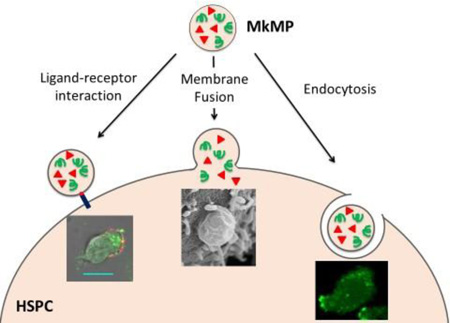
Abstract
Megakaryocytic microparticles (MkMPs), the most abundant MPs in circulation, can induce the differentiation of hematopoietic stem and progenitor cells (HSPCs) into functional megakaryocytes. This MkMP capability could be explored for applications in transfusion medicine but also for delivery of nucleic acids and other molecules to HSPCs for targeted molecular therapy. Understanding how MkMPs target, deliver cargo and alter the fate of HSPCs is important for exploring such applications. We show that MkMPs, which are distinct from Mk exosomes (MkExos), target HSPCs with high specificity since they have no effect on other ontologically or physiologically related cells, namely mesenchymal stem cells, endothelial cells or granulocytes. The outcome is also specific: only cells of the megakaryocytic lineage are generated. Observation of intact fluorescently-tagged MkMPs inside HSPCs demonstrates endocytosis as one mechanism of cargo delivery. Fluorescent labeling and scanning electron microscopy (SEM) imaging show that direct fusion of MkMPs into HSPCs is also engaged in cargo delivery. SEM imaging detailed the membrane-fusion process in four stages leading to full adsorption of MkMPs into HSPCs. Specifically, macropinocytosis and lipid raft-mediated were shown here as mechanisms of MkMP uptake by HSPC. In contrast, the ontologically related platelet-derived MPs (PMPs) cannot be taken up by HSPCs although they bind to and induce HSPC aggregation. We show that platelet-like thrombin activation is apparently responsible for the different biological effects of MkMPs versus PMPs on HSPCs. We show that HSPC uropods are the preferential site for MkMP binding, and that CD54 (ICAM-1), CD11b, CD18 and CD43, localized on HSPC uropods, are involved in MkMP binding to HSPCs. Finally, we show that MkMP RNA is largely responsible for HSPC programming into Mk differentiation.
Keywords: Microparticle, extracellular vesicle, stem cell, megakaryocytic
Graphical abstract
1. Introduction
Cell-derived microparticles (MPs) are small membrane-enclosed vesicles (0.1 to 1.0 µm in diameter) derived from direct budding off the plasma membrane of mammalian cells and mediate intercellular communication in various physiological and pathophysiological processes, including coagulation, inflammation, tumorigenesis, and differentiation [1–3]. MPs are different from the smaller (< 0.1 µm) exosomes that derive from the multivesicular endosome compartment of cells and have distinct physical and biological properties [4, 5]. Platelet-derived MPs (PMPs), discovered over 40 years ago, participate in many physiological processes, including hemostasis, maintenance of vasculature, immunity and inflammatory responses [6]. The most abundant (70% – 90%) circulating MPs express CD41 or CD61 [7]. For a long time, they were thought to originate from platelets and thus possess the biological functions of PMPs. However, it was shown that most circulating CD41+ MPs in healthy subjects derive from megakaryocytes (Mks), and these Mk-derived MPs (MkMPs) are different from PMPs [7, 8]. A role for MkMPs has not been known until our recent study, which documented a novel biological function of MkMPs, namely that they can induce differentiation of hematopoietic stem and progenitor cells (HSPCs) towards the Mk lineage without exogenous thrombopoietin (TPO) stimulation [9].
Several studies have examined mechanisms by which MPs interact with target cells. The interaction typically starts with a ligand-receptor mediated binding of MPs to target cells, and, in some cases, this initial interaction is sufficient to alter the fate of target cells [10–16]. Yet, in most cases, MPs exert their biological effect through transfer of signaling molecules (proteins, mRNAs, miRNAs or phospholipids), which requires uptake of MPs by the target cells [4, 17, 18]. It has been reported that two mechanisms used by target cells to integrate MPs are cell endocytosis and membrane fusion [4, 17, 18], and that cells use one of these two mechanisms to uptake MPs [19, 20]. For example, PMPs were internalized by human brain endothelial cells through active endocytosis as demonstrated by colocalization of MPs with endosomes and lysosomes [19]. Direct fusion of MPs with cells was claimed in other studies [20–23], whereby, MPs were first stained with a lipid-membrane or cytosol dye and then cocultured with target cells [20–23]. The evidence to support MP-cell fusion was the observed transfer of dye or MP-specific membrane proteins to target cells as detected by confocal or electron microscopy [20–23]. Yet, it is possible that endocytosis of MPs may contribute partially, if not fully, to the observed transfer of dyes or proteins. Thus, membrane fusion remains to be firmly established as a mechanism of MP uptake by cells.
Since MkMPs can be effectively frozen without loss of their biological activity (data not shown), it is possible that they can be used in transfusion medicine to enhance thrombopoiesis or for cargo delivery to HSPCs. Also, it is possible to engineer MkMPs to carry exogenous molecules for delivery to HSPCs, and/or identify key surface molecules that mediate the recognition of HSPCs for engineering synthetic or semisynthetic macro- or nano-particles for targeted drug delivery to HSPCs. Detailed understanding of how MkMPs recognize, target and deliver cargo to various HSPCs to alter their fate would be necessary prior to exploring such applications, and this is the main goal of this report. Here we first show that MkMP target the most primitive hematopoietic stem cells (HSCs) with higher effectiveness than more differentiated HSPCs. We show that both endocytosis and membrane fusion are responsible for delivery of MkMP cargo to HSPCs, and that MkMP RNA is partially, at least, responsible for changing the fate of HSPCs. Our data support the hypothesis that the platelet activation process necessary for PMP biogenesis explains the different biological effects of MkMP vs PMPs, and that MkMPs attach to and enter HSPCs preferentially through their uropods, with CD54, CD11b, CD18 and CD43 being involved in target-cell recognition.
2. Materials and methods
2.1 Materials and antibodies
All chemicals and protein reagents were obtained from Sigma-Aldrich or otherwise indicated. Recombinant human interleukin 3 (rhIL-3), rhIL-6, rhIL-9, rhIL-11, stem cell factor (rhSCF), thrombopoietin (rhTPO), Granulocyte colony-stimulating factor (rhG-CSF) were purchased from PeproTech Inc. Size standard fluorescent beads (0.22, 0.45, 0.88 and 1.34 µm) and AccuCount fluorescent particles (~5.0 µm) were from SpheroTech. Fluorescein isothiocyanate (FITC) - or phycoerythrin (PE)-conjugated anti-CD41 (GPαIIb), PE-conjugated anti-CD62P (P-selectin), allophycocyanin (APC)-conjugated anti-CD34, PE-conjugated anti-CD11b, APC-conjugated anti-CD235a, FITC-conjugated CD63, APC-conjugated CD81 and purified anti-CD41, anti-CD42b, anti-CD43, anti-CD50 antibodies as well as corresponding IgG isotype were all from BD Bioscience. APC-conjugated anti-CD133 antibody was obtained from Miltenyi Biotec. Purified anti-CD54 (ICAM-1) antibody was from Abcam. Anti-filamin A was from Santa Cruz Biotechnology.
2.2. Megakaryocytic (Mk) culture and kinetics of Mk microparticle (MkMP) formation
Frozen G-SCF mobilized human peripheral blood CD34+ cells were obtained from the Fred Hutchinson Cancer Research Center. CD34+ cells were cultured to Mks as described [24]. At d7, CD61+ cells (Mks) were enriched using anti-CD61 magnetic microbeads (Miltenyi Biotec) and were cultured as described [24]. From d8 to d12, CD41 and CD62P expression and concentration of MPs in the cell culture were measured by flow cytometer (FACSAria II, BD Biosciences) using AccuCount fluorescent particles (0.88 µm and 1.34 µm) as internal control.
2.3 Isolation of MkMPs and MkExos
MkMPs were isolated from d12 Mk culture as described [9]. Briefly, cells were removed from d12 culture medium at 150 ×g centrifugation for 10 min. Next, platelet-like particles (PLPs) and cell debris were removed by centrifugation at 1000 × g for 10 min [9]. Following that, MkMPs were enriched from the supernatant by ultracentrifugation at 25,000 rpm (38000 × g) for 1 hour at 4 °C using an Optima Max Ultracentrifuge and a TLA-55 rotor (Beckman Coulter). To isolate MkExos, we used a standard protocol [25], whereby the supernatant from previous step was filtered through 0.22 µm filters (CellTreat), and untracentrifuged at 100,000 × g for 90 min at 4 °C using an Optima L-90K Ultracentrifuge and a SW41 Ti Rotor (Beckman Coulter). MkMPs or MkExos were resuspended in IMDM or PBS and stored at −80 °C for further use.
2.4 Characterization of MkMPs and MkExos with dynamic light scattering (DLS)
For these experiments, MkMPs or MkExos isolated as above were suspended in PBS that had been filtered with 0.22 µm filters twice before being used. This filtered PBS contains no particles as confirmed by DLS measurement. DLS measurement of suspensions of MkMPs or MkExos were performed at 25 °C using a Zetasizer Nano ZS (Malvern Instruments) with 633 nm He-Ne laser, fixed angle of 173°, and automatic attenuator. Size distribution by number was collected with at least 3 measurements for each sample.
2.5 Characterization of MkMPs and MkExos with flow cytometry
To detect MkExos with flow cytometry, isolated MkExos were first captured with Dynabeads magnetic beads (4.5 µm, Thermo Fisher) coated with anti-CD63 antibody at 4 °C overnight, following the manufacturing protocol in Exosome - Human CD63 Isolation/Detection kit (Invitrogen). MkMPs or bead-bound MkExos were then incubated with isotype IgG, FITC anti-CD63, or APC anti-CD81 antibodies for 15 min at room temperature before flow cytometry analysis. The latter two antibodies target the common surface antigens of endosomal origin expressed by human exosomes [26], and which are not expressed on MkMPs.
2.6. Preparation of human platelets and PMPs
Blood for isolation of human platelets was collected by venipuncture from adult human volunteers after providing written informed consent as approved by the Institutional Review Board at the University of Delaware (IRB protocol # 622751-1). Blood was collected from healthy donors and PMPs were prepared as described [9]. Briefly, 50 mL of blood was collected into syringe with ACD buffer (trisodium citrate, 65 mM; citric acid, 70 mM; dextrose, 100 mM; pH 4.4) at a volume ratio of 1:6 (ACD:blood). Following that, blood were centrifuged at 250 × g for 10 minutes and the platelet rich plasma was isolated from the supernatant. Platelets were then pelleted at 750 × g for 10 minutes, followed by 1 wash with HEN buffer (10 mM HEPES, pH 6.5, 1mM EDTA, 150 mM NaCl) containing 0.05 U/ml apyrase. After that, platelets were resuspended in HEPES-Tyrode’s buffer (137 mM NaCl, 20 mM HEPES, 5.6 mM glucose, 1 g/l BSA, 1 mM MgCl2, 2.7 mM KCl, 3.3 mM NaH2PO4). To generate PMPs, platelets were activated by 2 U/mL human thrombin (Sigma), and removed by centrifugation at 1000 × g for 10 minutes. Lastly, PMPs were isolation by ultracentrifugation at 25,000 rpm for 1 hour at 4 °C, resuspended in IMDM medium, and stored at −80 °C.
2.7 Human umbilical vascular endothelial cells (HUVECs), mesenchymal stem cells (MSCs) and granulocytic cultures
Primary HUVECs were obtained from ATCC and cultured according to ATCC recommendation (growth medium: vascular cell basal medium (ATCC) supplemented with endothelial cell growth kit-VEGF (ATCC)). Passages 3–5 of HUVECs were used for following coculture experiment. Human MSCs (passages 2–4) were courtesy from Prof. Xinqiao Jia at the University of Delaware and cultured according to Lonza recommendation (growth medium: mesenchymal stem cell basal medium (Lonza) supplemented with MSCGM™ SingleQuots™ (Lonza)). Human granulocytes were differentiated from human CD34+ cells as previously described [27]. At d7 of granulocytic cell culture, CD15+ cells were enriched using MS column (Miltenyi Biotec) and CD15 microbeads (Miltenyi Biotec).
2.8. Coculture of human CD34+ lineage negative (Lin−) cells, HUVECs, human MSCs and human granulocytes with MkMPs
Lin+ (CD2+, CD3+, CD11b+, CD14+, CD15+, CD16+, CD19+, CD56+, CD123+, or CD235a+) cells were removed from CD34+ cells using the Miltenyi lineage-cell depletion kit. 60,000 CD34+ Lin− cells, cultured HUVECs, MSCs or CD15+ granulocytes were incubated with 10 MkMPs per cell in 50 µL IMDM medium for 1 hour at 37°C. After that, cocultures of MkMPs with CD34+ Lin− cells, MSCs and granulocytes were diluted in 550 µL IMDM supplemented with 5% BIT9500 and 50 ng/mL SCF; the coculture of MkMPs with HUVECs was diluted in 550 µL growth medium without any endothelial cell growth factors. Cocultures were maintained at 37°C and 20% O2 for 8 days before harvesting for ploidy and CD41 analyses. For some experiments, CD34+ Lin− cells at d0 and cells from coculture at d3, d5 and d8 were stained with anti-CD41, anti-CD34, anti-CD11b, anti-CD15 and anti-CD235a antibodies and analyzed by flow cytometry.
