Abstract
Background: The mouth cavity hosts various types of anaerobic bacteria including Porphyromonas gingivalis, which causes periodontal inflammatory diseases. P. gingivalis is a gram-negative oral anaerobe and is considered as a main etiological factor in periodontal diseases. Several studies have reported a relationship between P. gingivalis in individuals with periodontal diseases and a critical role of this bacterium in the pathogenesis of periodontal diseases. The present study aimed at estimating this probability using a meta-analysis.
Methods: We searched several databases including PubMed, Scopus, Google Scholar, and Web of Science to identify case-control studies addressing the relationship between P. gingivalis with periodontal diseases. A total of 49 reports published from different countries from 1993 to 2014 were included in this study. I² (heterogeneity index) statistics were calculated to examine heterogeneity. Data were analyzed using STATA Version 11.
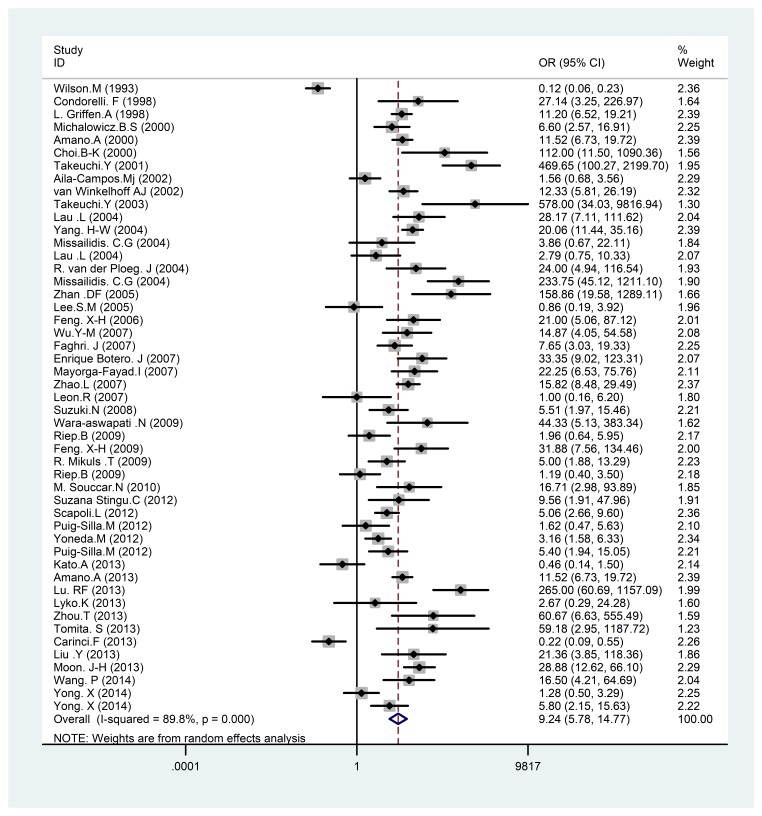
Results: After a detailed analysis of the selected articles, 49 case-control studies with 5924 individuals fulfilled the inclusion criteria for the meta-analysis. The healthy controls included 2600 healthy individuals with a Mean±SD age of 36.56±7.45 years. The periodontal diseases group included 3356 patients with a mean age of 43.62±8.35 years. There was a statistically significant difference between P. gingivalis in periodontal patients and healthy controls; 9.24 (95% CI: 5.78 to 14.77; P = 0.000). In the other word, there was a significant relationship between the presence of P. gingivalis and periodontal diseases.
Conclusion: Analyzing the results of the present study, we found a strong association between the presence of P. gingivalis and periodontal diseases. This result suggests that another research is needed to further assess this subject.
Keywords: Porphyromonas gingivalis, Periodontal Diseases, Chronic Periodontitis, Aggressive Periodontitis, Gingivitis
↑ What is “already known” in this topic:
More than 700 bacteria are found in samples taken from the oral cavity. In several studies, qualitative information of the p.gingivillis has been done in oral diseases.
→ What this article adds:
A variety of qualitative studies have been done on Gengivals, but there are just a few conducted quantitative studies on it which their results are poor and inconsistent as well. So a Meta Analysis method is used in this study to integrate the results of the all previous quantitative investigations into a unique and statistically valid conclusion.
Introduction
Periodontal diseases are complicated oral diseases, which are specified by bacterial- induced inflammatory destruction of tooth-supporting tissues (1). Periodontal diseases can be described as one of the predominant polymicrobial infections of humans, which can advance and lead to gum retrogression, soft tissue harm, weakness of the bone, eventual osteoporosis and tooth loss (severe periodontitis). Periodontal disease have many risk factors including smoking and diabetes, moreover, several bacteria have been connected to the intensity and progress of periodontitis (2, 3).
The oral cavity is a source of different microorganisms that cause a class of infections and inflammation inside the cavity (4). So far, more than 700 bacterial taxa have been identified in sample s taken from oral cavities (2). Evidence for periodontal etiology relies on the performance of several criteria described by Socransky (5).
Of the bacteria believed to be pathogenic in periodontal disease, P. gingivalis has been extensively studied due to its unique ability to evade the immune response (6). P .gingivalis is a gram-negative oral anaerobe and considered as a main etiological factor in periodontal diseases by producing a number of virulence factors and extracellular proteases such as lipopolysaccharide, fimbria, gingipain etc., resulting in destruction of periodontal tissues (7–11). The various surface components of P. gingivalis enable the bacterium to interact with the external medium and simplify its growth, nutrient gain, colonization, and formation of a biofilm that protects it against the host’s defense (12, 13). In addition to being painful, persistent dental disease is linked to diabetes, heart disease, high blood pressure, and MS in later years of life; therefore, extensive studies have been conducted to control the bacteria causing dental diseases (14).
The pathogenicity of P. gingivalis has been investigated in a variety of experimental animal models such as rat, mouse, rabbit, drosophila, and cell models, showing complicated mechanisms of P. gingivalis -host interplay in the expansion of periodontal diseases (15-19).
