Abstract
After their disappearance from the human population in 1968, influenza H2 viruses have continued to circulate in the natural avian reservoir. The isolation of this virus subtype from multiple bird species as well as swine highlights the need to better understand the potential of these viruses to spread and cause disease in humans. Here we analyzed the virulence, transmissibility and receptor-binding preference of two avian influenza H2 viruses (H2N2 and H2N3) and compared them to a swine H2N3 (A/swine/Missouri/2124514/2006 [swMO]), and a human H2N2 (A/England/10/1967 [Eng/67]) virus using the ferret model as a mammalian host. Both avian H2 viruses possessed the capacity to spread efficiently between cohoused ferrets, and the swine (swMO) and human (Eng/67) viruses transmitted to naïve ferrets by respiratory droplets. Further characterization of the swMO hemagglutinin (HA) by x-ray crystallography and glycan microarray array identified receptor-specific adaptive mutations. As influenza virus quasispecies dynamics during transmission have not been well characterized, we sequenced nasal washes collected during transmission studies to better understand experimental adaptation of H2 HA. The avian H2 viruses isolated from ferret nasal washes contained mutations in the HA1, including a Gln226Leu substitution, which is a mutation associated with α2,6 sialic acid (human-like) binding preference. These results suggest that the molecular structure of HA in viruses of the H2 subtype continue to have the potential to adapt to a mammalian host and become transmissible, after acquiring additional genetic markers.
Keywords: Influenza, Ferret, Transmission, Hemagglutinin
Introduction
The 1957 Asian influenza pandemic was caused by an influenza virus of the H2N2 subtype. H2N2 viruses established a stable presence in humans for 11 years and caused approximately 2 million deaths worldwide before they disappeared from human circulation following the emergence of the 1968 pandemic H3N2 virus (Simonsen et al., 1998). The reintroduction of the H2 subtype into the human population would pose a significant global health threat in a population where the majority of humans now lack anti-H2 hemagglutinin (HA) serologic immunity (Pyhala 1985; Webster 1997; Nabel et al., 2011). Avian influenza virus surveillance provides evidence for the continuing circulation of H2 viruses in wild migratory birds and poultry in live bird markets (Schafer et al., 1993; Piaggio et al., 2012; Chen et al., 2010). In some cases, contemporary H2 viruses have been found to be antigenically related to the early pandemic human viruses (Schafer et al., 1993; Piaggio et al., 2012; Chen et al., 2010). Moreover, currently circulating avian H2 virus reassortants containing gene segments from both Eurasian and North American lineages have been found in North American wild birds, indicating that genetic reassortment is occurring for this subtype and novel H2 viruses are being generated (Piaggio et al., 2012; Makarova et al., 1999; Liu et al., 2004; Kishida et al., 2008; Ferro et al., 2010; Ramey et al., 2010).
Zoonotic influenza viruses are constantly emerging from avian (H9N2, H5N1, H7N9) and swine species (H1N1, H1N2, H3N2) to sporadically infect humans, mainly through exposure to infected animals or contaminated environments (http://www.who.int/en/). Individuals exposed to avian influenza viruses may present a range of clinical manifestations from asymptomatic to more serious complications of the respiratory tract that can even lead to death (World Health Organization, 2013). Although H5 and H7 subtype viruses have received the greatest attention as potential pandemic threats, the concept of pandemic recycling in humans has been proposed and validated by the reemergence of swine origin H1N1 pandemic virus in 2009 (Dowdle, 2006). Should an avian influenza H2 virus emerge that possesses the ability to infect swine productively, the pandemic potential of such a virus would be substantial. In 2006, H2N3 subtype influenza viruses were isolated from pigs in two swine farms in the U.S. (Ma et al., 2007). These viruses were characterized as containing 6 internal genes (PB2, PB1, PA, NP, M and NS) derived from contemporary North American swine triple reassortant viruses and the remaining genes (HA and NA) derived from avian H2 influenza viruses. The HA of both H2N3 viruses were found to be antigenically related to the currently circulating avian influenza viruses (Ma et al., 2007; Killian et al., 2011). Experimentally, these viruses were pathogenic in mice and shown to transmit efficiently in ferrets and pigs in direct contact models (Ma et al., 2007). The swine H2N3 virus was also reported to cause severe pneumonia with elevated plasma cytokine responses compared to human H2N2 virus in nonhuman primates (Richt et al., 2012). In addition, the novel H2N3 virus contained a receptor binding change (Leu at position 226; H3 numbering) associated with both increased binding of H2 HA to human-like α2–6 sialic acid (SA) receptors and enhanced respiratory droplet transmissibility in ferrets (Pappas et al., 2010; Viswanathan et al., 2010; Xu et al., 2010). Previous studies of 1957 H2N2 HA suggested that both a hydrophobic switch from Gln to Leu at 226 and a slight elongation (~0.5 Å) of the receptor-binding site (RBS) were critical for avian H2 HA to acquire human receptor specificity (Xu et al., 2010; Liu et al., 2009a).