2.9. MkMP binding and uptake analyses
MkMPs were stained with 20 µM CFDA-SE dye (Life Technologies) for 20 minutes at 37°C and washed thrice with IMDM. Then, MkMPs were cocultured with hematopoietic stem/progenitor cells (HSPCs) from d3 Mk culture at a ratio of 30 MkMPs/cell for the indicated time. For the first hour, the coculture medium was 50 µL IMDM and after that, the coculture was diluted in IMDM supplemented with 5% BIT9500, 50 ng/mL SCF and 1% pen strep. Flow cytometry was used to measure binding of MkMPs to cells. Images of cocultured cells were collected via confocal microscopy (Zeiss 5 LIVE DUO highspeed/spectral confocal microscope).
2.10 Cell ploidy and CD41 expression analyses
Cells from various MkMP coculture and vehicle control cultures were harvested for ploidy assay as described [28]. Measurement of CD41 expression, Mk ploidy and numbers was performed on flow cytometry at d8 of coculture using AccuCount fluorescent particles as internal control.
2.11. Scanning electron microscopy (SEM) analysis
HSPCs from d3 of Mk culture were cocultured with MkMPs (30 MPs/cell) in 100 µL IMDM medium for 2 or 4 hours. Then HSPCs or MkMPs were let spread on round coverslips coated with 1 µg/mL human fibronectin and poly-L-lysine for another hour. 2% glutaraldehyde/PBS (Electron Microscopy Sciences, EM grade) was added to fix cells for at least 1 hour at room temperature or overnight at 4°C. After that, samples were postfixed for 1.5 hours in 1% OsO4 at room temperature. After rinsed with H2O, samples were dehydrated in a series of ascending ethanol concentrations for 10 min in each solution. After critical-point drying in Autosamdri-815B Critical Point Dryer (Tousimis), samples were sputter-coated with gold using a Benchtop Turbo III Sputter Coater (Denton Vacuum). The electron images were collected via Field-Emission Scanning Electron Microscope (Hitachi S4700) at a working distance of 8.5–10.5 mm and voltage of 3.0 kV.
2.12. Transmission electron microscopy (TEM) analysis
Transmission Electron Microscopy was performed on Zeiss Libra 120 Transmission Electron Microscope and images were acquired using a Gatan Ultrascan 1000 CCD. The d3 HSPCs were incubated with MPs (30 MPs/cell) in 100 µL medium for 5 hours and then fixed in 2% EM grade glutaraldehyde and 2% paraformaldehyde (Electron Microscopy Sciences, EM grade) in 0.2 M cacodylate buffer. Samples were then rinsed 3 times, 15 min each, in 0.1 M sodium cacodylate buffer containing 2 mM CaCl2 and fixed for 1 hour in 1.5% potassium ferrocyanide, 2 mM CaCl2, 2% OsO4 in 0.1 M sodium cacodylate buffer on ice. After 3 washes with H2O, the samples were post-fixed in 2% OsO4 for 1 hour at room temperature, followed by 4 washes in H2O. Then samples were stained en bloc overnight at 4°C with filtered 1% uranyl acetate. After 3 washes in H2O, the samples were dehydrated in a series of ascending acetone solutions. The samples were then infiltrated within n-BGE and then Quetol-NSA resin on a rotator. Samples were embeded in labeled BEEM capsules and polymerized at 60°C for 24–48 hours. The ultrathin sections were prepared using a Reichert Jung Ultracut E ultramicrotome, and were collected onto 200 mesh formvar/carbon coated copper grids. Grids were stained with 2% methanolic uranyl acetate and Reynolds’ lead citrate.
2.13. Thrombin treatment of MkMPs and generation of MkMPs from thrombin-treated Mks
MkMPs were isolated and enriched from d12 Mk culture as described above. After adding 1 mM CaCl2, half of MkMPs were treated with 10 U/mL human thrombin for 30 min at 37°C and the other half were treated in the same way without thrombin as the control for MkMP coculture. Mks, isolated from d12 Mk culture by centrifugation at 150 × g for 10 min, were treated with 10 U/mL human thrombin with 1 mM CaCl2 presence for 30 min at 37°C. MkMPs were isolated and enriched from thrombin-treated Mk as described. Thrombin-treated MkMPs and MkMPs generated from thrombin-treated Mks were washed with IMDM medium thrice before coculture with CD34+ cells. CD41 and CD62P expression of these MPs as well as PMPs and MkMPs were examined by flow cytometry.
2.14. Western analysis
MkMPs, thrombin-treated MkMPs, MkMPs from thrombin-treated Mks and PMPs were lysed in 1X SDS lysis buffer and the proteins were separated by SDS-polyacrylamide gel electrophoresis. Primary mouse anti-N-terminal filamin A (Santa Cruz #sc-17749) antibody and secondary Alexa Fluor®488 conjugated anti-mouse IgG antibody (Life Technologies #A11017) were used for filamin A detection. Images were captured by Typhoon FLA 9500 (GE Healthcare) and quantitative analysis of blotting band intensity was performed on ImageJ.
2.15. Antibody blocking assays
At d1 and d3 of CD34+ cell culture, 30,000 cells were incubated with 100 µg/mL anti-CD11a, anti-CD11b, anti-CD18, anti-CD34, anti-CD43, anti-CD44, anti-CD50, anti-CD54 or anti-CD133 antibodies or isotype IgG for 15 min at room temperature. Then MkMPs (30 per cell) stained with CFDA-SE were added to cells and more antibody was added to maintain the same concentration. MkMP-cell cocultures were incubated in the incubator for 1 hour. Cells were washed once with cold PBS before flow cytometric analysis. For some experiments, MkMP cocultures without any antibody blocking were incubated for 3 hours and anti-CD133 antibody was added to cell culture for 15 min before confocal microscopic analysis. For other experiments, MkMPs stained with CFDA-SE were incubated with anti-CD41, anti-CD42b antibodies or annexin V at 100 µg/mL first before coculture with d3 HSPCs.
2.16. Inhibitor studies for interrogating specific MkMP-uptake mechanisms
CD34+ cells were cultured as described above. At d1 or d3 of Mk culture, cells were pre-treated with 5 mM methyl-β-cyclodextrin (MβCD, Sigma), 10 µM dimetylamiloride (DMA, Sigma), 20 µM chlorpromazine (CH, Sigma), or 80 µM dynasore (Sigma) for 45 min at 37 °C, which were used to inhibit lipid raft-mediated endocytosis, macropinocytosis, clathrin-dependent endocytosis, and dynamin-dependent or independent endocytosis, respectively. Cells were then cocultured with CFDA-SE-stained MkMPs at the concentration of 30 MkMPs per cell, for 30 min at 37 °C. For other experiments, cells treated with inhibitors were coculture with labeled molecules known to be taken up by cells using specific mechanisms. This was to examine the impact of inhibitors on specific mechanisms and compare the impact of the inhibitors above on the uptake of these molecules (used as positive control) versus the uptake of MkMPs. Specifically, Alexa 647-conjugated cholera toxin subunit B (CT-B, Life Tech), Alexa-594-conjuagted dextran 10k (Dx, Life Tech), and Alexa 647-conjugated transferrin (Tf, Life Tech) were used as positive control, since these molecules were established to be taken up by cells through lipid-raft mediated endocytosis, macropinocytosis, and clathrin-dependent endocytosis, respectively [29]. Briefly, cells treated with inhibitors were cocultured with 10 µg/ml CT-B, 100 µg/ml Dx or 5 µg/ml Tf for 15 min at 37 °C. After being washed with cold PBS, the uptake of MkMP, CT-B, Dx or TF were analyzed by flow cytometry.
2.17 RNase treatment assay, RNA size distribution measurement and coculture experiments
MkMPs were isolated as above and resuspended in IMDM. For RNase treatment, MkMPs were incubated with 1 U/mL Ambion® RNase A/T1 cocktail (Life Technologies) or 10 U/mL RNase ONE™ (Promega) for 1 hour at 37°C. Following that, 1 U/mL RNase inhibitor, SUPERase-IN (Ambion) was used to stop the RNase reactions. To further remove RNase from each MkMP samples, MkMPs were washed with IMDM thrice through 25,000 rpm ultracentrifugation for 1 hour at 4 °C. The three washes result in a RNase dilution by at least 106 in each sample. This dilution eliminates any impact from RNase in downstream experiments. For some experiments, total RNA were isolated from control MkMPs (without RNase treatment) or RNase-treated MkMPs with miRNeasy micro kit (Qiagen), following the manufacturing protocol. Total RNA concentration from each sample were measured with Qubit RNA HS Assay kit (Invitrogen). Measurement of size distribution of RNA were performed by Bioanalyzer 2100 (Agilent) with RNA600 Pico kit (Agilent). For some other experiments, human CD34+ cells were cocultured with control MkMPs or RNase-treated MkMPs at 10 MPs/cell for 8 days. Cells were harvested at day 8 before ploidy analysis, as described above.
2.18. Statistical analysis
Paired student t test of all data was performed on Minitab 16 (Minitab).
3. Results
3.1. Characterization of MkMPs as distinct from Mk exosomes
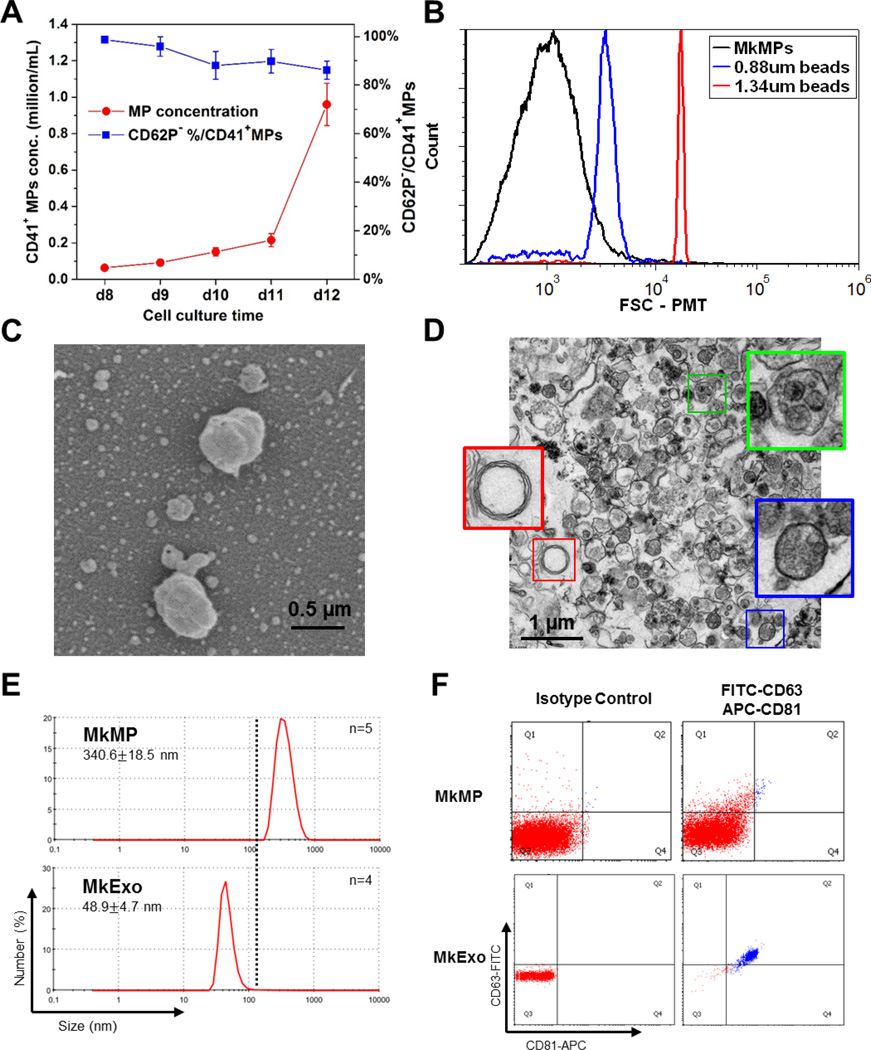
We first characterized the kinetics of formation, size distribution and intracellular texture of MkMPs using a variety of techniques. CD34+ HSCs were differentiated into Mks as described [24]. Flow cytometric analysis showed that, from d8 to d12, most CD41+ MPs were CD62P−, indicating that these MPs were derived from Mks rather than PLPs (Fig. 1A). Based on the concentration of CD41+ MPs, most MkMPs were released by mature Mks (d11 to d12) rather than immature Mks (d8 to d11) (Fig. 1A). TEM micrographs and flow cytometric analysis suggest that these MkMPs were a heterogeneous population with the diameter smaller than 0.88 µm (Fig 1B & 1D). MP heterogeneity is a common characteristic of all reported MPs and not unique to these MPs [30–32]. SEM micrographs show that MkMPs are not spherical and their membrane not smooth (Fig. 1C). This information was useful for the SEM studies reported below. To further validate our MkMP isolation protocol, and to show that our MkMP preparations are not contaminated by MkExos, we examined the MkMPs and MkExos, prepared as described in our Methods, using DLS and flow cytometry. In DLS measurements (Fig. 1E), the size distribution of our MkMP and MkExo preparations show a mean size of 340.6 nm and 48.9 nm, respectively, which are in the range of common sizes of MPs (100–1000 nm) and exosomes (<100 nm) as reported in the literature [4]. These size distributions show no population overlap between our MkMPs and MkExos. We further examined our particle preparation using two common exosomal markers, CD63 and CD81, through flow cytometry analysis [26]. As expected, MkExos expressed both CD63 and CD81, while MkMPs were CD63 and CD81 negative. Taken together, these data demonstrate that MkMPs isolated from this protocol are free from contamination from MkExos. Based on these studies, we subsequently used MkMPs from d12 Mk culture isolated through successive centrifugations.
Fig. 1. Kinetic of MkMP generation and characterization of MkMPs and MkExo.