Although many studies aimed to specify the macrobiotic dependents of specific disease types and the extent of periodontal destruction, there is yet no quantitative data on the levels of P. gingivalis in periodontal diseases. To authenticate the studies, performing a meta-analysis seemed necessary. Thus, this study aimed at evaluating the prevalence of P. gingivalis in patients with periodontal diseases.
Methods
Search Strategies
A database was built for the prevalence of P. gingivalis periodontal diseases from 1993 to 2014 using PubMed, Web of Science, Google scholar Medline, Embase, the Cochrane Library, and Scopus databases. The search was restricted to original articles published in English that presented the prevalence or incidence of P. gingivalis among patients with periodontal diseases. The following keywords from medical subject headings, titles, or abstracts were used with the help of Boolean operators (and, or): P. gingivalis, chronic periodontitis, aggressive periodontitis, and gingivitis. We also searched bibliographies of the retrieved articles for additional references. The titles from the search results were examined closely and determined to be suitable for potential inclusion into the study. In addition, the references from the selected articles were examined as a further search tool. Relevant trials noted in the reference lists of each selected article were also evaluated for inclusion. All papers whose keywords were present in their titles or abstracts were used in the initial list and other unrelated articles were eliminated.
Inclusion and Exclusion Criteria
All original articles presenting case-control studies on the prevalence of P. gingivalis in periodontal diseases were considered. The selection of articles for review was completed based on 3 stages: titles, abstracts, and full texts. When necessary, authors were contacted for additional information. Studies were excluded if they presented insufficient data, if they were not epidemiologic studies, and if they focused on the prevalence of P. gingivalis in diseases other than periodontal diseases. Review articles, congress abstracts, studies reported in languages other than English or Persian, meta-analyses or systematic reviews, and duplicate publication of the same study were also excluded. The STROBE (strengthening the reporting of observational studies in epidemiology) statement was used for quality control of the studies. We assessed the quality of studies according to variables related to the study objectives, characteristics of the study population, clearly explained inclusion/exclusion criteria, data collection method, as well as the validity, explicit findings, and appropriate data analysis methods of the studies.
Data Extraction
For all studies, the following data were extracted: first author, year of publication, location, sample size, sample age, P. gingivalis screening method, and sample specimens, percentage of P. gingivalis in patients and healthy individuals. Abstracts and full articles were reviewed independently by 2 of the authors, and if results were discordant, papers were reviewed jointly until the differences were resolved.
Data Synthesis and Analysis
Studies were combined based on the sample size, mean and standard deviation. The difference between the average variance of the normal distribution was calculated using the formula of 2 integrated variance. To assess the heterogeneity of the studies, Cochran test and the I2 (heterogeneity index) were used. Due to the significant heterogeneity in the studies, random effects model was used. The findings are described in forest plots (the point estimations and their 95% CI). To examine publication bias, Begg plot and regressions method were used. P value less than 5% was considered as a significant heterogeneity test. Sensitivity analyses were prespecified. Statistical analyses were performed using STATA version 11.
Results
Our initial search strategy yielded 172 potential articles for inclusion; in a secondary screening, 48 of them were excluded based on title and abstract evaluation, and 124 were retained for detailed full text evaluation. We excluded another 75 articles (9 review articles, 18 articles with other diseases, 13 for lack of enough information, 23 articles with percentage of P. gingivalis only in patients group, and 12 duplicates). After a detailed analysis of the selected articles, 49 case- control studies with 5924 individuals (3356 patients and 2600 healthy individuals) fulfilled the inclusion criteria for the meta-analysis (Fig. 1).The characteristics of the 49 trials and the quality scores included in the meta-analysis are summarized in Table 1.
Fig. 1.
The flow diagram of studies identified the systematic review and meta-analysis
Table 1. Study characteristics .
|
First author (Reference) |
Country (year) |
Case | Control | Mean age |
OR (95% CI) |
Mean difference | Type of disease | Sample specimens | Methods of Cupper measurement | ||
| Case | Control | Lower | Upper | ||||||||
| Michalowicz.B.S(52) | Jamaica(2000) | 35 | 65 | 14-18 | 14-18 | 2.75 | 16.91 | 6.60 | Periodontitis | subgingival plaque | PCR method |
