mites at 24 h after treatment in RNA-seq. Table S2. The differentially expressed genes between scopoletin- and solvent-treated mites at 48 h after treatment in RNA-seq. Table S3. Primers used for cloning, qPCR, and RNAi. Table S4. Sequences and relevant information for phylogenetic analysis of TcCHITs. Figure S1. Chemical structure of scopoletin (A) and diflubenzuron (DFB) (B). Figure S2. A picture of leaf-disc mediated dsRNA feeding.
References
- 1.Jeppson L. R., Keifer H. H., Baker E. W. Mites Injurious to Economic Plants. Berkeley, CA, USA; 1975. (University of California Press). [Google Scholar]
- 2.Zhang Z. Q. Mites of Greenhouses: Identification, Biology and Control. Oxon, UK: CABI Publishing; 2003. [DOI] [Google Scholar]
- 3.Zhang Y., Zhang Z., Yutaka S., Liu Q., Ji J. On the causes of mite pest outbreaks in mono- and poly-cultured moso bamboo forests. Chinese Journal of Applied Ecology. 2004;15(7):1161–1165. [PubMed] [Google Scholar]
- 4.Çakmak I., Başpinar H., Madanlar N. Control of the carmine spider mite Tetranychus cinnabarinus boisduval by the predatory mite Phytoseiulus persimilis (Athias-Henriot) in protected strawberries in Aydin, Tukey. Turkish Journal of Agriculture and Forestry. 2005;29(4):259–265. [Google Scholar]
- 5.Sarwar M. Management of spider mite Tetranychus cinnabarinus (Boisduval) (Tetranychidae) infestation in cotton by releasing the predatory mite Neoseiulus pseudolongispinosus (Xin, Liang and Ke) (Phytoseiidae) Biological Control. 2013;65(1):37–42. doi: 10.1016/j.biocontrol.2012.09.017. [DOI] [Google Scholar]
- 6.Cruz E. M. D. O., Costa-Junior L. M., Pinto J. A. O., et al. Acaricidal activity of Lippia gracilis essential oil and its major constituents on the tick Rhipicephalus (Boophilus) microplus. Veterinary Parasitology. 2013;195(1-2):198–202. doi: 10.1016/j.vetpar.2012.12.046. [DOI] [PubMed] [Google Scholar]
- 7.Dekeyser M. A. Acaricide mode of action. Pest Management Science. 2005;61(2):103–110. doi: 10.1002/ps.994. [DOI] [PubMed] [Google Scholar]
- 8.Marcic D. Acaricides in modern management of plant-feeding mites. Journal of Pest Science. 2012;85(4):395–408. doi: 10.1007/s10340-012-0442-1. [DOI] [Google Scholar]
- 9.Van Leeuwen T., Vontas J., Tsagkarakou A., Dermauw W., Tirry L. Acaricide resistance mechanisms in the two-spotted spider mite Tetranychus urticae and other important Acari: a review. Insect Biochemistry and Molecular Biology. 2010;40(8):563–572. doi: 10.1016/j.ibmb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Liu X., Wu D., Zhang Y., Zhou H., Lai T., Ding W. RNA-Seq Analysis Reveals Candidate Targets for Curcumin against Tetranychus cinnabarinus. BioMed Research International. 2016;2016:10. doi: 10.1155/2016/2796260.2796260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Zhang Z., Zhao Z. Pesticide resistance of Tetranychus cinnabarinus (Acari: Tetranychidae) in China: a review. Systematic and Applied Acarology. 1998;3(1):3–7. doi: 10.11158/saa.3.1.1. [DOI] [Google Scholar]
- 12.Van Leeuwen T., Van Pottelberge S., Tirry L. Biochemical analysis of a chlorfenapyr-selected resistant strain of Tetranychus urticae Koch. Pest Management Science. 2006;62(5):425–433. doi: 10.1002/ps.1183. [DOI] [PubMed] [Google Scholar]
- 13.Yang H.-Z., Hu J.-H., Li Q., et al. Acaricidal Activity of Boenninghausenia sessilicarpa Against Panonychus citri. Agricultural Sciences in China. 2009;8(9):1097–1102. doi: 10.1016/S1671-2927(08)60317-X. [DOI] [Google Scholar]
- 14.Feng R., Isman M. B. Selection for resistance to azadirachtin in the green peach aphid, Myzus persicae. Experientia. 1995;51(8):831–833. doi: 10.1007/BF01922438. [DOI] [Google Scholar]
- 15.Zhang Y.-Q., Yang Z.-G., Ding W., Luo J.-X. Synergistic inhibitory effect of scopoletin and bisdemethoxycurcumin on Tetranychus cinnabarinus (Boisduval) (Acari: Tetranychidae) Zeitschrift fur Naturforschung - Section C Journal of Biosciences. 2016;71(1-2):1–8. doi: 10.1515/znc-2014-4131. [DOI] [PubMed] [Google Scholar]
- 16.Yong X. J., Zhang Y. Q., Ding W. Repellent and oviposition deterrent properties of scopoletin to Tetranychus cinnabarinus. Chinese Journal of Applied Entomology. 2012;49(2):422–427. [Google Scholar]
- 17.Peterson J. K., Harrison H. F., Jackson D. M., Snook M. E. Biological Activities and Contents of Scopolin and Scopoletin in Sweetpotato Clones. HortScience. 2003;38(6):1129–1133. [Google Scholar]