(A) CD62P expression and concentration of CD41+ MPs in Mk culture from d8 to d12. Mk cultures of CD34+ cells were enriched in Mk cells at d7 and were cultured at a concentration of 200K cell/mL. From d8 to d12, 100 µL of cell culture medium was harvested every day for CD41 and CD62P analyses by flow cytometry. MP concentration was counted by flow cytometry using microbeads as a control. The data represent the average of three biological replicates ± standard error of mean. (B) Representative size distribution histogram of MkMPs from d12 cells analyzed by flow cytometry using microbeads (0.88 µm and 1.34 µm in diameter) as internal size standards. (C–D) Representative SEM (C) and TEM (D) micrographs of MkMPs from d12 cell culture. The very small particles in the background of panel C are artifacts from sample preparation for SEM analysis as such small particles (<40 nm) cannot be collected during our MkMP preparation protocol as also shown in panel E below. Panel (D) demonstrates that MkMPs are heterogeneous in content as judged by the TEM-image texture of the MP contents, with three MPs enlarged for better examination. (E) Representative size distribution of MkMPs and MkExo measured by DLS with 2 biological replicates and 4–5 measurements. (F) Flow cytometry analysis of CD63 and CD81 expression on MkMPs and MkExos, with negative isotype IgG control.
3.2 MkMPs promote Mk differentiation and CD34+-cell maintenance and expansion of primitive Lin− CD34+ stem cells
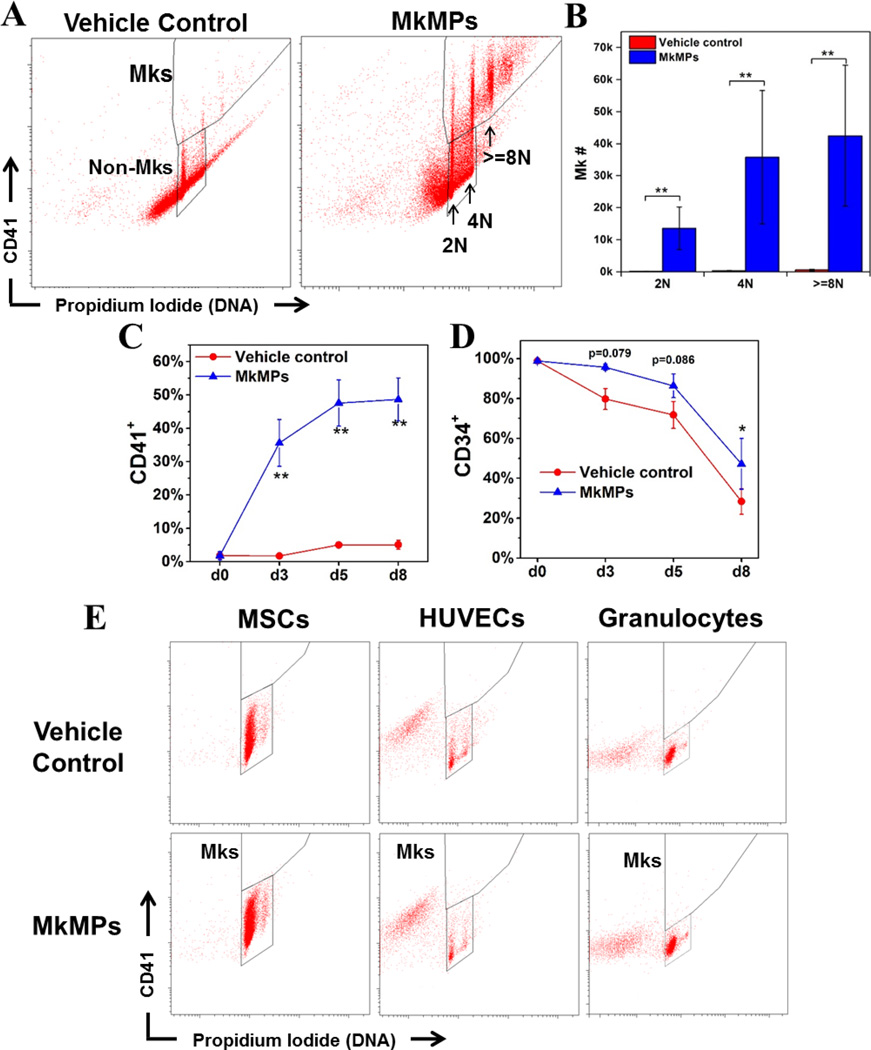
In order to better assess possible applications of MkMPs, we examined if MkMPs target HSPCs at all stages of differentiation. We had shown that MkMPs induce the differentiation of CD34+ HSPCs and partially differentiated HSPCs (d3 & d5 cells from cultures of CD34+ cells) towards the Mk lineage without TPO [9]. Mk cells from these MkMP cocultures are functional in that they project proplatelets and synthesize both α-and dense-granules [9]. Here, we investigated the impact of MkMPs on more primitive, Lin− CD34+ cells. CD34+ cells were depleted of Lin+ (CD2+, CD3+, CD11b+, CD14+, CD15+, CD16+, CD19+, CD56+, CD123+, or CD235a+) cells before being cocultured for 8 days with MkMPs. The coculture contained a significant number of Mks with 2N, 4N and >=8N ploidy classes, while very few Mks and none in higher ploidy classes were found in control cultures (Fig. 2A–B) (95-fold difference, P<0.01).
Fig. 2. MkMPs promote Mk differentiation of primitive Lin CD34+ stem cells and this effect is target specific for HSPCs.
60,000 primitive Lin HSPCs were enriched from CD34+ cells and cocultured with MkMPs at concentration of 10 MkMPs/cell for 8 days. (A) Representative flow cytometric CD41 expression and ploidy analyses of cells from vehicle control cultures and the MkMP cocultures at d8. (B) Numbers of Mks with different ploidy classes (2N, 4N and >=8N) in vehicle control cultures and the MkMP cocultures at d8. Percentages of (C) CD41+ and (D) CD34+ cells in vehicle control cultures and the MkMP cocultures at d0, d3, d5 and d8. (E) Representative CD41 expression and ploidy analyses of two sets of MkMP cocultures with MSCs, HUVECs or granulocytes. Human MSCs (passage 2–4), HUVECs (passage 3–5) and CD34+ cell-derived granulocytes (d7 of culture) were cocultured with MkMPs at the concentration of 10 MkMPs/cell for 8 days before being harvested for CD41 expression and ploidy analyses. The data in panels (B–D) are represented as the average of three biological replicates ± standard error of mean. *, P<0.05; **, P<0.01.
To characterize the CD41− cells in both the MkMP cocultures and control cultures, anti-CD34, anti-CD41, anti-CD11b, anti-CD15+ and anti-CD235a antibodies were used to identify CD34+ HSPCs, Mks, granulocytes, monocytes and erythrocytes, respectively. Very few cells differentiated into granulocytes or erythrocytes in either the control culture or the MkMP coculture, indicating that MkMPs could not induce HSPC differentiation to these two lineages (Fig. S1). In addition, MkMPs significantly inhibited differentiation of HSPCs to CD15+ cells by ca. 50% (Fig. S1). These data suggest that the effect of MkMPs is specific for the Mk lineage in that they do not induce differentiation to other myeloid cells, and notably the erythroid lineage, which has a common progenitor with the Mk lineage. In MkMP cocultures, the percentage of CD41+ cells increased from 0 at d0 to ~47% at d5 and plateaued after d5 (Fig. 2C), indicating that 5 days are sufficient for HSPCs to commit to megakaryopoiesis induced by MkMPs. Compared to control cultures, MkMP cocultures had a higher percentage of CD34+ cells at d8 (47% vs. 28%, Fig. 2D, P<0.05) and more CD34+ cells (although not statistically significantly so: 108K vs. 27K, P=0.23), thus suggesting that MkMPs can maintain a higher percentage of CD34+ cells compared to control.
We also compared the present data (Fig. 2B) against our previous data (Figs. 5B&5E of Ref [9]) to examine any differential impact of MkMPs on Lin−CD34+, CD34+ and d3 HSPC target cells. The number of Mks at d8 that resulted from the cocultures of 60K Lin−CD34+, CD34+ or d3 HSPC target cells with MkMPs at 10 MkMPs/cell were ca. 90K, 52K and 38K, respectively, suggesting that MkMPs target primitive HSPCs with greater effectiveness.
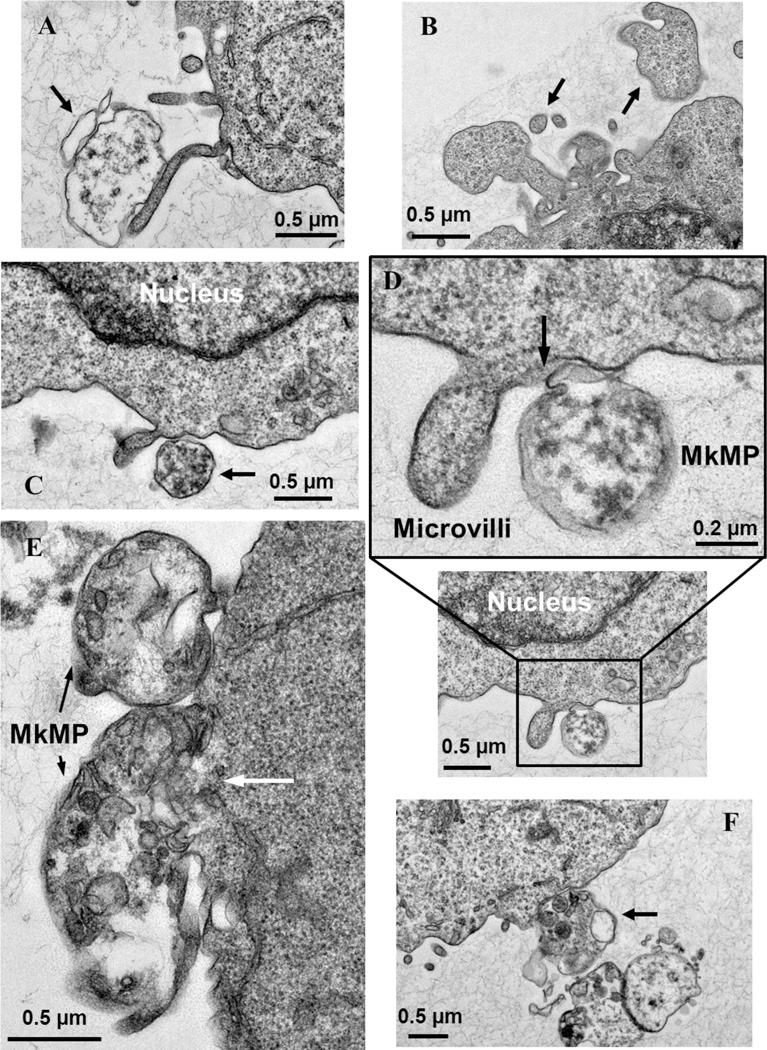
Fig. 5. TEM details the fusion of MkMPs into HSPC membranes.
d3 HSPCs from Mk cultures of CD34+ cells were cocultured with MkMPs for 3 ~ 5 hours and examined using TEM. (A, C-F) MkMPs with different internal TEM textures bind to an HSPC. (A) An electron-light MkMP (arrow) makes contact with a microvillus emanating from the cell surface. (B) Control for image interpretation: the particles (arrow) have the same TEM texture as the cell and were thus part of the cell and not MkMPs. (C) An MkMP with electron-dense texture makes contact with the cell surface near a microvillus. (D) An MkMP interacting with the cell membrane near a microvillus displays partial membrane fusion (arrow). (E) Two MkMPs interacting with one cell display different degrees of membrane fusion; the top, smaller MkMP shows an early stage of interaction, while the interaction between the larger MkMP and the cell is at an advance stage where the membrane between the cell and the MkMP is barely visible (white arrow). (F) A cluster of MkMPs make contact with a cell via an MkMP containing small organelle-like particles.
3.3. Target specificity: MkMPs cannot transdifferentiate human granulocytes, MSCs or HUVECs into Mk cells
Certain types of cells can transdifferentiate into other unrelated types of mature cells [33–37], and some of these events could be mediated by MPs. For example, MPs from lung endothelial cells induced transdifferentiation of bone marrow cells into endothelial cells [38]. Here, to investigate if MkMPs can induce Mk differentiation of related cell types, we cocultured MkMPs with the ontologically or physiologically related granulocytes (that contain several related white blood cells, including neutrophils), MSCs or HUVECs, all of which are encountered by MkMPs in the bone-marrow microenvironments or in circulation. Flow cytometric analysis (Fig. 2E) showed that no CD41+ or polyploid cells were identifiable in these cocultures, indicating that MkMPs could not transdifferentiate these cells into Mks, and thus the action of MkMPs is specific to HSPC targets. Although MkMPs may interact with some of these cells (e.g., as neutrophils interact with platelets, they are likely to also interact with MkMPs), the interaction does not lead to fate-changing phenotype.
3.4. Endocytosis is a first mechanism by which MkMPs deliver their cargo to HSPCs
As discussed, the two key mechanisms by which MPs are presumed to bind to and be taken up by target cells are membrane fusion and endocytosis [18]. To inform the mechanistic investigations, we first examined the time scale of the interaction between MkMPs and HSPCs. MkMPs were stained with the cytoplasmic tracker dye CFDA-SE and cocultured with HSPCs from d3 Mk culture. Cells from the coculture were analyzed by flow cytometry to examine the kinetics of MkMP-cell binding based on the mean fluorescence intensity (MFI) of cell CFDA-SE. The cell CFDA-SE MFI increased quickly within 10 min from the start of the coculture and reached the maximum at 60 min, dropping quickly after 2 hours and reaching a basal low level after 24 hours (Fig. S2). These data demonstrate that MkMPs bind to cell surface quickly to start delivering cargo to target cells. After one hour of coculture, the decreased cell CFDA-SE MFI could be caused by dilution due to cell proliferation.