| Kato.A (38) | Japan(2013) | 85 | 20 | 57.4±13.1 | 45.9±17.0 | 0.14 | 1.50 | 0.46 | Chronic periodontitis | plaque samples | PCR method |
| Lee.S.M (53) | Korea(2005) | 17 | 19 | 52 ± 11.1 | 49 ± 10.2 | 0.19 | 3.92 | 0.86 | Gingivitis | subgingival plaque | PCR method |
| R. Mikuls .T(54) | USA(2000) | 39 | 40 | 52 ± 11.1 | 49. ± 10.2 | 1.88 | 13.29 | 5.0 | Periodontitis | subgingival plaque | stabilization P. gingivalis antibody seropositivity |
| L. Griffen.A (55) | Ohio State (1998) | 130 | 181 | 51.4 ± 9.3 | 49.2 ± 9 | 6.52 | 19.21 | 11.20 | Periodontitis | subgingival plaque | PCR method |
| Avila-Campos .Mj(40) | Brazil (2002) | 50 | 50 | 45.5 ±9.7 | 32.3 ±8.9 | 0.68 | 3.56 | 1.56 | Periodontitis | subgingival plaque | PCR method |
| Winkelhoff AJ(27) | Netherlands (2002) | 116 | 94 | 42.9 ± 9.8 | 40.4 ± 11.9 | 5.81 | 26.19 | 12.33 | Periodontitis | subgingival plaque | Anaerobic cultivation |
| Ruben.L (56) | Chile (2007) | 20 | 6 | 27 ± 5.2 | 22.7 ± 4.9 | 0.16 | 6.20 | 1.00 | Periodontitis/ gingivitis | subgingival plaque | Bacterial culture |
| Amano.A(33) | Japan (2013) | 139 | 380 | 6.73 | 19.72 | 11.52 | periodontitis | Dental plaque | PCR method | ||
| Takeuchi.Y(57) | Japan (2001) | 103 | 20 | 100.27 | 21997 | 469.65 | periodontitis | Saliva and subgingival plaque | PCR method | ||
| Missailidis. C.G(44) | Brazil (2004) | 57 | 25 | 45.12 | 1211.10 | 233.75 | periodontal attachment loss | subgingival plaque | PCR method | ||
| Missailidis. C.G (44) | Brazil (2004) | 20 | 25 | 0.67 | 22.11 | 3.86 | gingivitis | subgingival plaque | PCR method | ||
| Zhao.L (58) | Chile (2007) | 115 | 136 | 8.48 | 29.49 | 15.82 | Chronic periodontitis | subgingival samples | PCR method | ||
| Yang. H-W (59) | Taiwan (2004) | 407 | 91 | 11.44 | 35.16 | 20.06 | periodontal disease | subgingival plaque | indirect immunofluorescent assay | ||
| Choi.B-K (60) | Korea (2000) | 29 | 20 | 11.5 | 1090.36 | 112.0 | periodontitis | subgingival plaque | PCR method | ||
| Lyko.K (39) | Brazil (2013) | 48 | 24 | 11.1 ±3.52 | 10.54 ±3.75 | 0.29 | 24.28 | 2.67 | periodontitis | saliva samples | PCR method |
| Carinci.F (37) | Italy (2013) | 66 | 46 | 48.9±18.2 | 31.6 ±18.6 | 0.09 | 0.55 | 0.22 | periodontitis | periodontal pocket | PCR method |
| Wilson.M (41) | USA (1993) | 28 | 18 | 18-59 | 18-59 | 0.06 | 0.23 | 0.12 | periodontitis | subgingival plaque | ELISA |
| M. Souccar.N (61) | Lebanon (2010) | 20 | 20 | 34.3±5.36 | 26.10 ±4.57 | 2.98 | 93.89 | 16.71 | periodontitis | Oral plaque | PCR method |
| Riep.B (20) | Germany (2000) | 46 | 21 | 55.2± 11.2 | 66.6 ± 1.5 | 0.64 | 5.95 | 1.96 | Chronic periodontitis | subgingival plaque | PCR method |
| Riep.B (20) | Germany (2000) | 44 | 21 | 34.4 ± 6.5 | 66.6 ± 1.5 | 0.40 | 3.50 | 1.19 | aggressive periodontitis | subgingival plaque | PCR method |
| Yoneda.M (62) | Japan (2013) | 150 | 60 | 54.6 ± 1.2 | 52.9 ± 2.4 | 1.58 | 6.33 | 3.16 | periodontitis | saliva samples | PCR method |
| Feng. X-H (63) | Chine (2000) | 48 | 25 | 38.9 ± 9.9 | 23.6 ± 1.8 | 7.56 | 134.46 | 31.88 | periodontitis | subgingivalplaque | PCR method |
| Zhou.T (32) | Chine (2013) | 27 | 20 | 6.63 | 555.49 | 60.67 | Chronic periodontitis | subgingival | PCR and reverse hybridization assay | ||
| Lu. RF (34) | Chine (2013) | 80 | 56 | 60.69 | 1157.09 | 256.0 | Aggressive periodontitis | gingival crevicular fluid | PCR method | ||
| Condorelli. F (64) | Italy (1998) | 33 | 21 | 3.25 | 226.97 | 27.14 | severe periodontal disease | subgingival plaque | culture | ||
| Wu.Y-M(65) | Chine (2007) | 61 | 30 | 42.4±8.7 | 37.35 ± 7.3 | 4.05 | 54.58 | 14.87 | chronic periodontitis | subgingival plaque | PCR method |
| Puig-Silla.M (12) | Spain (2013) | 33 | 37 | 43.39 ± 7.4 | 40.68 ± 7.1 | 1.94 | 15.05 | 5.40 | chronic periodontitis | subgingival plaque | PCR method |
| Puig-Silla.M (12) | Spain (2013) | 16 | 37 | 38.81 ± 6.9 | 40.68 ± 7.1 | 0.47 | 5.63 | 1.62 | Gingivitis | subgingival plaque | PCR method |
| Liu .Y (15) | China (2013) | 25 | 20 | 3.58 | 118.36 | 21.36 | periodontitis | gingival crevicular fluid | PCR method | ||
| Mayorga-Fayad.I (66) | Colombia (2007) | 143 | 40 | 39.5±9.85 | 32.6±10.6 | 6.53 | 75.76 | 22.25 | periodontitis | subgingival plaque | culture |
| Scapoli.L (35) | Italy (2013) | 127 | 66 | 48.9 ±18.2 | 31.6 ±18.6 | 2.66 | 9.60 | 5.06 | periodontitis | periodontal pocket microbiota | PCR method |
| Wang. P (30) | China(2014) | 25 | 29 | 4.21 | 64.69 | 16.50 | periodontitis | subgingival plaque | PCR method | ||
| Wara-aswapati .N (47) | Thailand (2000) | 20 | 20 | 5.13 | 383.34 | 44.33 | chronic periodontitis | subgingival plaque | PCR method | ||
| Faghri. J (67) | Iran (2007) | 61 | 40 | 43 ± 11 | 41.35 ± 9.8 | 3.03 | 19.33 | 7.65 | chronic periodontitis | subgingival plaque | PCR method |
| Feng. X-H (68) | China (2000) | 55 | 17 | 5.06 | 87.12 | 21.0 | Aggressive periodontitis | subgingival plaque | PCR method | ||
| Zhan .DF (69) | China (2005) | 152 | 30 | 19.58 | 1289.11 | 158.86 | chronic periodontitis | Periodontal pocket and gingival sulcus | PCR method | ||