- 18.Tripathi A. K., Bhakuni R. S., Upadhyay S., Gaur R. Insect feeding deterrent and growth inhibitory activities of scopoletin isolated from Artemisia annua against Spilarctia obliqua (Lepidoptera: Noctuidae) Insect Science. 2011;18(2):189–194. doi: 10.1111/j.1744-7917.2010.01350.x. [DOI] [Google Scholar]
- 19.Kunnumakkara A. B., Anand P., Aggarwal B. B. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Letters. 2008;269(2):199–225. doi: 10.1016/j.canlet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 20.van Leeuwen T., Dermauw W., Grbic M., Tirry L., Feyereisen R. Spider mite control and resistance management: does a genome help? Pest Management Science. 2013;69(2):156–159. doi: 10.1002/ps.3335. [DOI] [PubMed] [Google Scholar]
- 21.Arakane Y., Hogenkamp D. G., Zhu Y. C., et al. Characterization of two chitin synthase genes of the red flour beetle, Tribolium castaneum, and alternate exon usage in one of the genes during development. Insect Biochemistry and Molecular Biology. 2004;34(3):291–304. doi: 10.1016/j.ibmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Lehane M. J. Peritrophic matrix structure and function. Annual Review of Entomology. 1997;42(42):525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- 23.Merzendorfer H., Zimoch L. Chitin metabolism in insects: structure, function and regulation of chitin synthases and chitinases. Journal of Experimental Biology. 2003;206(4):4393–4412. doi: 10.1242/jeb.00709. [DOI] [PubMed] [Google Scholar]
- 24.Xia W.-K., Ding T.-B., Niu J.-Z., et al. Exposure to diflubenzuron results in an up-regulation of a chitin synthase 1 gene in citrus red mite, panonychus citri (Acari: Tetranychidae) International Journal of Molecular Sciences. 2014;15(3):3711–3728. doi: 10.3390/ijms15033711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakuda S., Isogai A., Matsumoto S., Suzuki A. Search for Microbial Insect Growth Regulators II. Allosamidin, A Novel Insect Chitinase Inhibitor. The Journal of Antibiotics. 1987;40(3):296–300. doi: 10.7164/antibiotics.40.296. [DOI] [PubMed] [Google Scholar]
- 26.Wang D., Zhang B. C., Ding W. Effects of curcumin on the expression of chitinase genes from the carmine spider mite, Tetranychus cinnabarinus (Boisduval) Chinese Journal of Pesticide Science. 2016;18(2):165–176. [Google Scholar]
- 27.Zhang J., Zhang X., Arakane Y., et al. Comparative genomic analysis of chitinase and chitinase-like genes in the African malaria mosquito (Anopheles gambiae) PLoS ONE. 2011;6(5) doi: 10.1371/journal.pone.0019899.e19899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito S., Odagiri M., Furuya S., Suzuki S., Takayanagi T. Inhibitory effect of chitinases isolated from Semillon grapes (Vitis vinifera) on growth of grapevine pathogens. Journal of Plant Biochemistry and Biotechnology. 2011;20(1):47–54. doi: 10.1007/s13562-010-0025-2. [DOI] [Google Scholar]
- 29.Õmura S., Arai N., Yamaguchi Y., et al. Argifin, a new chitinase inhibitor, produced by Gliocladium sp. FTD-0668. I. Taxonomy, fermentation, and biological activities. The Journal of Antibiotics. 2000;53(6):603–608. doi: 10.7164/antibiotics.53.603. [DOI] [PubMed] [Google Scholar]
- 30.Nitoda T., Usuki H., Kurata A., Kanzaki H. Macromolecular insect chitinase inhibitors produced by fungi: Screening and partial characterization. Journal of Pesticide Science. 2003;28(1):33–36. doi: 10.1584/jpestics.28.33. [DOI] [Google Scholar]
- 31.Hill C. B., Li Y., Hartman G. L. Resistance to the Soybean Aphid in Soybean Germplasm. Crop Science. 2004;44(1):98–106. doi: 10.2135/cropsci2004.0098. [DOI] [Google Scholar]
- 32.Yong X. J., Zhang Y. Q., Ding W. Sublethal effects of scopoletin on the experimental population of the carmine spider mite, Tetranychus cinnabarinus (Boisduval) (Acari: Tetranychidae) Acta Entomological Sinica. 2011;54(12):1377–1383. [Google Scholar]
- 33.Zhu Q., Arakane Y., Beeman R. W., Kramer K. J., Muthukrishnan S. Functional specialization among insect chitinase family genes revealed by RNA interference. Proceedings of the National Acadamy of Sciences of the United States of America. 2008;105(18):6650–6655. doi: 10.1073/pnas.0800739105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia W.-K., Shen X.-M., Ding T.-B., et al. Functional analysis of a chitinase gene during the larval-nymph transition in Panonychus citri by RNA interference. Experimental and Applied Acarology. 2016;70(1):1–15. doi: 10.1007/s10493-016-0063-0. [DOI] [PubMed] [Google Scholar]