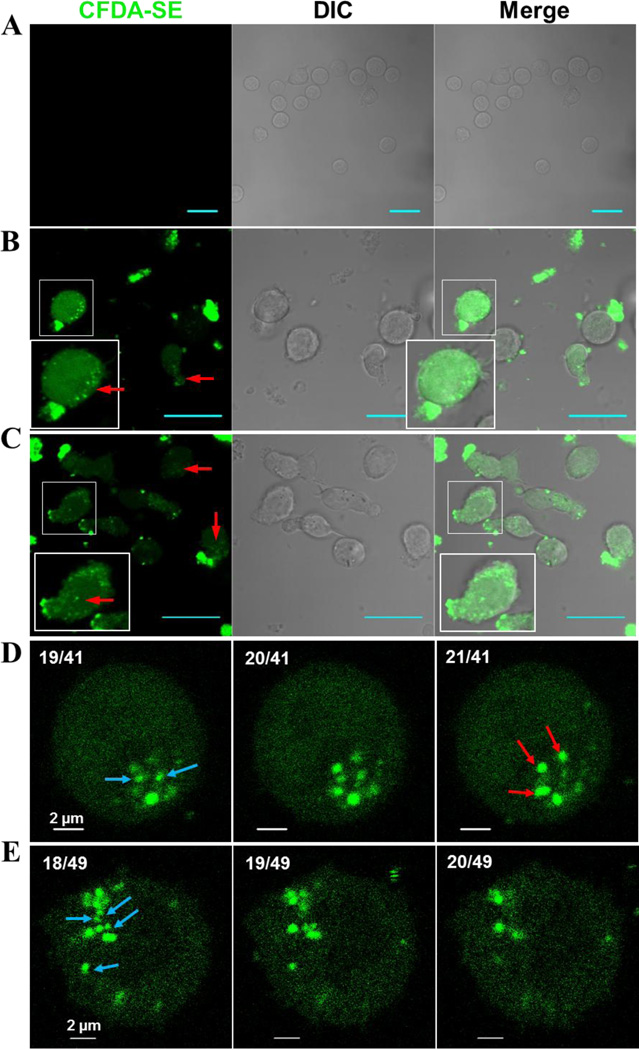
To investigate if MkMPs are just binding to cells or taken up by them, MkMP cocultures were examined using confocal microscopy. After 3 to 5 hours of coculture, most HSPCs contained CFDA-SE dye of variable intensity (Fig. 3A–C), indicating that uptake of MkMPs and/or transfer of dye (labeled cargo) from MkMPs to cells took place. Further analyses demonstrated that uptake of MkMPs took place through both endocytosis and membrane fusion. We discuss endocytosis first.
Figure 3. MkMPs are quickly endocytosed by HSPCs.
MkMPs were stained with CFDA-SE (green) dye and then cocultured for 3–5 hours with HSPCs from d3 of CD34+ -cell Mk cultures. Fluorescent and Differential Interference Contrast (DIC) images were collected via confocal microscopy. (A) Vehicle control cultures without fluorescent MkMPs display no background green signal. (B–C) MkMP cocultures (~ 4 hours) contain cells with intact fluorescence particles (MkMPs, red arrow) inside them, diffuse fluorescence staining, or fluorescent MkMPs on their surface. Inserts amplify images to show more details. Scale bar in panels (A–C) represents 20 µm. (D–E) Fluorescent images of two cells from the MkMP cocultures (~4 hours) at different confocal planes with 0.4 µm apart. The numbers on the top left of each image represent (image slice number)/(total number of slices). As the image slice number increases, intact MkMPs indicated by red arrows appear while the other MkMPs indicated by blue arrow disappear, thus demonstrating that these MkMPs are inside the cell.
In MkMP coculture, some cells contained variable numbers of distinct CFDA-SE fluorescence dots representing intact MkMPs, several of which were not associated with cell membranes (Fig. 3B–C, arrow). This suggested that these MkMPs were inside the cells, and, thus, that MkMP uptake took place through endocytosis. To confirm this, images of single cells from the MkMP coculture at different confocal planes, 0.4 µm apart, were collected to construct a 3D image of cells interacting with MkMPs.
To identify the MkMP location (inside, out or on the surface of the cell) these images were rotated around an axis through the center of the cell to examine the distance with which the various MkMPs (concentrated green dots) “move” around this axis (supplemental video 1). Some intact MkMPs rotated around this central axis at a shorter distance than the cell surface (delineated by a dim green signal), thus indicating that these MkMPs were inside the cell rather than on the cell membrane. This can also be identified on the image slices (Figs. 3D–E), by focusing on MkMPs that are clearly within the image boundaries (delineated by a dim uniform green signal) of a cell: some of these MkMPs (blue arrow) disappear and others (red arrows) show up as the image slice “moves up”. These data demonstrate that intact MkMPs were internalized by cells through endocytosis, similar to what was reported for PMP internalization by endothelial cells [19].
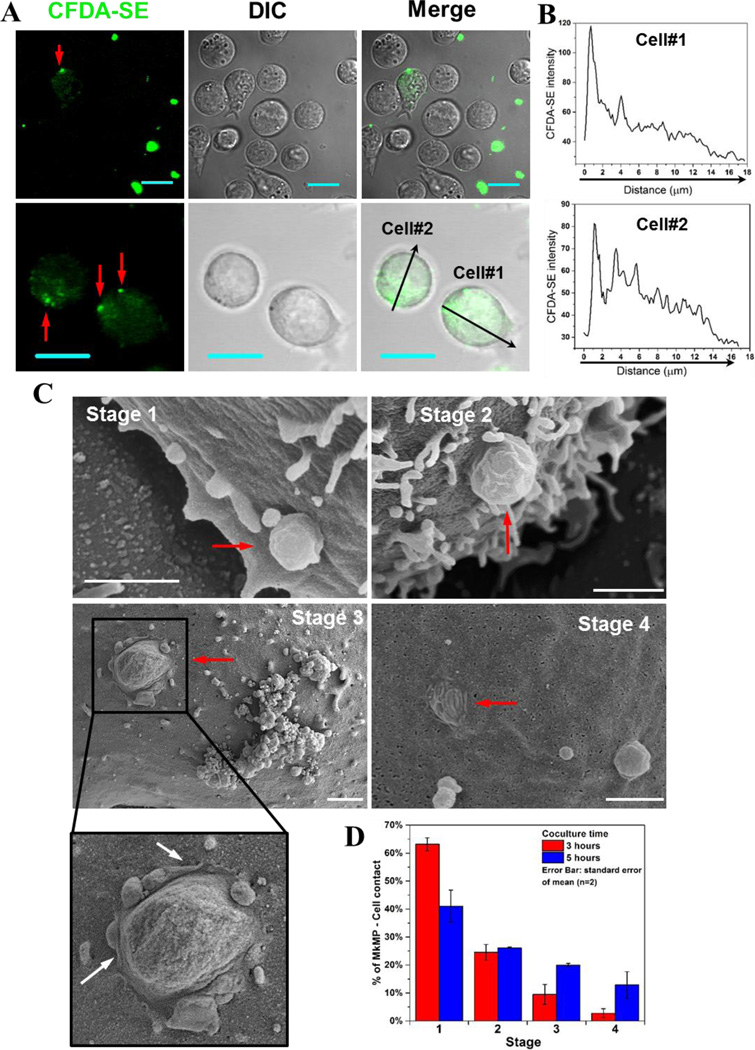
3.5. Cell fusion is a second mechanism by which MkMPs deliver cargo to HSPCs
In addition to observing intact MkMPs inside HSPCs, we also observed gradients of CFDA-SE dye (Fig. 4A–B) in some cells emanating from individual MkMPs on cell membranes as indicated by the highly concentrated dye (Fig. 4A, red arrows). These CFDA-SE gradients appear to result from direct discharge of the dye-stained MkMP content into the HSPC cytoplasm after they fuse with cells, and not from endocytosed MkMPs. To confirm membrane fusion, cells from MkMP cocultures were examined using SEM. Examination of a large number of SEM micrographs easily identified several events demonstrating fusion of MkMPs into HSPC membranes (Fig. 4C–D). We identified 4 sequential stages through which MkMPs are fused into cells based on the relative volume of MkMPs left on cell membranes (Fig. 4C). At stage 1, following MkMP/target-cell contact and recognition, MkMPs appear to be just attached to cell surfaces, retaining their shape as an intact particle. At stage 2, less than half of the MkMP body is fused into the cell membrane. This stage is characterized by a visible bridge firmly connecting the MkMP to the HSPC membrane. At stage 3, MkMPs are half fused into cells, and this consistently results in the formation of microvillar-like membrane-ruffle structures emanating from the contact area between the MkMP and the cell (Fig. 4C, white arrows). These membrane structures appear to be remnants of the process by which MkMPs are fused into HSPCs. At stage 4, MkMPs are almost completely fused into cells, with HSPC membranes appearing almost smooth, with just a few “wrinkles” (remnants of the fused MkMP) on the cell surface. Note the presence of the typical microvilli on the surface of HSPCs near the location where the MkMPs interact with the HSPCs [39]. As we discuss below, these microvilli may be important in the process of how MkMPs deliver cargo to HSPCs.
Fig. 4. MkMPs can fuse into HSPC membranes and release their content into the target HSPCs.
(A–B) MkMPs were stained with CFDA-SE (green) dye and then cocultured with for 3–5 hours with d3 HSPCs from Mk cultures of CD34+ cells. Fluorescent and DIC images were collected via confocal microscopy. (A) Images of cells from the MkMP cocultures demonstrate CFDA-SE dye gradients inside the cells emanating from one or few fluorescent particles (red arrow) on the cell surface; Scale bar, 20 µm. (B) CFDA-SE dye intensity profiles quantitating the dye gradient along the black arrows of cells #1 and #2 in the lower panel (A). (C–D) HSPCs as above were cocultured with MkMPs for 3 and 5 hours and examined using SEM. (C) Representative electron micrographs demonstrate four consecutive stages through which MkMPs (red arrow) were fused into HSPC membranes; scale bar, 1 µm. (D) Percentages of MkMP-HSPC interactions at each stage after 3 and 5 hours of coculture. The data represent the average of two biological replicates ± standard error of mean.
We also quantified the percentage of MkMP-cell interaction events at each stage after 3 and 5 hours of coculture (Fig. 4D). Most (~63%) fusion events were at the 1st stage and very few at the 3rd (9.7%) and the 4th (2.8%) stages at hour 3. The fusion events at the 1st stage decreased to ~41% while the percentages at the 3rd and 4th stages increased to ~20% and ~12.9%, respectively, after 5 hours (Fig. 4D), thus indicating that the fusion process takes place continuously in MkMP cocultures.
TEM analysis was also used to examine interactions between MkMPs and HSPCs. As a basis for unambiguously identifying MkMPs interacting with target HSPCs, we used differences in the texture and internal structures of the particles (see also Fig. 1D) in TEM images in relation to the texture of cells. In Fig. 5A, one observes large texture differences between cells and the cell-emanating microvilli [39] and an MkMP (arrow) that has an electron-light texture: this image shows an MkMP interacting with an HSPC microvillus-like structure [39]. In contrast, the particles in Fig. 5B (arrows) have the same texture as the cell, and are thus viewed as microvilli or microvillar-like membrane extensions and not MkMPs. Fig. 5C shows an early-stage interaction of an MkMP with a HSPC near the base of a microvillus. Fig. 5D shows a more advanced interaction (that includes a partial membrane fusion) of an MkMP with a HSPC near the base of a microvillus. In Fig. 5E, one observes two MkMPs at different stages of fusing into a target cell: the upper, smaller MkMP is apparently only binding to the cell membrane. In contrast, for the lower, larger MkMP, the membrane (white arrow) between the MkMP and the cell body is greatly diminished, suggesting that the MkMP is partially fused into the cell. Fig. 5F displays an image of a cluster of MkMPs near an HSPC membrane at an early stage of interaction. We should note that TEM analysis of the vehicle control culture did not identify any fusion such events, and extensive literature search displaying TEM and SEM images of HSPCs did not identify such fusion events.
These images are then uniquely associated with the cocultures of MkMP and HSPCs, complement the fluorescence microscopy images of Fig. 4A, support the concept that MkMPs can fuse into HSPCs, and provide evidence for the likely importance of an early interaction between cellular microvilli and MkMPs. Although the fusion process by which cells take up MkMPs or other MPs has not been yet dissected, it is known that in generic membrane fusion, lipid rafts regulate exocytosis and are present at the membrane fusion site [40]. It is possible then that lipid rafts may also play a role in MkMP uptake via membrane fusion.