| Amano.A (70) | Japan (2000) | 139 | 380 | 6.73 | 19.72 | 11.52 | periodontitis | Dental plaque | PCR method | ||
| Lau .L (45) | Spain (2004) | 32 | 30 | 49.4 ±8.99 | 37.8 ±7.5 | 7.11 | 111.62 | 28.17 | periodontitis | subgingival plaque | PCR method |
| Lau .L (45) | Spain (2004) | 30 | 30 | 46.6 ±9.82 | 37.8 ±7.5 | 0.75 | 10.33 | 2.79 | Gingivitis | ||
| Yong. X (1) | China (2014) | 42 | 32 | 0.50 | 3.29 | 1.28 | chronic gingivitis | Gingival crevicular fluid | PCR method | ||
| Yong. X (1) | China (2014) | 95 | 32 | 2.15 | 15.63 | 5.80 | chronic periodontitis | Gingival crevicular fluid | PCR method | ||
| Tomita. S (31) | Japan (2013) | 20 | 10 | 43.6 ±11.1 | 28.7 ± 3.2 | 2.95 | 1187.72 | 59.18 | chronic periodontitis | subgingival plaque | PCR method |
| Moon. J-H (7) | Korea (2013) | 284 | 128 | 48.3±9.5 | 42.3±13.5 | 12.62 | 66.10 | 28.88 | chronic periodontitis | subgingival plaque | PCR method |
| SuzanaStingu.C (36) | Germany (2013) | 33 | 20 | 33.39 ±10.47 | 37.65 ±10.88 | 1.91 | 47.96 | 9.56 | chronic periodontitis | subgingival plaque | PCR method |
| Enrique Botero. J (71) | Colombia (2007) | 80 | 30 | 33.91 ±9.32 | 26.90 ±7.17 | 9.02 | 123.31 | 33.35 | periodontitis | subgingival plaque | PCR method |
| R.van der Ploeg. J (72) | Switzerland(2004) | 17 | 33 | 53.1 ±8.53 | 26.8 ± 5.3 | 4.94 | 116.54 | 24.0 | chronic periodontitis | subgingival plaque | PCR method |
| Takeuchi.Y (73) | Japan (2003) | 35 | 18 | 51.8 ±7.29 | 27.3 ±3.71 | 34.03 | 9816.94 | 578.0 | chronic periodontitis | subgingival plaque | PCR method |
| Suzuki.N (74) | Japan (2008) | 21 | 305 | 43.8±12.1 | 45.4±14.9 | 1.97 | 15.46 | 5.51 | chronic periodontitis | saliva samples | PCR method |
The healthy controls comprised 2600 healthy individuals, with the age range of 14 to 67 years and a mean age of 36.56±7.45 years. The periodontal diseases group included 3356 patients with periodontal diseases, ranging from 14 to 59 years, and with a mean age of 43.62±8.35 years.
In the present study, a statistically significant difference was found between P. gingivalis in periodontal patients and healthy controls; 9.24 (95% CI: 5.78 to 14.77; p<0.001) (Fig. 2). In the other word, on analyzing the results of the present study, a strong association was found between the
Fig. 2.
Meta-analysis of the association of P. gingivalis with periodontal diseases. Square represents effect estimate of individual studies with their 95 % confidence intervals with size of squares proportional to the weight assigned to the study in the meta-analysis. In this chart, studies are stored in order of the year of publication and author’s names, based on a random effects model.
presence of P. gingivalis and periodontal diseases (Fig. 2).
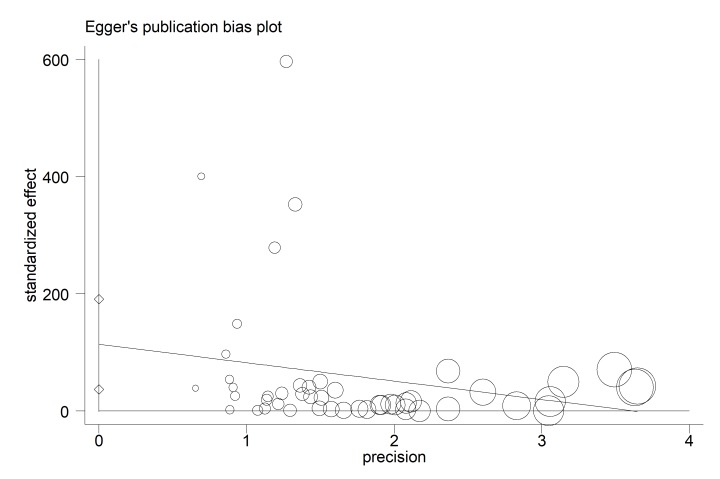
According to publication bias figure, the effect of bias in these studies was not significant. In fact, most studies were located inside the Funnel Plot, and thus the results of most relevant studies, considering the title, were included into the analysis (p= 0.005) (Fig. 3).
Fig. 3.
Egger’s funnel plot for publication bias. The diameter of each circle represents the weight in the meta-analysis. Each circle represents the RDs according to the standard error of each RDs. The diameter of each circle represents the weight in the meta-analysis.
Discussion
It is widely accepted that the etiology of periodontal diseases is polymicrobial in nature. Worsening or improvement of periodontal situation goes along by a change in the bacterial composition of subgingival plaque (20). It has, therefore, been suggested that microbial testing be used to diagnose and optimize periodontal therapy and assess its outcome. However, this strategy may be confusing as the beginning and the progress of periodontal diseases are impressed by interaction of myriad genetic, environmental, host, and microbial factors (20-23). Furthermore, molecular studies reveal an unexpectedly high diversity of microorganisms and progression of disease remains to be investigated. Nevertheless, current microbiological testing mostly involves the classic doubtful oral pathogens (20).
The aim of this study was to evaluate the association between the presence of P. gingivalis and periodontal diseases. P. gingivalis has been known to be a risk factor for periodontal diseases although its exact roles in the initiation and progression of the oral diseases remain unclear (15, 24).
In the present study, a significantly strong positive correlation was found between the presence of P. gingivalis and periodontal diseases. Published data suggests that the flora associated to chronic periodontal diseases is dominated by the presence of Gram-negative anaerobic bacteria, and especially P. gingivalis (25). Slots and Ting described a synergistic association of P. gingivalis in presentations with evolutive lesions (26). Van Winkelhoff et al. concluded that P. gingivalis was the marker of a destructive lesion (27). Authors of previous studies reported similar results. The authors of various studies on P. gingivalis have reported that these pathogens are able to pollute soft tissues and flee the surgical debridement of periodontal lesions (28, 29).