The continuous virologic surveillance of avian and swine species in the wild, in farms and in live bird markets has been critical for detection of newly emerging influenza viruses and for assessment of potential public health risks (World Health Organization, 2013). In addition to the genetic and antigenic characteristics and antiviral susceptibility properties of the viruses, assessments of emergent influenza virus properties such as molecular markers that contribute to receptor binding specificity, pathogenicity and transmissibility have provided substantial information related to the overall risks that viruses may pose for the human population (Pappas et al., 2010; Maines et al., 2011; Pearce et al., 2012; Belser et al., 2013). In a recent report, Jones et al. characterized a number of historic avian H2N2 viruses in two mammalian models and concluded that these viruses constitute a potential risk for human infection (Jones et al., 2014).
The molecular markers associated with increased adaptation and transmission of H2 viruses are not well understood. Previous results reveal a good correlation between binding affinity to α2,6 SA (human-like) glycan receptors and efficient respiratory droplet transmission for H2N2 viruses (Pappas et al., 2010). During a respiratory droplet transmission experiment in ferrets there was an apparent selection of a virus variant with a receptor binding site (Gln226Leu) mutation in the HA, which rendered the virus transmissible (Pappas et al., 2010). This data shows that these viruses have the capability to adapt in the mammalian host and become transmissible when favorable genetic factors are present. The structural analyses of these viruses’ HAs suggest that key amino acids in both human and avian viruses provide a framework that allows facile switching of receptor binding preferences (Pappas et al., 2010). Since avian H2 viral HAs are structurally similar to the human pandemic H2 viruses, adaptation of these viruses to the human host is plausible and of public health concern. Here we evaluate the relative virulence and transmissibility of genetically distinct H2 viruses in the ferret model. The present work also evaluated their genetic potentials for HA adaptation to the mammalian host.
Results
Genetic and phenotypic characteristics of the H2 viruses used in this study
For this study we chose viruses that represent each of the three major phylogenetic lineages of the H2 subtype (Liu et al., 2009b). The HA protein sequences of both mallMD (H2N3) and swMO (H2N3) viruses belong to the Western Hemisphere (North American) sub-lineage. The avian ckPA (H2N2) HA sequences are closely related to viruses from the Eastern Hemisphere (Eurasian) sub-lineage and Eng67 (H2N2) virus belongs to the human pandemic sub-lineage, which is most closely related to Eastern Hemisphere viruses. Genetic analysis of the entire genome of all four viruses used in this study shows that they are genetically distinct due to a variety of factors such as host origin, isolation year and reassortment. The percentage of amino acid identity between swMO virus and the other three H2 viruses ranges from 80% (NS1, non-structural protein) to 99% (PA, polymerase acidic) (Supplementary Table 1).
The amino acid makeup of the four H2 viruses at key positions associated with virulence and transmission in mammalian hosts is shown in Table 1. The human Eng67 (H2N2) virus contains the Leu226/Ser228 constellation in the HA RBS corresponding to an expected binding preference to α2–6 SA (human-like) glycan receptors. This was experimentally confirmed in hemagglutination assays using resialylated turkey red blood cells containing α2–6 SA (Table 1). Eng67 also contains Lys627 in the PB2 gene which has been shown to be important for respiratory droplet transmission (Pappas et al., 2010; Van Hoeven et al., 2009). In contrast, both avian H2 (ckPA and mallMD) viruses possess the Gln226/Gly228 constellation in the HA RBS and Glu627 in the PB2 gene. Both are typical markers of avian viruses that confer α2–3 SA receptor binding and poor transmissibility via aerosolized droplets in mammalian animal models, respectively (Pappas et al., 2010; Viswanathan et al., 2010; Maines et al., 2006; Glaser et al., 2006). In comparison to the avian H2 HAs, the swine (swMO) virus possesses a mutation, Gln226Leu, which correlates with increased binding affinity to α2–6 SA (Rogers et al., 1983; Connor et al., 1994); swMO HA has Leu226/Gly228 at the RBS and was found to agglutinate both α2–6 and α2–3 sialylated RBCs (Table 1). swMO virus also contains Glu627 in the PB2 gene. With respect to other key residues in the PB2 subunit, avian influenza viruses normally contain Gly590/Gln591 sequences in the PB2 gene; however mallMD has a Ser rather than a Gly at position 590, the same amino acid found in swine origin viruses. The swMO virus PB2 gene displays the Ser590/Arg591 constellation, also observed in viruses of swine origin and 2009 H1N1 [A(H1N1)pdm09] viruses, which confers increased polymerase activity in human cells (Mehle and Doudna, 2009). In addition, swMO virus displays Ala271 in PB2, which, along with the Ser590/Arg591 constellation, has been reported to be critical for viral replication and virulence of swine viruses (Liu et al., 2012). The other markers for pathogenesis, such as Ser66 in the PB1-F2 gene and the presence of the PDZ domain in the NS gene, have been related to highly pathogenic phenotypes in other virus subtypes (Conenello et al., 2007; Jackson et al., 2008).