3.6. Role of macropinocytosis and lipid rafts in MkMP uptake by HSPC
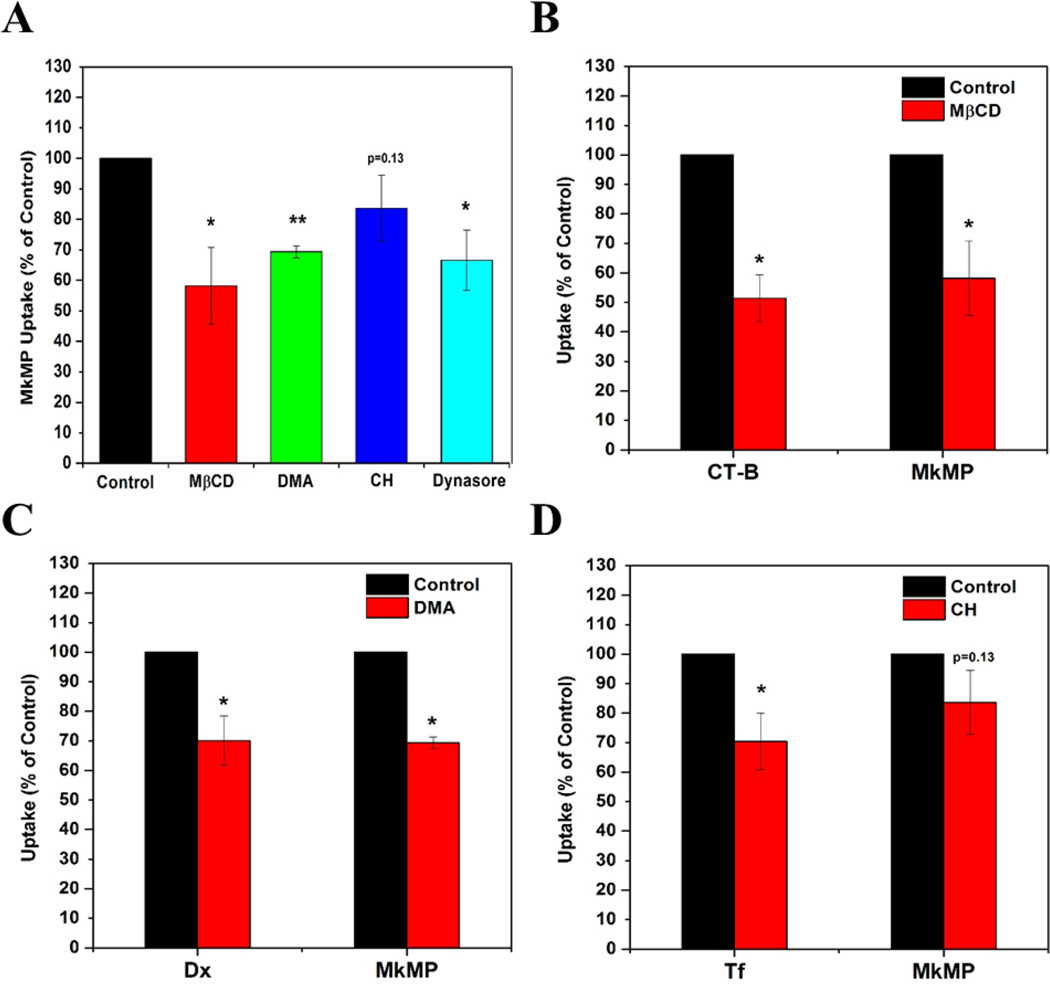
To investigate the uptake of MkMP by HSPCs, we treated cells with specific inhibitors before being cocultured with MkMPs. The inhibitors were MβCD, DMA, CH and dynasore, which were used for targeting mechanisms involving lipid rafts, macropinocytosis, clathrin-dependent endocytosis and dynamin-dependent or independent endocytosis. To access the impact of inhibitors on MkMP uptake, Alexa 647-conjugated cholera toxin subunit B (CT-B), Alexa-594-conjuagted dextran 10k (Dx), and Alexa 647-conjugated transferrin (Tf) were used for positive control, since these molecules are established to be taken up by cells through lipid-raft mediated endocytosis, macropinocytosis, and clathrin-dependent endocytosis, respectively [29]. Based on the MFI from flow cytometry analysis, we found that the MβCD treatment significantly decreased MkMP uptake by 42% (Fig. 6A), while the uptake of CT-B, a positive control for the engagement of lipid rafts, was inhibited by MβCD by 49% (Fig. 6B). DMA, a Na+/H+ exchange inhibitor, decreased MkMP and Dx uptake by 31% and 30%, respectively (Fig. 6A, 6C). These results show that these inhibitors inhibited the uptake of known molecules (positive controls) and MkMPs to a similar extent and thus these data suggest that lipid rafts and macropinocytosis play a role in MkMP uptake. Although the treatment with the CH inhibitor significantly inhibited the clathrin-dependent uptake of Tf by 30%, it had only a small effect (16% inhibition) on MkMP uptake (Fig.6A, 6D). Thus, clathrin-dependent endocytosis in not a major player in the MkMP uptake.
Figure 6. Macropinocytosis, lipid-raft and dynamin are engaged in uptake of MkMP by HSPCs.
HSPCs were pre-incubated with specific inhibitors impacting MkMP uptake by HSPCs. The inhibitors used were methyl-β-cyclodextrin (MβCD), dimetylamiloride (DMA), chlorpromazine (CH), and dynasore, which target lipid raft, macropinocytosis, clathrin-dependent endocytosis and dynamin-dependent or independent endocytosis, respectively. To assess the impact of the inhibitors on MkMP uptake, we compared the outcomes from the inhibitor studies against the uptake of the following molecules (positive controls), cholera toxin subunit B (CT-B), dextran 10k (Dx), and transferrin (Tf), which are established to be taken up by lipid-raft mediated endocytosis, macropinocytosis, and clathrin-dependent endocytosis, respectively. Specifically, d3-HSPCs were pre-incubated with or without 5 mM MβCD, 10 µM DMA, 20 µM CH, or 80 µM dynasore for 45 min at 37 °C, before the coculture with (A–D) CFDA-SE-stained MkMP for 30 min, (B) CT-B, (C) Dx, or (D) Tf for 15 min at 37 °C. Uptake of MkMP, CT-B, Dx, or Tf were analyzed by flow cytometry. Data represent averages of 3 biological replicates ± standard error of mean. *, P<0.05; **, P<0.01; ***, P<0.001
We also found that dynasore, a dynamin inhibitor, decreased MkMP uptake by 33% (Fig. 6A). As detailed in the discussion, the inhibitory effect of dynasore, combined with the lack of inhibition by CH, can be interpreted to support micropinocytosis as a mechanism of MkMP uptake. In addition to the inhibitors mentioned above, Filipin and Latrunculin B (Lat B) were also used for examine MkMP uptake, which are established-inhibitors for targeting cholesterol and inhibiting actin polymerization, respectively. Filipin showed no inhibitory effect on either MkMP or CT-B uptake (data no shown), indicating that Filipin is not an appropriate inhibitor to target cholesterol in this system. Lat B also showed no effect on MkMP uptake (data no shown), thus suggesting that actin might not be involved in early steps at least of the MkMP uptake.
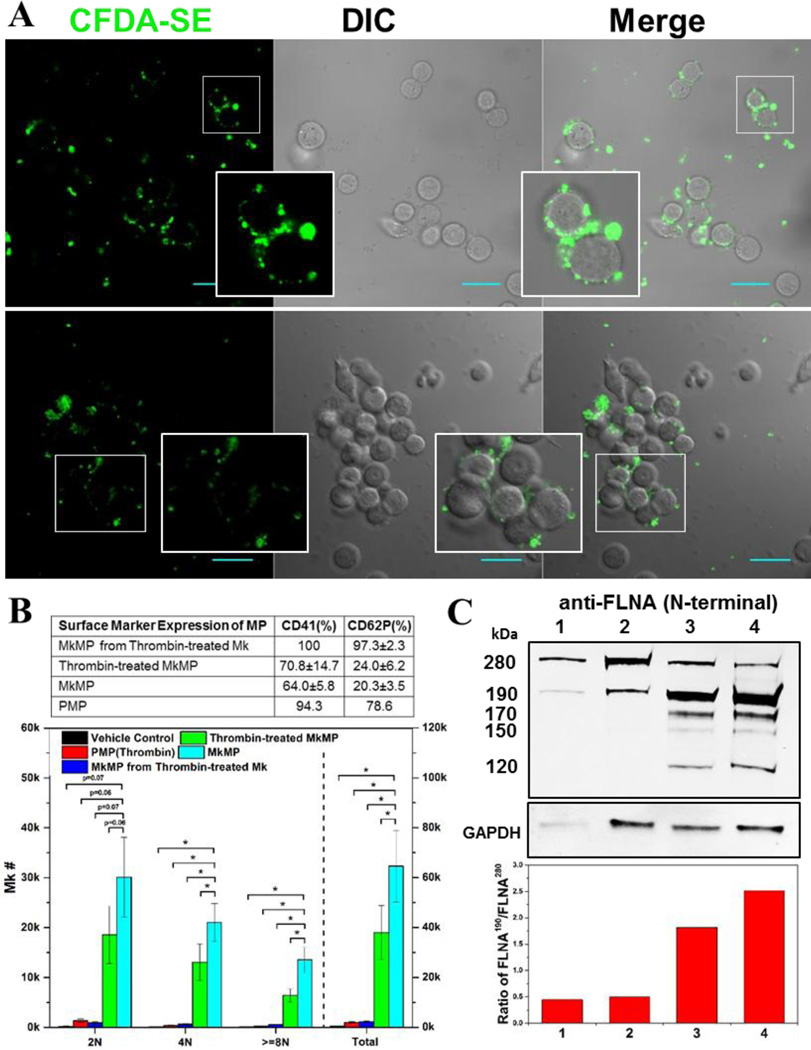
3.7. PMPs interact with and induce aggregation of HSPCs, but cannot apparently deliver cargo to HSPCs; thrombin-mediated platelet-like activation may account for the different effects of MkMPs vs PMPs
Since PMPs originate from activated platelets, and platelets derive from mature Mks, PMPs are ontologically similar to MkMPs. Both PMPs and MkMPs are CD41+CD42b+ but, unlike PMPs, MkMPs are CD62P− and LAMP-1− [7]. PMPs promote survival, proliferation, adhesion, chemotaxis and in vivo engraftment efficiency of HSPCs [10, 16]. In our previous study [9], we reported that PMPs were unable to promote Mk differentiation of HSPCs. Here, we examined if PMPs interact with HSPCs like MkMPs do, and what might explain the aforementioned differences of the two types of MPs. PMPs were stained with CFDA-SE dye and then cocultured with d3 HSPCs. We did not observe uptake of PMPs by HSPCs even after 24 hours of coculture (Fig. 7A), thus suggesting that PMPs cannot deliver, macroscopically at least, cargo to HSPCs. However, PMPs could bind to and induce aggregation of HSPCs (Fig. 7A); MkMPs did not induce HSPC aggregation. These data suggest that although both MkMPs and PMPs can recognize HSPCs, small differences, possibly related to platelet activation which gives rise to PMPs, are responsible for these profound differences. To test this hypothesis, we used thrombin treatment aiming to achieve a platelet-like activation of Mks prior to MkMP generation and also of MkMPs. Like PMPs, most (97.3%) of MkMPs from thrombin-activated Mks were CD62P+ while directly thrombin treatment on MkMPs only slightly increased the percentage of CD62P+ MkMPs from 20.3% to 24% (Fig. 7B). These findings were confirmed by Western analysis (Fig. 7C) of the characteristic activation-induced cleavage of filamin A [7, 41, 42].
Fig. 7. PMPs are not taken up by HSPCs and a thrombin-mediated platelet-like activation may account for the different effects of MkMPs vs PMPs.
(A) PMPs were stained with CFDA-SE (green) dye and cocultured with d3 HSPCs for 3–5 hours. Fluorescent and DIC images were collected using confocal microscopy. PMPs were not taken up by HSPCs, but induced HSPC aggregation. There was no aggregation observed in the control cultures of HSPCs without PMPs (data not shown). Inserts amplify images to show more details. Scale bar, 20 µm. (B) Thrombin treatment activates Mk cells giving rise to MkMPs incapable of inducing Mk differentiation HSPCs, but direct MkMPs activation by thrombin is ineffective and attenuates but does not abolish the impact of MkMPs. Table displays CD41 and CD62P expression levels of various MP types to assess the impact of thrombin activation. The effect of thrombin treatment on the biological effects of MkMPs and PMPs was assessed by coculture with CD34+ cells (10 MPs/cell) for 7 days. Ploidy distribution of Mks at d7 was analyzed by flow cytometry. Error bars: standard error of mean (n=3); * P<0.05. (C) Assessment of thrombin activation via Western-blot examination of filamin A (FLNA) truncation as demonstrated by the relative amount of truncated filamin A (190 kDa; full size is 280 kDa) in MkMPs (lane 1), thrombin-treated MkMPs (lane 2), MkMPs from thrombin-treated Mks (lane 3), and PMPs (lane 4). GAPDH expression was used as loading control. The low panel displays the ratio of FLNA190 /FLNA280 estimated by quantitation of normalized band intensity of displayed Western blot and this ratio represents the level of filamin A cleavage. The majority of filamin A in MkMPs and thrombin-treated MkMPs was full-length, while most filamin A was cleaved in MkMPs from thrombin-treated Mks and PMPs. The 190 kDa N-terminal fragment of filamin A results from calpain-dependent cleavage, while the 170, 150, 120 kDa N-terminal fragments are products of caspase-3-dependent cleavage.
Consistent with the CD62P and Western data, coculture of MkMPs from thrombin-treated Mks with CD34+ cells did not generate any Mks and thrombin treatment of MkMPs reduced the numbers of Mks at all ploidy classes in the coculture by 41% compared to coculture with untreated MkMPs (Fig. 7B). These results suggest that the inability of PMPs to target and induce Mk differentiation of HSPCs is caused by the changes, including changes in protein expression, brought about by thrombin activation.
3.8. HSPC uropods are the preferred area of interaction with MkMPs, with the uropod-enriched CD54 (ICAM-1), CD43, CD11b, and CD18 mediating binding and/or uptake of MkMPs
We next investigated what surface molecules might mediate binding to and/or uptake of MkMPs by HSPCs. We first examined the characteristic surface molecules of MkMPs, including CD41, CD42b and phosphatidylserine (PS). MkMPs were stained with CFDA-SE dye and CD41, CD42b or PS on MkMPs were blocked by either antibodies or annexin V before the coculture with HSPCs. Blocking did not change the CFDA-SE MFI of HSPCs (data not shown), suggesting that CD41, CD42b or PS are not involved in binding and/or uptake of MkMPs.