- 35.Ding L.-J., Ding W., Zhang Y.-Q., Luo J.-X. Bioguided fractionation and isolation of esculentoside P from Phytolacca americana L. Industrial Crops and Products. 2013;44(1):534–541. doi: 10.1016/j.indcrop.2012.09.027. [DOI] [Google Scholar]
- 36.Busvine J. R. Recommended Methods for Measurement of Pest Resistance to Pesticides. Food and Agriculture Organization of the United Nations; 1980. [Google Scholar]
- 37.Bansal R., Hulbert S., Schemerhorn B., et al. Hessian fly-associated bacteria: Transmission, essentiality, and composition. PLoS ONE. 2011;6(8) doi: 10.1371/journal.pone.0023170.e23170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bairoch A. The PROSITE dictionary of sites and patterns in proteins, its current status. Nucleic Acids Research. 1993;21(13):3097–3103. doi: 10.1093/nar/21.13.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: signalP 3.0. Journal of Molecular Biology. 2004;340(4):783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 40.Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 41.Gupta R., Birch H., Rapacki K., Brunak S., Hansen J. E. O-GLYCBASE version 4.0: A revised database of O-glycosylated proteins. Nucleic Acids Research. 1999;27(1):370–372. doi: 10.1093/nar/27.1.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel A. P., Mian M. A. R., Davila-Olivas N. H., Caas L. A. Detached leaf and whole plant assays for soybean aphid resistance: Differential responses among resistance sources and biotypes. Journal of Economic Entomology. 2010;103(3):949–957. doi: 10.1603/EC09337. [DOI] [PubMed] [Google Scholar]
- 44.Misener S., Krawetz S. A. Bioinformatics Methods and Protocols. 1, no. 2. Humana Press; 2010. [DOI] [Google Scholar]
- 45.Sun W., Jin Y., He L., Lu W.-C., Li M. Suitable reference gene selection for different strains and developmental stages of the carmine spider mite, Tetranychus cinnabarinus, using quantitative real-time PCR. Journal of Insect Science. 2010;10, article 208 doi: 10.1673/031.010.20801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Livak K. J., Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Manuele M. G., Ferraro G., Barreiro Arcos M. L., López P., Cremaschi G., Anesini C. Comparative immunomodulatory effect of scopoletin on tumoral and normal lymphocytes. Life Sciences. 2006;79(21):2043–2048. doi: 10.1016/j.lfs.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 48.Happi E. N., Tcho A. T., Sirri J. C., et al. Tirucallane triterpenoids from the stem bark of Araliopsis synopsis. Phytochemistry Letters. 2012;5(3):423–426. doi: 10.1016/j.phytol.2012.03.014. [DOI] [Google Scholar]
- 49.Kramer K. J., Corpuz L., Choi H. K., Muthukrishnan S. Sequence of a cDNA and expression of the gene encoding epidermal and gut chitinases of Manduca sexta. Insect Biochemistry and Molecular Biology. 1993;23(6):691–701. doi: 10.1016/0965-1748(93)90043-R. [DOI] [PubMed] [Google Scholar]
- 50.Arakane Y., Muthukrishnan S. Insect chitinase and chitinase-like proteins. Cellular and Molecular Life Sciences. 2010;67(2):201–216. doi: 10.1007/s00018-009-0161-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W.-J., Xu K.-K., Zhang R.-Y., Dou W., Wang J.-J. Transcriptional regulation of a chitinase Gene by 20-hydroxyecdysone and starvation in the oriental fruit fly, Bactrocera dorsalis. International Journal of Molecular Sciences. 2013;14(10):20048–20063. doi: 10.3390/ijms141020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Merzendorfer H., Kim H. S., Chaudhari S. S., et al. Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum. Insect Biochemistry and Molecular Biology. 2012;42(4):264–276. doi: 10.1016/j.ibmb.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding X., Gopalakrishnan B., Johnson L. B., et al. Insect resistance of transgenic tobacco expressing an insect chitinase gene. Transgenic Research. 1998;7(2):77–84. doi: 10.1023/A:1008820507262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The differentially expressed genes between scopoletin- and solvent-treated mites at 24 h after treatment in RNA-seq. Table S2. The differentially expressed genes between scopoletin- and solvent-treated mites at 48 h after treatment in RNA-seq. Table S3. Primers used for cloning, qPCR, and RNAi. Table S4. Sequences and relevant information for phylogenetic analysis of TcCHITs. Figure S1. Chemical structure of scopoletin (A) and diflubenzuron (DFB) (B). Figure S2. A picture of leaf-disc mediated dsRNA feeding.