Many epidemiological studies report a positive association between the presence of P. gingivalis and periodontal diseases (7, 25, 30-36), whereas in some studies, no association was found between the presence of P. gingivalis and periodontal diseases (20, 37-41). This has also been shown by Kumar et al. (42). The fact that all types of bacteria studied could be detected, even in healthy controls, might indicate that the presence of periodontal pathogens does not necessarily lead to periodontal diseases (1). However, these species show just a small percentage of whole bacteria, and the open-ended approach of their study may not be geared to indicate an association of this species with the disease (20). In another study, Kumar et al. demonstrated a significant association of P. gingivalis with chronic periodontitis using PCR amplification of 16S rRNA genes (43).
Some studies have reported a strong association between the presence of P. gingivalis and periodontitis, but did not find any association between the presence of P. gingivalis and gingivitis (1, 12, 44, 45). Although a number of putative bacteria are considered to be associated with chronic periodontitis, it has become clear that other factors are involved in the etiology of several types of periodontitis. In the other word, P. gingivalis is one of the risk factors responsible for periodontitis.
P. gingivalis is one of the bacteria that form the classic ‘red complex’ described by Socransky et al. (46). It is the bacteria most frequently found in patients with periodontal disease (12). Wara-aswapati et al. (47) found that the mean age of patients was higher than that of the healthy participants. P. gingivalis has been reported to be related to adult periodontitis (48, 49). In a study by Liu et al (15), they analyzed the correlation of patients’ age and occurrence of P. gingivalis and found that the age of P. gingivalis positive and negative was statistically different, implying that the prevalence of P. gingivalis may increase as the patients’ age increase.
The association of certain putative pathogens with periodontal diseases shown in many studies may be explained in part by deeper pockets in these patients. In the present study, the association between probing severely and the occurrence of periodontal disease has not studied because most previous studies conducted on the association of particular bacteria with periodontitis did not test the influence of probing depth. Some studies found that the search depth had a much greater impact on the species than did the diagnosis. In study of Riep et al. (20), the prevalence of P. gingivalis was highly associated (p< 0.001) with pocket depth. Ali et al. (50) studied the existence of P. gingivalis in very deep pockets (≥ 6 mm) and deep pockets (4–5 mm) of 36 Romanian patients presenting with chronic periodontitis. P. gingivalis was recognized in 75.8% of the patients and 63.6% of the sampled sites.
Listgarten and Loomer (51) also questioned whether microbial identification should be considered as a strategy in managing this disease. They found no strong evidence supporting the benefit of microbial testing, partly due to a lack of standardization among diagnostic methods.
In summary, on analyzing the results of the present study, a strong association was found between the presence of P. gingivalis and periodontal diseases. Furthermore, the results of the present study indicated that P.gingivalis is one of the risk factors responsible for periodontitis. This result suggests that further research is needed to investigate their pathogenicity.
Acknowledgments
The authors extend their gratitude to the Student Research Committee, Ilam University of Medical Sciences for its support.
Conflict of Interests
The authors declare that they have no competing interests.
Cite this article as: Rafiei M, Kiani F, Sayehmiri F, Sayehmiri K, Sheikhi A, Zamanian Azodi M. Study of Porphyromonas gingivalis in periodontal diseases: A systematic review and meta-analysis. Med J Islam Repub Iran. 2017 (12 Sep);31:62. https://doi.org/10.18869/mjiri.31.62
References
- 1.Yong X, Chen Y. Periodontopathogens and human b-defensin-2 expression in gingival crevicular fluid from patients with periodontal disease in Guangxi, China. J Periodontal Res. 2015;50(3):403–410. doi: 10.1111/jre.12220. [DOI] [PubMed] [Google Scholar]
- 2.Tomita S, Komiya-Ito A. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microb Pathogene. 2013;61-62:11–15. doi: 10.1016/j.micpath.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 3.S Michaud.D. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34(10):2193–2197. doi: 10.1093/carcin/bgt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varghese J, Kumar Tumkur V, Ballal V, Subraya Bhat G. Antimicrobial effect of Anacardium occidentale leaf extract against pathogens causing periodontal disease. Adv Biosci Biotechno. 2013;4(8):15–18. [Google Scholar]
- 5.Socransky SS. Relationship of bacteria to the etiology of periodontal disease. J Dent Res. 1970;49:203–222. doi: 10.1177/00220345700490020401. [DOI] [PubMed] [Google Scholar]
- 6.Tribble G, E Kerr J, Wang B-Y. Genetic diversity in the oral pathogen Porphyromonas gingivalis: molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607–620. doi: 10.2217/fmb.13.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moon JH, Herr Y, Lee HW, Shin SI, Kim Ch, Amano A, Lee JY. Genotype analysis of Porphyromonas gingivalis fimA in Korean adults using new primers. J Med Microb. 2013;62:1290–1294. doi: 10.1099/jmm.0.054247-0. [DOI] [PubMed] [Google Scholar]