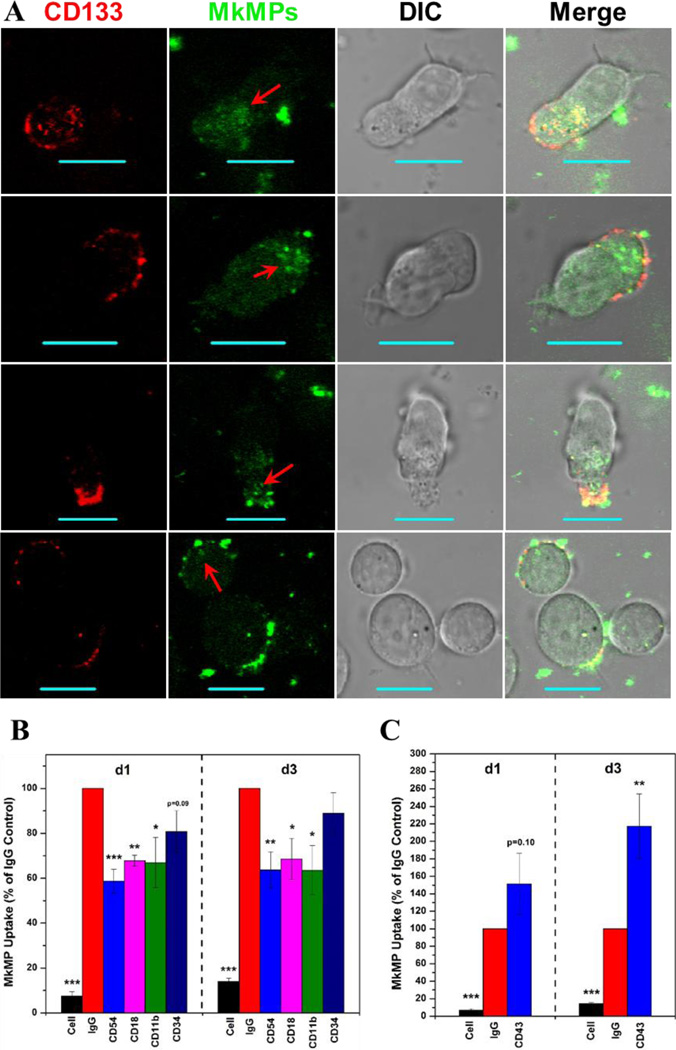
Examining images of cells interacting with MkMPs, we found several polarized HSPCs (e.g., three cells on the top row of Fig. 4A), forming uropod structures at their rear edge [43], and that MkMPs preferentially bound to uropods compared to the leading side of polarized HSPCs. To investigate this observation, we identified HSPC uropods using the surface marker CD133, which is concentrated on uropods of polarized HSPCs [43]. More than 67% of CD133+ cells (30% of total cells) had localized CD133 expression, including all polarized HSPCs with uropods, and significantly, MkMPs only bound to the plasma membrane regions of cells, which had localized CD133 expression (Fig. 8A). The cytoplasm near membrane regions with concentrated CD133 contained some intact MkMPs (apparently from endocytosis) and had higher overall green signal (Fig. 8A; red arrow). These observations suggest that proteins localized on HSPC uropods may mediate MkMP binding and uptake. To investigate this hypothesis, we used antibodies to block surface proteins enriched on HSPC uropod membranes: CD43 (leukosialin), CD44 (HCAM), CD50 (ICAM-3), CD54 (ICAM-1) and CD133 [43]. CD43, a transmembrane sialoglycoprotein expressed exclusively on hematopoietic cells [44] and a potential CD54 ligand, interacts with cytoskeletal proteins and is involved in signaling regulating cell activation, proliferation and survival [45]. CD44, CD50 and CD54 are glycoproteins with roles in cell-to-cell adhesion [46]. Blocking with either anti-CD44, anti-CD50 or anti-CD133 antibodies did not affect MkMP binding and/or uptake (data not shown). However, CD54 blocking significantly reduced CFDA-SE MFI of cells by >40% (Fig. 8B). Potential receptors for CD54 on the MkMPs are the leukocyte function-associated antigen 1 (LFA-1, CD11a/CD18), CD43 and macrophage-1 antigen (Mac-1, CD11b/CD18). The former two are expressed on Mks and platelets [47, 48], while Mac-1 is expressed at low level on Mks [49]. Mks and platelets also express CD54 [47], thus suggesting that binding of MkMPs to HSPCs could be also mediated by CD54 on MkMPs and its receptors LFA-1, Mac-1 and CD43 on CD34+ cells [50]. To further test this hypothesis, cells from d1 or d3 CD34+ cell culture were blocked with anti-CD11a, anti-CD11b, and anti-CD18. Blocking of CD18 and CD11b significantly decreased MkMP uptake (Fig. 8B), while blocking of CD11a showed no effect (data not shown). This suggests Mac-1 and CD54 might be the pair mediating the interaction between MkMP and HSCs/HSPCs. In contrast, CD43 blocking increased, instead of decreasing, CFDA-SE MFI of d3 HSPCs (Fig. 8C). Moreover, we found that HSPCs aggregate when treated with anti-CD43 antibody and this was confirmed by confocal microscopy (Fig. S3). This and other anti-CD43 antibodies have been shown to activate CD43 [51–55]. Thus, a possible reason for the observed increased binding of MkMPs to HSPCs and HSPC aggregation is that the anti-CD43 antibody activates instead of blocking CD43. This would suggest that CD43 plays also a role in the interaction between MkMPs and HSPCs. We also blocked CD34 (hematopoietic progenitor cell antigen), since it is the most common marker for HSC/HSPC. However, there was no significant impact of anti-CD34 antibody on MkMP uptake (Fig. 8B).
Fig. 8. CD54 (ICAM-1), CD18/CD11b (Mac-1) and CD43 mediate the uptake of MkMPs, with the HSPC uropods being the preferred interaction area.
(A) MkMPs were stained with CFDA-SE (green) dye and then cocultured with HSPCs for 3 hours. Anti-CD133 antibody (red) was added to the coculture before microscopic examination to identify the uropod area. Fluorescent and DIC images were collected using confocal microscopy. Internalized MkMPs in uropods are indicated by red arrows. The scale bar represents 10 µm. (B–C) d1 HSPCs and d3 HSPCs were blocked with 10 µg/mL anti-CD54, anti-CD 18, anti-CD 11b, anti-CD34 antibodies or isotype IgG for 30 min before cocultured with MkMPs stained with CFDA-SE. After 1 hour, cells were harvested for analysis of CFDA-SE MFI by flow cytometry. “Cell” represents samples without fluorescent MkMPs. The data in panels (B–C) represent averages of 3 to 6 biological replicates ± standard error of mean. *, P<0.05; **, P<0.01; ***, P<0.001
3.9. RNA transfer mediates the impact of MkMPs on HSPCs
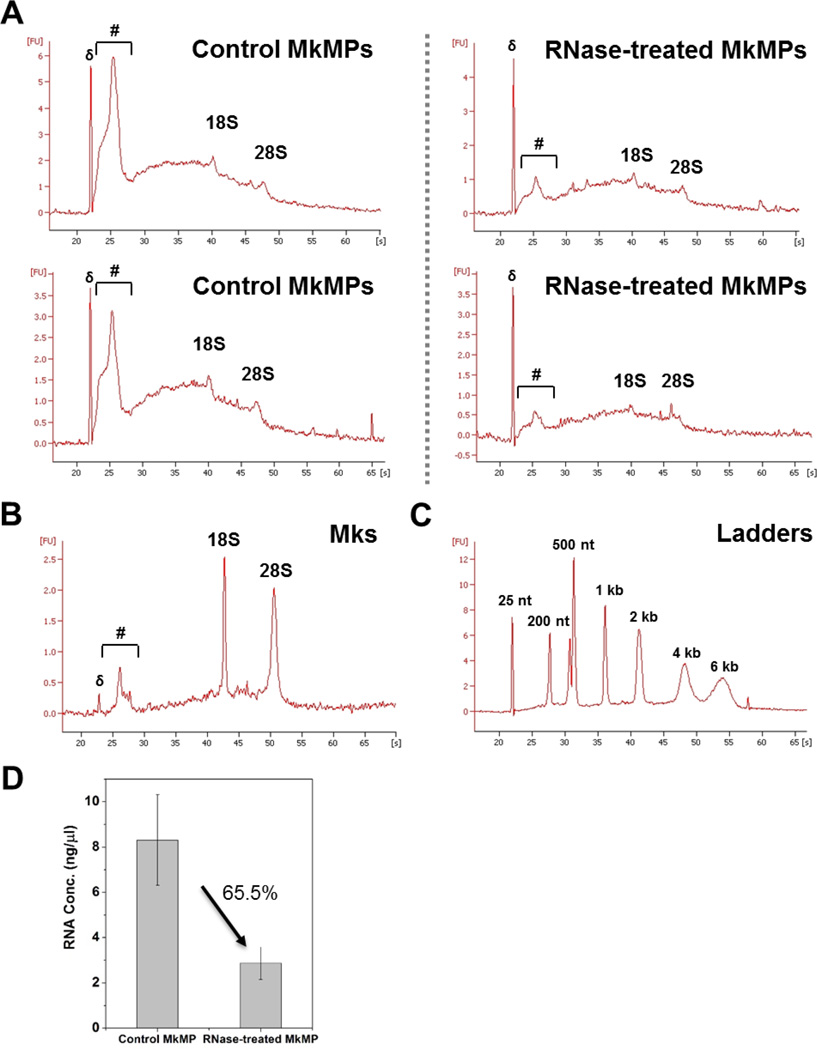
Using treatment with RNases, it has been reported that several MPs exert their biological function through RNA transfer [56–58]. Here, we used two RNases, RNase A/T1 and RNase ONE™, to examine the effect of RNase treatment of the RNA content of MkMPs and the ability of RNase treated MkMPs to induce Mk differentiation of HSPCs. First, we examined the impact of RNase treatment on the RNA content of MkMPs (Fig. 9). As shown in Fig. 9A, the RNA profiles of two biological replicate experiments before and after RNase treatment are remarkably reproducible. The RNA profiles of MkMPs (Fig. 9A), when compared to the RNA profile of Mks (Fig. 9B), show an enrichment in small RNAs and a low concentration of rRNAs; the RNA ladder of Fig. 9C enables the identification of rRNAs and the identification of the range of small RNAs, such as RNAs <200 nts. RNase treatment degrades RNAs of all sizes, but apparently more so of small RNAs, the range that would include miRNAs. RNase treatment degrades 65.5% of all RNA in MkMPs (Fig. 9D). These data suggest that RNases can enter these MkMPs to degrade their RNA content. While it is firmly established that RNase A, RNase 1 and likely most RNases are actively endocytosed by mammalian cells (so much so that RNase A has been termed a “cell-penetrating protein” [59, 60]), the mechanism by which RNases enter MPs like these MkMPs remains unexplored.
Fig. 9. RNA profile of Mk and MkMP with or without RNase treatment.
(A) MkMPs were first treated with or without RNase I for 2 hours at 37 °C. After incubated with RNase inhibitors and washed with IMDM thrice, total RNA were isolated from MkMPs and RNA profiles were analyzed by Bioanalyzer 2100. (B) RNA profile of d12 Mks. (A) presents the data from two biological replicates to demonstrate the exquisite reproducibility of the RNA profiles before and after RNase treatment. The symbol (#) in (A) and (B) marks the range of small RNAs. 18S and 28S represent rRNA, while (δ) in (A) and (B) marks the ladder of 25 nt. (C) RNA profile of ladders for size verification. (D) Total RNA concentration from MkMPs treated with or without were measured by Qubit RNA HS kit. The measurement shows 65.5% of RNA being digested after RNA treatment. The data represent two biological replicates ± standard deviation.
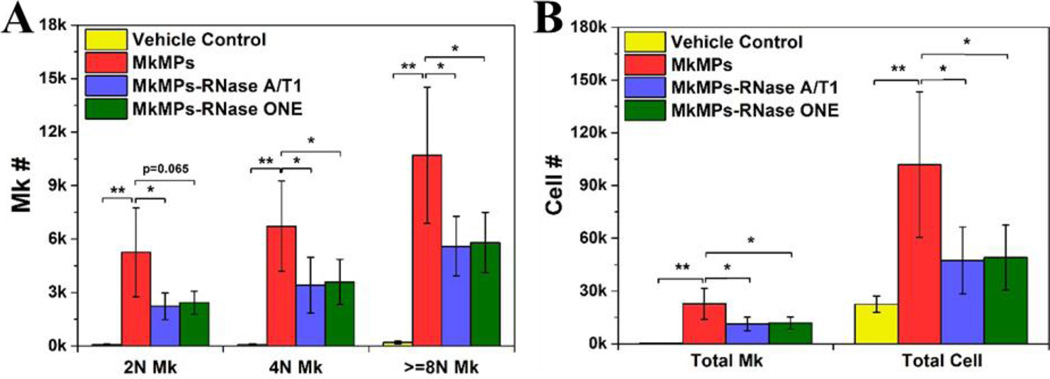
Next we examined the impact of MkMP RNase treatment on their ability to program HSPCs into megakaryopoiesis. Our data show that the number of 2N, 4N and >=8N Mks and of total Mks decreased by ~50% (Fig. 10), suggesting that RNase treatment attenuates but does not abolish the effect of MkMPs. This could be due to incomplete digestion of MkMP RNA by RNases, especially of RNase-resistant miRNAs, which have been proposed as important effector molecules in other MPs [61–63]. It is also possible that some proteins carried by MkMPs are involved in inducing Mk differentiation, as in the case of ESC MPs and tumor MPs whose biological functions are dependent on both protein and RNA transfer [56, 58].