Table 1.
Viral and protein markers and features of H2 viruses used in this study considered relevant for transmission and pathogenesisa.
| Virus original host and subtypeb | HA receptor binding amino acids | PB2 | PB1-F2 | NS1 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
||||||||||
| Receptor binding amino acids | Sialic acid binding preference (HA titer)c | Amino acid positions | Protein length | Position 66 | Protein length | PDZ domain | |||||||
|
|
|
||||||||||||
| 226 | 228 | 271 | 590 | 591 | 627 | 701 | |||||||
| Eng67 | huH2N2 | L | S | α2–6 (8) | A | G | Q | K | D | 90 | N | 237 | No ESEV |
| ckPA | avH2N2 | Q | G | α2–3 (1024) | T | G | Q | E | D | 90 | S | 230 | ESEV |
| mallMD | avH2N3 | Q | G | α2–3 (128) | T | S | Q | E | D | 90 | N | 230 | ESEV |
| swMO | swH2N3 | L | G | α2–6/α2–3 (256/128) | A | S | R | E | D | 57 | – | 219 | Not present |
This table does not display all the amino acid differences these viruses may contain.
hu: human, av: avian, sw: swine.
The viruses (HA=8 or 16) were assayed by hemagglutination using resialylated turkey red blood cells (RBCs).Values between parenthesis show the HA titers obtained with corresponding treated RBC.
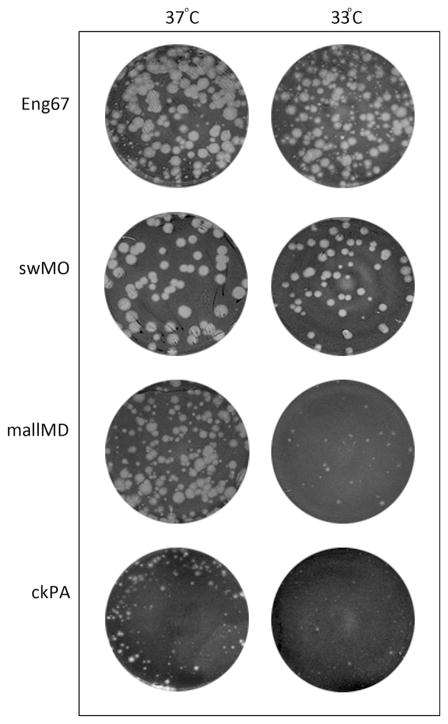
As the optimal replication temperature of influenza viruses is a determinant for virus host range, we evaluated the growth of the H2 viruses at either 33 °C or 37 °C. A plaque assay was performed using MDCK cells in order to assess the sensitivity of each virus to replicate at temperatures that simulate the upper (33 °C) and lower (37 °C) respiratory tracts of mammals (Fig. 1). As expected for a human-adapted virus, Eng67 formed visible plaques at both 33 °C and 37 °C, indicating its ability to replicate efficiently at both temperatures. Similarly, swMO virus was able to replicate efficiently at both temperatures but plaque sizes appeared slightly reduced at 33 °C. However, mallMD and ckPA viruses, which replicated well at 37 °C failed to form significant plaques at 33 °C. These results demonstrate that the avian H2 viruses do not replicate as well the human (H2N2) and swine (H2N3) viruses at the lower temperatures found in the upper airway of mammals.
Fig. 1.
Plaque morphology of MDCK cells (London) infected with H2 viruses of human, avian and swine origin. For every virus, the same dilution was used to inoculate a confluent cell monolayer and was incubated at either 37 °C or 33 °C for 65 h.