- 8.Decaillet F, Giannopoulou C, Cionca N, Almaghlouth A, Mombelli A. Microbial profiles of patients seeking treatment for periodontitis. Influence of origin, smoking and age? Schweiz Monatsschr Zahnmed. 2012;122(3):198–204. [PubMed] [Google Scholar]
- 9.Bostanci N, Belibasakis GN. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;331(1):1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi F, Okada M, Oda Y, Kojima T, Kozai K. Prevalence of Porphyromonas gingivalis fimA genotypes in Japanese children. J Oral Sci. 2012;54(1):77–83. doi: 10.2334/josnusd.54.77. [DOI] [PubMed] [Google Scholar]
- 11.Perez-Chaparro PJ, Lafaurie GI, Gracieux P, Meuric V, Tamanai-Shacoori Z, Castellanos JE. et al. Distribution of Porphyromonas gingivalis fimA genotypes in isolates from subgingival plaque and blood sample during bacteremia. Biomedica. 2009;29(2):298–306. [PubMed] [Google Scholar]
- 12.Puig-Silla M, Dasi-Fernandez F, Montiel-Company JM, Almerich-Silla JM. Prevalence of fimA genotypes of Porphyromonas gingivalis and other periodontal bacteria in a Spanish population with chronic periodontitis. Med Oral Patol Oral Cir Bucal. 2012;17(6):e1047–53. doi: 10.4317/medoral.17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoshimura F, Murakami Y, Nishikwa K, Hasegawa Y, Kawaminami S. Surface components of Porphyromonas gingivalis. J Periodontal Res. 2009;44:1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 14.Patra JK, Kim ES, Oh K, Kim HJ, Dhakal R, Kim Y. et al. Bactericidal effect of extracts and metabolites of Robinia pseudoacacia L on Streptococcus mutans and Porphyromonas gingivalis causing dental plaque and periodontal inflammatory diseases. Molecules. 2015;20(4):6128–6139. doi: 10.3390/molecules20046128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, Zhang Y, Wang L, Guo Y, Xiao S. Prevalence of porphyromonas gingivalis four rag locus genotypes in patients of orthodontic gingivitis and periodontitis. PLoS ONE. 2013;8(4):e61028. doi: 10.1371/journal.pone.0061028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Igboin CO, Moeschberger ML, Griffen AL, Leys EJ. Porphyromonas gingivalis virulence in a Drosophila melanogaster model. Infect Immun. 2011;79(1):439–448. doi: 10.1128/IAI.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzpatrick RE, Aprico A, Wijeyewickrema LC, Pagel CN, Wong DM, Potempa J. et al. High molecular weight gingipains from porphyromonas gingivalis induce cytokine responses from human mcrophage-like cells via a nonproteolytic mechanism. J Innate Immun. 2009;1(2):109–117. doi: 10.1159/000181145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haruyama K, Yoshimura A, Naito M, Kishimoto M, Shoji M, Abiko Y. et al. Identification of a gingipain-sensitive surface ligand of porphyromonas gingivalis that induces Toll-like recepotor 2- and 4- independent NF-kappaB activation in CHO cells. Infect Immun. 2009;77(10):4414–4420. doi: 10.1128/IAI.00140-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grenier D, Tanabe S. Porphyromonas gingivalis gingipains trigger a proinflammatory response in human monocyte-derived macrophages through the p38a mitogen-activated protein kinase signal transduction pathway. Toxins (Basel) 2010;2(3):341–352. doi: 10.3390/toxins2030341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riep B, Edesi-Neuss L, Claessen F, Skarabis H, Ehmke B, Flemmig TF. et al. Are putative periodontal pathogens reliable diagnostic markers? J Clin Microbiol. 2009;47(6):1705–1711. doi: 10.1128/JCM.01387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nibali L, Ready DR, Parkar M, Brett PM, Wilson M, Tonetti MS. et al. Gene polymorphisms and the prevalence of key periodontal pathogens. J Dent Res. 2007;86(5):416–420. doi: 10.1177/154405910708600505. [DOI] [PubMed] [Google Scholar]
- 22.Page RC, Kornman KS. The pathogenesis of human periodontitis: an introduction. Periodontol. 1997;2000(14):9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 23.Umeda M, Chen C, Bakker I, Contreras A, Morrison J L, SlotsJ SlotsJ. Risk indicators for harboring periodontal pathogens. J Periodontol. 1998;69:1111–1118. doi: 10.1902/jop.1998.69.10.1111. [DOI] [PubMed] [Google Scholar]
- 24.Shi X, Hanley SA, Kele MF, Fawell SC, Aduse-Opoku J, Whiley RA. et al. The rag locus of Porphyromonas gingivalis contributes to virulence in a murine model of soft tissue destruction. Infect Immun. 2007;75(4):2071–2074. doi: 10.1128/IAI.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benrachadi L, Bouziane A, Azziman Z, Bouziane-Ouartini F, Ennibi O. Screening for periodontopathogenic bacteria in severe chronic periodontitis in a Moroccan population. Médecine et maladies infectieuses. 2012;42(12):599–602. doi: 10.1016/j.medmal.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Slots J, Ting M. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in human periodontal disease: occurrence and treatment. Periodontol. 2000;22:82–121. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 27.Van Winkelhoff AJ, Loos BG, van der Reijden WA, van der Velden U. Porphyromonas gingivalis, Bacteroides forsythus and other putative periodontal pathogens in subjects with and without periodontal destruction. J Clin Periodontol. 2002;29:1023–8. doi: 10.1034/j.1600-051x.2002.291107.x. [DOI] [PubMed] [Google Scholar]
- 28.Meyer DH, Lippmann JE, Five-Taylor PM. Invasion of epithelial cells by Actinobacillus actinomycetemcomitans: a dynamic, multistep process. Infect Immun. 1996;64:2988–97. doi: 10.1128/iai.64.8.2988-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haffajee AD, Cugini MA, Dibart S, Smith C, Kentjr RL, Socransky SS. The effect of SRP on the clinical and microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–34. doi: 10.1111/j.1600-051x.1997.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Duan D, Zhou X, Li X, Yang J, Deng M. et al. Relationship between expression of human gingival beta-defensins and levels of periodontopathogens in subgingival plaque. J Periodontal Res. 2014;50(1):113–122. doi: 10.1111/jre.12187. [DOI] [PubMed] [Google Scholar]