Fig. 10. RNase treatment reduces the biological effect of MkMPs on HSPCs.
Human CD34+ cells were cocultured with MkMPs (10 MkMPs/cell) treated with RNase (Ambion® RNase A/T1 cocktail or RNase ONE™) for 8 days before harvested for ploidy analysis by flow cytometry. (A) Numbers of Mk cells with different ploidy classes (2N, 4N, >=8N). (B) Numbers of total Mks and total cells. The data are represented as the averages of four biological replicates ± standard error of mean. *, P<0.05; **, P<0.01.
4. Discussion
4.1. MkMPs target and induce the megakaryocytic differentiation of primitive Lin−CD34+ and all HSPCs but not so of MSCs, HUVECs or granulocytes
We demonstrated that MkMPs generated by mature Mks induces Mk differentiation of Lin−CD34+ primitive HSPCs. Combined with our previous report [9], these data show that MkMPs target all types of HSPCs. Targeting appears to be specific, as MkMPs could not transdifferentiate MSCs, HUVECs and granulocytes, three types of cells that are ontologically and physiologically related to HSPCs, and which are in contact with MkMPs in vivo. This could be either because these cells are not capable of taking up MkMPs or and the signaling molecules in MkMPs could not transdifferentiate these cells. It will be interesting to examine if MkMPs could induce embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs) into Mk differentiation given that both ESCs and iPSCs are capable of giving rise to all types of cells. It was also interesting that MkMPs appeared to promote better maintenance and expansion of the CD34+-cell compartment compared to control. These findings make a good argument for examining if ex-vivo generated MkMPs, the most abundance MPs in circulation, can be used in transfusion medicine to support enhanced thrombopoiesis and also if they can be used as a vehicle to deliver nucleic acids, proteins and organic molecules to HSPCs.
4.2. MkMPs are taken up by HSPCs via endocytosis and cell fusion, whereby macropinocytosis and lipid rafts play an important role
Three mechanisms have been suggested in the literature to explain how MPs and target cells interact: receptor-mediated binding, endocytosis, and membrane fusion [4, 17, 18]. The interaction always starts from MP binding to cells, which is apparently a receptor mediated process [15], and may largely account for the MP target specificity [64]. MP binding could be unstable, leading to dissociation of MPs from cell surface, or stable, ending in uptake of MPs by the cells [4], and signaling from temporary or persistent binding could be sufficient for altering cell fate [10–16, 65]. Through confocal microscopy, SEM and TEM analyses, we demonstrated that both direct fusion and endocytosis are involved in the uptake of MkMPs by HSPCs. Our SEM and TEM analyses provide the first direct observation of the membrane fusion process (as it proceeds in distinct stages), which engages microvillar-like membrane structures of the target cell (Fig. 4C, stage 3) during the fusion process. While previous studies have provided indirect evidence for a membrane fusion process, this is the first report of direct, detailed microscopic evidence of the process.
In the studies to interrogate specific mechanisms involved in MkMP uptake by HSPCs, the level of the inhibitory effect of MβCD and DMA on MkMP uptake was similar to that of the corresponding molecules that serve as positive controls for the specific mechanisms (Fig. 6B, 6C). This strengthens the conclusion that implicates macropinocytosis and the involvement of lipid rafts (through either lipid-raft dependent endocytosis or through the role of lipid rafts in the membrane-fusion process) in the uptake of MkMPs by HSPC. We also found that MkMP uptake was inhibited by dynasore. Dynasore inhibits dynamin, a GTPase that plays an important role in the formation of clathrin-dependent vesicles via fission [66]. Dynasore also reduces cholesterol in plasma membrane to disrupt lipid-raft formation and membrane ruffling, which is a dynamin-independent mechanism [67]. Our data from the effect of CH suggest that MkMP uptake is not related to clathrin-dependent endocytosis. This then leads to the hypothesis of the dynamin-independent inhibitory effects on MkMP uptake by dynasore [67]. Preta G et al demonstrated that dynasore can target processes dependent on the use of lipid rafts [67], while Park et al. showed that dynasore can inhibit membrane ruffling, which is the essential process of macropinocytosis [68]. Taken together, these data support our hypothesis that MkMP uptake is associated with macropinocytosis and lipid raft-mediated endocytosis.
4.3. MkMP RNA is in part at least responsible for their biological effects
We showed here that the ontologically similar PMPs cannot deliver cargo to or induce Mk differentiation of HSPCs, despite the fact that they express a similar set of receptors. This would argue against the possibility that the impact of MkMPs is purely receptor mediated. Furthermore, after we treated MkMPs with two different RNases to digest about 65% of the RNA carried by MkMPs, the numbers of Mks with different ploidy levels decreased by half in the coculture with treated MkMPs compared to MkMPs without treatment. This demonstrates that horizontal transfer of RNA is in part at least responsible for the observed biological effect of MkMPs. Transfer of RNA requires uptake of MkMPs by cells following stable binding and thus the RNase treatment results provide additional evidence that MkMPs are taken up by cells.
Sequencing of the MkMP, Mk-cell, platelet and PMP RNomes shows distinct differences, and notably the enrichment of the MkMP RNA in small non-coding RNAs (miRNA, piRNAs, and long-non-coding RNAs (lncRNAs)) (data not shown), but the information cannot yet be interpreted to explain the impact of MkMPs on HSPCs. Some of these differences are also shown in the data of Fig. 9, where the remarkably reproducible RNA profile is very distinct from the RNA profile of the parent Mk cells. While it is possible that molecules other than RNAs are responsible for the observed effects, the observed 50% reduction in the biological effect of MkMPs by the RNase treatment could be due to the inability to completely digest the MkMP RNA (as shown in Fig. 9), thus leaving as a distinct possibility that the impact of MkMP on HSPCs is solely RNA mediated.
4.4. A role for HSPC uropods, CD54 and CD43, and why PMPs cannot act on HSPCs like MkMPs do
We have shown that MkMPs interact with HSPCs largely through their uropods, which are enriched in CD54 and CD43 surface proteins. We showed that blocking CD54, CD11b, and CD18 reduces uptake of MkMPs to HSPCs by ca. 30–40%, and also that the use of an antibody that stimulates CD43 expression promotes MkMP uptake. Since CD54 is a ligand of Mac-1 (CD11b/CD18), this would suggest an important role of pairing MkMP with HSPCs. Although blocking CD43 increased MkMP uptake, since CD54 is also a ligand for CD43 on HSPCs, this still suggests a role for CD43 in mediating the interaction between MkMP with HSPCs. Our data involving thrombin induced platelet-like activation of Mks prior to MkMP generation, which abolishes their biological effect on HSPCs, suggest that the inability of PMPs to target and induce Mk differentiation of HSPCs is caused by the thrombin activation.
5. Conclusions
We have demonstrated that MkMPs are taken up by HSPC via endocytosis and membrane fusion. Specifically, we showed that macropinocytosis and lipid raft-mediated endocytosis or fusion play an important role in MkMP uptake. Several surface proteins, CD54, CD11b, CD18 and CD43 were identified as mediating the interaction between MkMP and HSPCs. Additionally, we have provided data that suggest that thrombin activation may account for the inability of HSPCs to take up PMPs. Finally, using RNase treatment, we demonstrated that RNA is an important MkMP cargo mediating the biological effect of MkMPs on HSPCs. These finding pave the way for exploring potential applications of MkMP in cell therapies to treat thrombocytopenias but also for delivering nucleic acids, proteins and drugs to HSPCs for applications in gene therapies and targeted molecular therapy.
Supplementary Material
Acknowledgments
We thank Jeffrey Caplan and members of Bioimaging Center (University of Delaware; UD) for assistance with immunofluorescence and electron microscopy, and Dr. Dongjun Li (UD) and Blood Bank of Delmarva for assistance with platelet collection.
Funding: This work was supported by National Institutes of Health grant [R21HL106397]; State of Delaware CAT grant [#15A01570], and, for supporting the DLS analysis, a grant from the National Institute of General Medical Sciences – NIGMS (5 P30 GM110758-02) from the National Institutes of Health.
Abbreviations
- HSC(s)
hematopoietic stem cell(s)
- HSPC(s)
hematopoietic stem and progenitor cell(s)
- HUVECs
human umbilical vascular endothelial cells
- LFA-1
leukocyte function-associated antigen 1
- Mac-1
macrophage-1 antigen
- MFI
mean fluorescence intensity
- Mk(s)
megakaryocyte
- MkExo(s)
megakaryocytic exosome(s)
- MkMP(s)
megakaryocytic microparticle
- MP(s)
microparticle
- MSCs
mesenchymal stem cells
- PMP(s)
platelet-derived microparticles
- PS
phosphatidylserine
- SEM
scanning electron microscopy
- TEM
transmission electron microscopy
- TPO
thrombopoietin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions.
E.T.P. and J.J. designed the study and analyzed the data; J.J. carried out most of the experiments. CY.K. conducted several coculture experiments, some characterization experiments on MkMPs and MkExos, the endocytosis inhibitor experiments and some Ab blocking experiments and provided data for Figs 1, 6, 7 and 8.; E.T.P., J.J. and CY.K. wrote the manuscript.
Conflict-of-interest. The authors declare that they have no conflicts of interest with the contents of this article. A PCT patent application that includes some of the data reported in this article is pending.
References
- 1.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends in cell biology. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 2.Barteneva NS, Fasler-Kan E, Bernimoulin M, Stern JN, Ponomarev ED, Duckett L, Vorobjev IA. Circulating microparticles: square the circle. BMC cell biology. 2013;14:23. doi: 10.1186/1471-2121-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratajczak MZ, Kucia M, Jadczyk T, Greco NJ, Wojakowski W, Tendera M, Ratajczak J. Pivotal role of paracrine effects in stem cell therapies in regenerative medicine: can we translate stem cell-secreted paracrine factors and microvesicles into better therapeutic strategies? Leukemia. 2012;26:1166–1173. doi: 10.1038/leu.2011.389. [DOI] [PubMed] [Google Scholar]
- 4.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. The Journal of cell biology. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gyorgy B, Szabo TG, Pasztoi M, Pal Z, Misjak P, Aradi B, Laszlo V, Pallinger E, Pap E, Kittel A, Nagy G, Falus A, Buzas EI. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aatonen M, Gronholm M, Siljander PR. Platelet-derived microvesicles: multitalented participants in intercellular communication. Semin Thromb Hemost. 2012;38:102–113. doi: 10.1055/s-0031-1300956. [DOI] [PubMed] [Google Scholar]
- 7.Flaumenhaft R, Dilks JR, Richardson J, Alden E, Patel-Hett SR, Battinelli E, Klement GL, Sola-Visner M, Italiano JE., Jr Megakaryocyte-derived microparticles: direct visualization and distinction from platelet-derived microparticles. Blood. 2009;113:1112–1121. doi: 10.1182/blood-2008-06-163832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamouard P, Desprez D, Hugel B, Kunzelmann C, Gidon-Jeangirard C, Lessard M, Baumann R, Freyssinet J-M, Grunebaum L. Circulating Cell-Derived Microparticles in Crohn’s Disease. Dig Dis Sci. 2005;50:574–580. doi: 10.1007/s10620-005-2477-0. [DOI] [PubMed] [Google Scholar]
- 9.Jiang J, Woulfe DS, Papoutsakis ET. Shear enhances thrombopoiesis and formation of microparticles that induce megakaryocytic differentiation of stem cells. Blood. 2014;124:2094–2103. doi: 10.1182/blood-2014-01-547927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baj-Krzyworzeka M, Majka M, Pratico D, Ratajczak J, Vilaire G, Kijowski J, Reca R, Janowska-Wieczorek A, Ratajczak MZ. Platelet-derived microparticles stimulate proliferation, survival, adhesion, and chemotaxis of hematopoietic cells. Exp Hematol. 2002;30:450–459. doi: 10.1016/s0301-472x(02)00791-9. [DOI] [PubMed] [Google Scholar]
- 11.Pluskota E, Woody NM, Szpak D, Ballantyne CM, Soloviev DA, Simon DI, Plow EF. Expression, activation, and function of integrin alpha(M)beta(2) (Mac-1) on neutrophil-derived microparticles. Blood. 2008;112:2327–2335. doi: 10.1182/blood-2007-12-127183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinez MC, Larbret F, Zobairi F, Coulombe J, Debili N, Vainchenker W, Ruat M, Freyssinet JM. Transfer of differentiation signal by membrane microvesicles harboring hedgehog morphogens. Blood. 2006;108:3012–3020. doi: 10.1182/blood-2006-04-019109. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Lorenzo MJ, Anel A, Gamen S, Monle n I, Lasierra P, Larrad L, Pineiro A, Alava MA, Naval J. Activated human T cells release bioactive Fas ligand and APO2 ligand in microvesicles. Journal of immunology. 1999;163:1274–1281. [PubMed] [Google Scholar]