Transmissibility and pathogenicity of H2 viruses in the ferret model
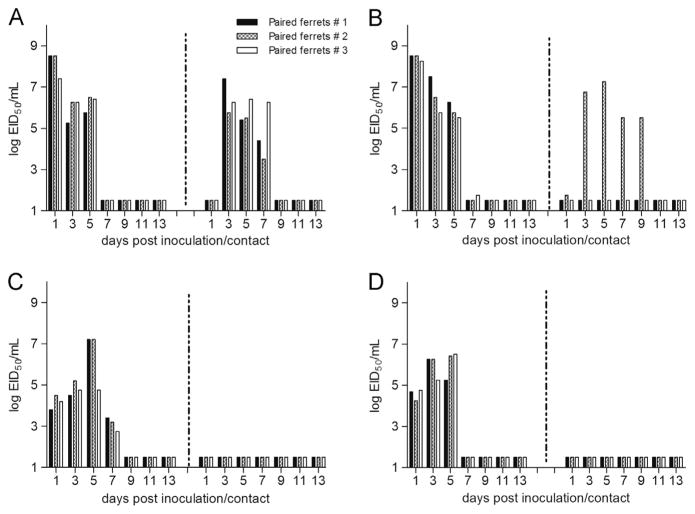
For direct contact (DC) transmission experiments, three animals were inoculated i.n. with 106 EID50 of virus. Twenty-four hours later, a naïve contact ferret was introduced into the same cage with each inoculated ferret. Transmission was assessed by titration of infectious virus in nasal washes and measurement of HI antibody titers in convalescent sera of contact ferrets. Signs of influenza illness were also observed through the presence of fever, lethargy, and weight loss. As expected, Eng67 virus transmitted efficiently in the DC model; virus titers in the nasal washes of inoculated ferrets achieved peak mean titers of 108.1 EID50/ml on day 1 post inoculation (p.i.) and infectious virus was detected in the nasal washes of all contact animals as early as day 1 post direct contact (p.d.c.) (Suppl. Fig. 1A, Table 2). Contact ferrets had detectable virus up to day 7 p.d.c. and all ferrets developed high titers of H2N2-specific HI antibodies, confirming efficient DC transmission of Eng67 virus. Eng67-inoculated ferrets exhibited 5% mean maximum weight loss and a maximum temperature increase of 1.7 °C over baseline (Table 2), consistent with seasonal influenza virus infections in ferrets (Maines et al., 2006).
Table 2.
Clinical signs, virus replication and transmission of H2 influenza viruses in ferrets.
| Mode of transmission | Virus | Inoculated ferrets | Contact ferrets | |||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Mean maximum weight loss (%) | Mean maximum temperature increase (°C) | Lethargya | Mean nasal wash titerb | Seroconversion (geometric mean titer) | Virus detection | Seroconversion (geometric mean titer) | ||
| Direct contact | Eng67 | 5 | 1.7 | 1.1 (2/3) | 8.1 (1) | 5120 | 3/3 | 3/3 (5120) |
| swMO | 15.3 | 1.7 | 1.5 (3/3) | 7.4 (1,5) | 640 | 3/3 | 3/3 (508) | |
| mallMD | <1 | 2.3 | 1.0 (0/3) | 7.1 (3,5) | 1016 | 3/3 | 3/3 (1016) | |
| ckPA | 1.1 | 1.7 | 1.0 (0/3) | 7.0 (3,5) | 50 | 2/3 | 2/3 (113) | |
| Respiratory droplet | Eng67 | 10.1 | 1 | 1.1 (1/3) | 8.3 (1) | 6451 | 3/3 | 3/3 (5120) |
| swMO | 14.1 | 1.7 | 1.2 (3/3) | 8.4 (1) | 1016 | 1/3 | 2/3 (640)c | |
| mallMD | <1 | 1.6 | 1.1 (1/3) | 7.0 (3,5) | 320 | 0/3 | 0/3 | |
| ckPA | 2.5 | 0.9 | 1.0 (0/3) | 6.4 (3,5) | 40 | 0/3 | 0/3 | |
Relative inactivity index of ferrets during the first 9 days p.i.
Peak mean log10EID50/mL (peak day p.i.).
HI titer in serum of ferret with no virus detection was 320.