- 31. Tomitaa S, Komiya-Itoa A, Imamuraa K, Kitaa D, Otaa K, Takayamaa S. et al. Prevalence of Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythia in Japanese patients with generalized chronic and aggressive periodontitis. Microbial Pathogenesis. 2013;61-62:11–15. doi: 10.1016/j.micpath.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 32.Ting Zh, Hong X, Zhaohui Y. Relationships of five periodontal pathogens causing subgingival plaque in patients with chronic periodontitis under different periodontal conditions. West China J Stomatol. 2013;5:518–521. [PubMed] [Google Scholar]
- 33.Amano A, Kuboniwa A, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79(9):1664–8. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 34.Lu R, Feng L, Gao X, Meng H, Feng X. Relationship between volatile fatty acids and Porphyromonas gingivalis and Treponema denticola in gingival crevicular fluids of patients with aggressive periodontitis. Beijing Da Xue Xue Bao. 2013;45(1):12–16. [PubMed] [Google Scholar]
- 35.Scapoli L, Girardi A, Palmieri A, Testori T, Zuffetti F, Monguzzi R. et al. Microflora and periodontal disease. Dent Res J (Isfahan) 2012;9(2):S202–S206. doi: 10.4103/1735-3327.109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stingu CS, Jentsch H, Eick S, Schaumann R, Knofler G, Rodloff A. Microbial profile of patients with periodontitis compared with healthy subjects. Quintessence Int. 2012;43(2):e23–31. [PubMed] [Google Scholar]
- 37.Carinci F, Scapoli L, Girardi A, Cura F, Lauritano D, Nardi GM. et al. Oral microflora and periodontal disease: new technology for diagnosis in dentistry. Ann Stomatol. 2013;5(2):170–173. [PMC free article] [PubMed] [Google Scholar]
- 38.Kato A, Imai K, Ochiai K, Ogata Y. Higher Prevalence of Epstein–Barr Virus DNA in Deeper Periodontal Pockets of Chronic Periodontitis in Japanese Patients. PLoS ONE. 2013;8(8):e71990. doi: 10.1371/journal.pone.0071990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kato A, Imai K, Ochiai K, Ogata Y. Salivary detection of periodontopathic bacteria in Fanconi’s anemia patients. Anaerobe. 2013;24:32–35. doi: 10.1016/j.anaerobe.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Avila-Campos MJ, Velasquez-Melendez G. Prevalence of putative periodontopathogens from periodontal patients and healthy subjects in São Paulo SP Brazil Rev Inst Med trop S. Paulo. 2002;44(1):1–5. doi: 10.1590/s0036-46652002000100001. [DOI] [PubMed] [Google Scholar]
- 41.Wilson M, Lopatin D, Osborne G, Kieser JB. Prevalence of Treponema denticola and Porphyromonas gingivalis in plaque from periodontally- healthy and periodontally-diseased sites. J Med Microbiol. 1993;38:406–410. doi: 10.1099/00222615-38-6-406. [DOI] [PubMed] [Google Scholar]
- 42.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 44.Missailidis CG, Umeda JE, Ota-Tsuzuki C, Anzai D, Mayer MP. Distribution of fimA genotypes of Porphyromonas gingivalis in subjects with various periodontal conditions. Oral Microb Immunol. 2004;19(4):224–229. doi: 10.1111/j.1399-302X.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 45.Lau L, Sanz M, Herrera D, Morillo JM, Martín C, Silva A. Quantitative real-time polymerase chain reaction versus culture: a comparison between two methods for the detection and quantification of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Tannerella forsythensis in subgingival plaque samples. J Clin Periodontol. 2004;31:1061–1069. doi: 10.1111/j.1600-051X.2004.00616.x. [DOI] [PubMed] [Google Scholar]
- 46.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25(2):134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 47.Wara-aswapati N, Pitiphat W, Chanchaimongkon L, Taweechaisupapong S, Boch JA, Ishikawa I. Red bacterial complex is associated with the severity of chronic periodontitis in a Thai population. Oral Dis. 2009;15:354–9. doi: 10.1111/j.1601-0825.2009.01562.x. [DOI] [PubMed] [Google Scholar]
- 48.Décaillet F, Giannopoulou C, Cionca N, Almaghlouth AA, Mombelli A. Microbial profiles of patients seeking treatment for periodontitis. Influence of origin, smoking and age? Schweiz Monatsschr Zahnmed. 2012;122(3):198–204. [PubMed] [Google Scholar]
- 49.Hayashi F, Okada M, Oda Y, Kojim T, Kozai K. Prevalence of Porphyromonas gingivalis fimA genotypes in Japanese children. J Oral Sci. 2012;54(1):77–83. doi: 10.2334/josnusd.54.77. [DOI] [PubMed] [Google Scholar]
- 50. Ali RW, Velcescu C, Jivanescu MC, Lofthus B. Prevalence of putative periodontal pathogens in subgingival plaque samples from adult periodontitis patients. J Clin Periodontol. 1996;23:133. doi: 10.1111/j.1600-051x.1996.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 51.Listgarten MA, Loomer PM. Microbial identification in the management of periodontal diseases A systematic review. Ann Periodontol. 2003;8:182–192. doi: 10.1902/annals.2003.8.1.182. [DOI] [PubMed] [Google Scholar]
- 52.Michalowicz BS, Ronderos M, Camara-Silva R, Contreras A, Slots J. Human herpesviruses and Porphyromonas gingivalis are associated with Juvenile periodontitis. J Periodontol. 2000;71:981–988. doi: 10.1902/jop.2000.71.6.981. [DOI] [PubMed] [Google Scholar]
- 53.Lee SM, Yoo SY, Kim HS, Kim KW, Yoon YJ, Lim SH. et al. Prevalence of Putative Periodontopathogens in Subgingival Dental Plaques from Gingivitis Lesions in Korean Orthodontic Patients. The J Microb. 2005:260–265. [PubMed] [Google Scholar]