- 14.Ansa-Addo EA, Lange S, Stratton D, Antwi-Baffour S, Cestari I, Ramirez MI, McCrossan MV, Inal JM. Human Plasma Membrane-Derived Vesicles Halt Proliferation and Induce Differentiation of THP-1 Acute Monocytic Leukemia Cells. J Immunol. 2010;185:5236–5246. doi: 10.4049/jimmunol.1001656. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh A, Li W, Febbraio M, Espinola RG, McCrae KR, Cockrell E, Silverstein RL. Platelet CD36 mediates interactions with endothelial cell-derived microparticles and contributes to thrombosis in mice. The Journal of clinical investigation. 2008;118:1934–1943. doi: 10.1172/JCI34904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janowska-Wieczorek A, Majka M, Kijowski J, Baj-Krzyworzeka M, Reca R, Turner AR, Ratajczak J, Emerson SG, Kowalska MA, Ratajczak MZ. Platelet-derived microparticles bind to hematopoietic stem/progenitor cells and enhance their engraftment. Blood. 2001;98:3143–3149. doi: 10.1182/blood.v98.10.3143. [DOI] [PubMed] [Google Scholar]
- 17.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. 2014 doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 18.Mause SF, Weber C. Microparticles: protagonists of a novel communication network for intercellular information exchange. Circ Res. 2010;107:1047–1057. doi: 10.1161/CIRCRESAHA.110.226456. [DOI] [PubMed] [Google Scholar]
- 19.Faille D, El-Assaad F, Mitchell AJ, Alessi MC, Chimini G, Fusai T, Grau GE, Combes V. Endocytosis and intracellular processing of platelet microparticles by brain endothelial cells. Journal of cellular and molecular medicine. 2012;16:1731–1738. doi: 10.1111/j.1582-4934.2011.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obregon C, Rothen-Rutishauser B, Gitahi SK, Gehr P, Nicod LP. Exovesicles from human activated dendritic cells, fuse with resting dendritic cells allowing them to present alloantigens. The American journal of pathology. 2006;169:2127–2136. doi: 10.2353/ajpath.2006.060453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, Farber DB. Transfer of microRNAs by embryonic stem cell microvesicles. PloS one. 2009;4:e4722. doi: 10.1371/journal.pone.0004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Conde I, Shrimpton CN, Thiagarajan P, Lopez JA. Tissue-factor-bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood. 2005;106:1604–1611. doi: 10.1182/blood-2004-03-1095. [DOI] [PubMed] [Google Scholar]
- 23.Mause SF, Ritzel E, Liehn EA, Hristov M, Bidzhekov K, Muller-Newen G, Soehnlein O, Weber C. Platelet microparticles enhance the vasoregenerative potential of angiogenic early outgrowth cells after vascular injury. Circulation. 2010;122:495–506. doi: 10.1161/CIRCULATIONAHA.109.909473. [DOI] [PubMed] [Google Scholar]
- 24.Panuganti S, Schlinker AC, Lindholm PF, Papoutsakis ET, Miller WM. Three-stage ex vivo expansion of high-ploidy megakaryocytic cells: toward large-scale platelet production. Tissue Eng Part A. 2013;19:998–1014. doi: 10.1089/ten.tea.2011.0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. Journal of extracellular vesicles. 2015;4:27031. doi: 10.3402/jev.v4.27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys Acta. 2012;1820:940–948. doi: 10.1016/j.bbagen.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Hevehan DL, Papoutsakis ET, Miller WM. Physiologically significant effects of pH and oxygen tension on granulopoiesis. Exp Hematol. 2000;28:267–275. doi: 10.1016/s0301-472x(99)00150-2. [DOI] [PubMed] [Google Scholar]
- 28.Lindsey S, Papoutsakis ET. The aryl hydrocarbon receptor (AHR) transcription factor regulates megakaryocytic polyploidization. British journal of haematology. 2011;152:469–484. doi: 10.1111/j.1365-2141.2010.08548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713–17724. doi: 10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51. doi: 10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalli J, Montero-Melendez T, Norling LV, Yin X, Hinds C, Haskard D, Mayr M, Perretti M. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Mol Cell Proteomics. 2013;12:2205–2219. doi: 10.1074/mcp.M113.028589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boilard E, Duchez AC, Brisson A. The diversity of platelet microparticles. Curr. Opin. Hematol. 2015;22:437–444. doi: 10.1097/MOH.0000000000000166. [DOI] [PubMed] [Google Scholar]
- 33.Bjornson CRR, Rietze RL, Reynolds BA, Magli MC, Vescovi AL. Turning brain into blood: A hematopoietic fate adopted by adult neural stem cells in vivo. Science. 1999;283:534–537. doi: 10.1126/science.283.5401.534. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Ramos J, Song S, Cardozo-Pelaez F, Hazzi C, Stedeford T, Willing A, Freeman TB, Saporta S, Janssen W, Patel N, Cooper DR, Sanberg PR. Adult bone marrow stromal cells differentiate into neural cells in vitro. Exp Neurol. 2000;164:247–256. doi: 10.1006/exnr.2000.7389. [DOI] [PubMed] [Google Scholar]
- 35.Zhang S, Wang DC, Estrov Z, Raj S, Willerson JT, Yeh ET. Both cell fusion and transdifferentiation account for the transformation of human peripheral blood CD34-Positive cells into cardiomyocytes in vivo. Circulation. 2004;110:138–138. doi: 10.1161/01.CIR.0000150796.18473.8E. [DOI] [PubMed] [Google Scholar]
- 36.Frid MG, Kale VA, Stenmark KR. Mature vascular endothelium can give rise to smooth muscle cells via endothelial-mesenchymal transdifferentiation - In vitro analysis. Circ Res. 2002;90:1189–1196. doi: 10.1161/01.res.0000021432.70309.28. [DOI] [PubMed] [Google Scholar]
- 37.Badorff CM, Brandes RP, Popp R, Rupp S, Urbich C, Aicher A, Fleming I, Busse R, Zeiher AM, Dimmeler S. Transdifferentiation of blood-derived human adult endothelial progenitor cells into functionally active cardiomyocytes. Circulation. 2002;106:138–139. doi: 10.1161/01.cir.0000051460.85800.bb. [DOI] [PubMed] [Google Scholar]
- 38.Aliotta JM, Pereira M, Johnson KW, de Paz N, Dooner MS, Puente N, Ayala C, Brilliant K, Berz D, Lee D, Ramratnam B, McMillan PN, Hixson DC, Josic D, Quesenberry PJ. Microvesicle entry into marrow cells mediates tissue-specific changes in mRNA by direct delivery of mRNA and induction of transcription. Exp Hematol. 2010;38:233–245. doi: 10.1016/j.exphem.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fonseca AV, Freund D, Bornhauser M, Corbeil D. Polarization and Migration of Hematopoietic Stem and Progenitor Cells Rely on the RhoA/ROCK I Pathway and an Active Reorganization of the Microtubule Network. Journal of Biological Chemistry. 2010;285:31661–31671. doi: 10.1074/jbc.M110.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salaun C, James DJ, Chamberlain LH. Lipid rafts and the regulation of exocytosis. Traffic. 2004;5:255–264. doi: 10.1111/j.1600-0854.2004.0162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gorlin JB, Yamin R, Egan S, Stewart M, Stossel TP, Kwiatkowski DJ, Hartwig JH. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. The Journal of cell biology. 1990;111:1089–1105. doi: 10.1083/jcb.111.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Umeda T, Kouchi Z, Kawahara H, Tomioka S, Sasagawa N, Maeda T, Sorimachi H, Ishiura S, Suzuki K. Limited proteolysis of filamin is catalyzed by caspase-3 in U937 and Jurkat cells. J Biochem. 2001;130:535–542. doi: 10.1093/oxfordjournals.jbchem.a003016. [DOI] [PubMed] [Google Scholar]
- 43.Giebel B, Corbeil D, Beckmann J, Hohn J, Freund D, Giesen K, Fischer J, Kogler G, Wernet P. Segregation of lipid raft markers including CD133 in polarized human hematopoietic stem and progenitor cells. Blood. 2004;104:2332–2338. doi: 10.1182/blood-2004-02-0511. [DOI] [PubMed] [Google Scholar]
- 44.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ostberg JR, Barth RK, Frelinger JG. The Roman god Janus: a paradigm for the function of CD43. Immunol. Today. 1998;19:546–550. doi: 10.1016/s0167-5699(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 46.Murray P, Frampton G, Nelson PN. Cell adhesion molecules. Sticky moments in the clinic. BMJ. 1999;319:332–334. doi: 10.1136/bmj.319.7206.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagiwara T, Nagasawa T, Nagahisa H, Takizawa M, Osada M, Abe T. Expression of adhesion molecules on cytoplasmic processes of human megakaryocytes. Exp Hematol. 1996;24:690–695. [PubMed] [Google Scholar]
- 48.Remold-O’Donnell E, Zimmerman C, Kenney D, Rosen F, Expression on blood cells of sialophorin. the surface glycoprotein that is defective in Wiskott-Aldrich syndrome. Blood. 1987;70:104–109. [PubMed] [Google Scholar]
- 49.Nagata Y, Nagahisa H, Aida Y, Okutomi K, Nagasawa T, Todokoro K. Thrombopoietin induces megakaryocyte differentiation in hematopoietic progenitor FDC-P2 cells. J Biol Chem. 1995;270:19673–19675. doi: 10.1074/jbc.270.34.19673. [DOI] [PubMed] [Google Scholar]
- 50.Gunji Y, Nakamura M, Hagiwara T, Hayakawa K, Matsushita H, Osawa H, Nagayoshi K, Nakauchi H, Yanagisawa M, Miura Y, et al. Expression and function of adhesion molecules on human hematopoietic stem cells: CD34+ LFA-1- cells are more primitive than CD34+ LFA-1+ cells. Blood. 1992;80:429–436. [PubMed] [Google Scholar]
- 51.Cho JY, Chain BM, Vives J, Horejsi V, Katz DR. Regulation of CD43-induced U937 homotypic aggregation. Exp Cell Res. 2003;290:155–167. doi: 10.1016/s0014-4827(03)00322-7. [DOI] [PubMed] [Google Scholar]
- 52.Bazil V, Brandt J, Chen S, Roeding M, Luens K, Tsukamoto A, Hoffman R. A monoclonal antibody recognizing CD43 (leukosialin) initiates apoptosis of human hematopoietic progenitor cells but not stem cells. Blood. 1996;87:1272–1281. [PubMed] [Google Scholar]
- 53.Serrador JM, Nieto M, Alonso-Lebrero JL, del Pozo MA, Calvo J, Furthmayr H, Schwartz-Albiez R, Lozano F, Gonzalez-Amaro R, Sanchez-Mateos P, Sanchez-Madrid F. CD43 interacts with moesin and ezrin and regulates its redistribution to the uropods of T lymphocytes at the cell-cell contacts. Blood. 1998;91:4632–4644. [PubMed] [Google Scholar]
- 54.Anzai N, Gotoh A, Shibayama H, Broxmeyer HE. Modulation of integrin function in hematopoietic progenitor cells by CD43 engagement: possible involvement of protein tyrosine kinase and phospholipase C-gamma. Blood. 1999;93:3317–3326. [PubMed] [Google Scholar]
- 55.Seveau S, Lopez S, Lesavre P, Guichard J, Cramer EM, Halbwachs-Mecarelli L. Leukosialin (CD43, sialophorin) redistribution in uropods of polarized neutrophils is induced by CD43 cross-linking by antibodies, by colchicine or by chemotactic peptides. Journal of cell science. 1997;110(Pt 13):1465–1475. doi: 10.1242/jcs.110.13.1465. [DOI] [PubMed] [Google Scholar]
- 56.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, Ratajczak MZ. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 57.Bruno S, Grange C, Deregibus MC, Calogero RA, Saviozzi S, Collino F, Morando L, Busca A, Falda M, Bussolati B, Tetta C, Camussi G. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. Journal of the American Society of Nephrology : JASN. 2009;20:1053–1067. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chao TY, Raines RT. Mechanism of Ribonuclease A Endocytosis: Analogies to Cell-Penetrating Peptides. Biochemistry. 2011;50:8374–8382. doi: 10.1021/bi2009079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eller CH, Lomax JE, Raines RT. Bovine Brain Ribonuclease Is the Functional Homolog of Human Ribonuclease 1. Journal of Biological Chemistry. 2014;289:25996–26006. doi: 10.1074/jbc.M114.566166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K, Shah P, Wisler J, Eubank TD, Tridandapani S, Paulaitis ME, Piper MG, Marsh CB. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood. 2013;121:984–995. doi: 10.1182/blood-2011-08-374793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Njock MS, Cheng HS, Dang LT, Nazari-Jahantigh M, Lau AC, Boudreau E, Roufaiel M, Cybulsky MI, Schober A, Fish JE. Endothelial cells suppress monocyte activation through secretion of extracellular vesicles containing antiinflammatory microRNAs. Blood. 2015;125:3202–3212. doi: 10.1182/blood-2014-11-611046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smallwood DT, Apollonio B, Willimott S, Lezina L, Alharthi A, Ambrose AR, De Rossi G, Ramsay AG, Wagner SD. Extracellular vesicles released by CD40/IL-4-stimulated CLL cells confer altered functional properties to CD4(+) T cells. Blood. 2016;128:542–552. doi: 10.1182/blood-2015-11-682377. [DOI] [PubMed] [Google Scholar]
- 64.Losche W, Scholz T, Temmler U, Oberle V, Claus RA. Platelet-derived microvesicles transfer tissue factor to monocytes but not to neutrophils. Platelets. 2004;15:109–115. doi: 10.1080/09537100310001649885. [DOI] [PubMed] [Google Scholar]
- 65.Mause SF, von Hundelshausen P, Zernecke A, Koenen RR, Weber C. Platelet microparticles: a transcellular delivery system for RANTES promoting monocyte recruitment on endothelium. Arterioscler Thromb Vasc Biol. 2005;25:1512–1518. doi: 10.1161/01.ATV.0000170133.43608.37. [DOI] [PubMed] [Google Scholar]
- 66.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preta G, Lotti V, Cronin JG, Sheldon IM. Protective role of the dynamin inhibitor Dynasore against the cholesterol-dependent cytolysin of Trueperella pyogenes. FASEB J. 2015;29:1516–1528. doi: 10.1096/fj.14-265207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park RJ, Shen H, Liu L, Liu X, Ferguson SM, De Camilli P. Dynamin triple knockout cells reveal off target effects of commonly used dynamin inhibitors. J Cell Sci. 2013;126:5305–5312. doi: 10.1242/jcs.138578. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.