- 54.Mikuls TR, Payne JB, Reinhardt RA, Thiele GM, Maziarz E, Cannella AC. et al. Antibody responses to Porphyromonas gingivalis (P gingivalis) in subjects with rheumatoid arthritis and periodontitis. Int Immunopharmacol. 2009;9(1):38–42. doi: 10.1016/j.intimp.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffen AL, Becker MR, Lyons SR, Moeschberger ML, Leys EJ. Prevalence of Porphyromonas gingivalis and Periodontal Health Status. J Clin Microbiol. 1998;36(11):3239–3242. doi: 10.1128/jcm.36.11.3239-3242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leon R, Silva N, Ovalle A, Chaparro A, Ahumada A, Gajardo M. et al. Detection of Porphyromonas gingivalis in the Amniotic Fluid in Pregnant Women With a Diagnosis of Threatened Premature Labor. J Periodontol. 2007;78:1249–1255. doi: 10.1902/jop.2007.060368. [DOI] [PubMed] [Google Scholar]
- 57.Takeuchi Y, Umeda M, Sakamoto M, Benno Y, Huang Y, Ishikawa I. Treponema socranskii, Treponema denticola, and Porphyromonas gingivalis Are Associated With Severity of Periodontal Tissue Destruction. J Periodontol. 2001;72(10):1354–1363. doi: 10.1902/jop.2001.72.10.1354. [DOI] [PubMed] [Google Scholar]
- 58.Zhao L, Wu YF, Meng S, Yang H, OuYang YL, Zhou XD. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J Periodont Res. 2007;42(6):511–517. doi: 10.1111/j.1600-0765.2007.00975.x. [DOI] [PubMed] [Google Scholar]
- 59.Yang HW, Huang YF, Chou MY. Occurrence of Porphyromonas gingivalis and Tannerella forsythensis in Periodontally Diseased and Healthy Subjects. J Periodontol. 2004;75(8):1077–1083. doi: 10.1902/jop.2004.75.8.1077. [DOI] [PubMed] [Google Scholar]
- 60.Choi BK, Park SH, Yoo YJ, Choi SH, Chai JK, Cho KS. et al. Detection of Major Putative Periodontopathogens in Korean Advanced Adult Periodontitis Patients Using a Nucleic Acid-Based Approach. J Periodontol. 2000;71(9):1387–1394. doi: 10.1902/jop.2000.71.9.1387. [DOI] [PubMed] [Google Scholar]
- 61.Souccar NM, Chakhtoura M, Ghafari JG, Abdelnoor AM. Porphyromonas gingivalis in dental plaque and serum C-reactive protein levels in pregnancy. J Infect Dev Ctries. 2010;4(6):362–366. [PubMed] [Google Scholar]
- 62.Yoneda M, Naka S, Nakano K, Wada K, Endo H, Mawatari H. et al. Involvement of a periodontal pathogen, Porphyromonas gingivalis on the pathogenesis of non-alcoholic fatty liver disease. BMC Gastroenterology. 2012;12:16. doi: 10.1186/1471-230X-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng XH, Zhang L, Meng HX, Xu L, Chen ZB, Shi D. et al. Detection of 3 anaerobic microorganisms in salvia and subgingival plaque of patients with periodontitis 2009. J Peking Uni. 2009;41(1):44–48. [PubMed] [Google Scholar]
- 64.Condorelli F, Scalia G, Cali G, Rossetti B, Nicoletti G, Lo Bue AM. Isolation of Porphyromonas gingivalis and Detection of Immunoglobulin A Specific to Fimbrial Antigen in Gingival Crevicular Fluid. J Clin Microbiol. 1998;36(8):2322–2325. doi: 10.1128/jcm.36.8.2322-2325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu YM, Yan J, Chen LL, Gu ZY. Association between infection of different strains of Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in subgingival plaque and clinical parameters in chronic periodontitis. J Zhejiang Uni Sci B. 2007;8(2):121–131. doi: 10.1631/jzus.2007.B0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mayorga-Fayad I, Lafaurie GI, Contreras A, Castillo DM, Baron A, Aya Mdel R. Subgingival microbiota in chronic and aggressive periodontitis in Bogotá, Colombia: an epidemiological approach. Biomédica. 2007;27:21–33. [PubMed] [Google Scholar]
- 67.Faghri J, Moghim S, Abed AM, Rezaei F, Chalabi M. Prevalence of Porphyromonas gingivalis and Bacteroides forsythus in chronic periodontitis by multiplex PCR. Pak J Biol Sci. 2007;15 10(22):4123–7. doi: 10.3923/pjbs.2007.4123.4127. [DOI] [PubMed] [Google Scholar]
- 68.Feng XH, Zhang L, Meng HX, Xu L, Chen ZB, Shi D. Prevalence of putative periodontal microorganisms in Chinese patients with aggressive periodontitis. Zhonghua Kou Qiang Yi Xue Za Zhi. 2006;41(6):344–7. [PubMed] [Google Scholar]
- 69.Zhan D, Liu Z, Xia X, Hu J, Chen L, Yan J. Study on the detection of P gingivalis, A actinomycetemcomitans and T denticola and the correlation between coinfections of the microbes and levels of chronic periodontitis lesion. Zhonghua Liu Xing Bing Xue Za Zhi. 2005;26(2):120–3. [PubMed] [Google Scholar]
- 70.Amano A, Kuboniwa A, Nakagawa I, Akiyama S, Morisaki I, Hamada S. Prevalence of specific genotypes of Porphyromonas gingivalis fimA and periodontal health status. J Dent Res. 2000;79(9):1664–8. doi: 10.1177/00220345000790090501. [DOI] [PubMed] [Google Scholar]
- 71.Botero JE, Contreras A, Lafaurie G, Jaramillo A, Betancourt M, Arce RM. Occurrence of Periodontopathic and Superinfecting Bacteria in Chronic and Aggressive Periodontitis Subjects in a Colombian Population. J Periodontol. 2007;78:696–704. doi: 10.1902/jop.2007.060129. [DOI] [PubMed] [Google Scholar]
- 72.van der Ploeg JR, Giertsen E, Lüdin B, Mörgeli Ch, Zinkernagel AS, Gmür R. Quantitative detection of Porphyromonas gingivalis ¢mA genotypes in dental plaque. FEMS Microbiology Letters. 2004;232:31–37. doi: 10.1016/S0378-1097(04)00064-3. [DOI] [PubMed] [Google Scholar]
- 73.Takeuchi Y, Umeda M, Ishizuka M, Huang Y, Ishikawa I. Prevalence of periodontopathic bacteria in Aggressive Periodontitis patients in a Japanese population. J Periodontol. 2003;74(10):1460–1469. doi: 10.1902/jop.2003.74.10.1460. [DOI] [PubMed] [Google Scholar]
- 74.Suzuki N, Yoneda M, Naito T, Iwamoto T, Masuo Y, Yamada K. et al. Detection of Helicobacter pylori DNA in the saliva of patients complaining of halitosis. J Med Microb. 2008;57:1553–1559. doi: 10.1099/jmm.0.2008/003715-0. [DOI] [PubMed] [Google Scholar]