As was observed with the human H2N2 (Eng67) virus, the swine H2N3 (swMO) virus replicated efficiently in inoculated ferrets (peak mean nasal wash titers 107.4 EID50/ml) and transmitted to all 3 DC ferrets (Supplementary Fig. 1B, Table 2). However, swMO virus-infected ferrets displayed greater weight loss (mean maximum weight loss of 15.3%) and lethargy (relative inactivity score of 1.5) compared to human H2N2 (Eng67) virus-infected ferrets. Among the three contact ferrets, virus was detected in nasal washes on days 1–9, and 3/3 contact animals seroconverted to swMO virus. The avian H2 (mallMD and ckPA) viruses also transmitted by DC, although ckPA had fewer (2/ 3) transmission events than the other three H2 viruses tested and the kinetics of transmission was delayed by 5 to 7 days p.d.c. (Supplementary Fig. 1C and D, Table 2). Both mallMD and ckPA viruses replicated efficiently; infectious virus in nasal washes peaked on days 3 and 5 p.i. (mean maximum 107.1 and 107.0 EID50/ml, respectively). Although the peak temperatures (1.7–2.3 °C) among ckPA- and mallMD-inoculated ferrets were relatively similar to that observed with the other two H2 viruses, lethargy and the weight loss was negligible (<1% to 1.1%, mean maximum weight loss).
We next tested the H2 viruses in the respiratory droplet (RD) transmission model, which is a more stringent test of virus transmissibility because it requires virus to be transmitted only through the exchange of air containing virus particles. The human H2N2 (Eng67) virus transmitted efficiently by RD, to all 3 of the 3 indirect contact ferrets, as early as day 3 post indirect contact (p.i.c.) (Fig. 2A). The peak mean titers of 108.3 EID50/ml on day 1 p.i. were similar to that observed in the DC transmission experiments. The swMO virus spread to 2 out of 3 indirect contact ferrets and all three inoculated ferrets displayed clinical signs similar to that observed in the DC transmission studies (Fig. 2B and Table 2). RD transmission to the indirect contact ferrets was evidenced through detection of virus titers in one ferret (ferret#2) and seroconversion to swMO virus in a second ferret (ferret#3). Negative results obtained by RT-PCR using oligonucleotide primers for HA and M genes confirmed that RD transmission to the two indirect contact ferrets #1 and #3 did not occur (Fig. 2B). It is interesting that the HI titers in the sera of indirect contact ferret #3 were four times lower than the titer obtained for the ferret that had virus detected in the NW (ferret #2, Fig. 2B, Table 2). No RD transmission was observed with the avian H2 viruses, mallMD and ckPA; infectious virus was not detected in indirect contact ferret NWs, and no seroconversion of indirect contact animals was observed (Fig. 2C and D). The mallMD- and ckPA-inoculated ferrets presented clinical signs similar to those observed in the direct contact experiment (<3% mean maximum weight loss, mean nasal wash titers 106.4–7.0 EID50/ml, and 0.9–1.6 °C mean temperature increase) (Table 2).
Fig. 2.
Respiratory droplet transmission experiments using H2 viruses of human, avian and swine origin. Each experiment was done in triplicate; ferrets were i.n. inoculated with 106 EID50 of virus and placed in individual cages. Twenty four hours later a naïve ferret was housed in adjacent cage to allow exchange of respiratory droplets through perforations on the adjoining walls. Nasal wash samples were collected from inoculated and indirect ferrets on alternating days, starting on day 1 post inoculation/post indirect contact (p.i./p.i.c.). (A) Ferrets inoculated with human virus Eng67; (B) swMO; (C) mallMD. Bars with the same pattern represent nasal wash titers of inoculated/indirect contact ferret pair, the bars on the left side correspond to inoculated ferret and on the right side to the indirect contact ferret.
We also examined the ability of the H2 viruses to replicate in respiratory tract tissues and organs outside of the respiratory tract, including the brain. None of the four H2 viruses could be detected in the brain, spleen, or heart tissues on day 3 p.i. (data not shown). Examination of virus titers in the upper and lower respiratory tracts of ferrets revealed that the virus titers levels in the nasal turbinates were relatively similar for all four viruses (P>0.1) (Fig. 3). swMO and mallMD viruses were recovered from ferret tracheas and lung tissues at the highest levels compared with that observed for Eng67 and ckPA virus infections.
Fig. 3.
Influenza virus titers recovered from respiratory tract ferret tissues. Three ferrets were i.n. inoculated with 106 EID50 of Eng67, swMO, mallMD and ckPA, and tissues were collected on day 3 p.i. Tissues homogenates were titrated in 10-day embryonated eggs. The limit of detection is 1.5 Log10 EID50/g or mL. Each bar represents the average titer for three ferrets, plus standard deviation. Significance was determined by student t-test and compared against swMO values: *p<0.05, **p<0.1, ***p<0.01.
Glycan binding and crystal structure of swMO HA
The RD transmission of swMO virus prompted us to perform a more detailed glycan array approach to analyze the repertoire of sialylated glycans recognized by swMO virus, as well as determine the crystal structure of the swMO HA. Glycan microarray results for swMO recombinant HA revealed a strong binding preference for the α2–6-linked sialosides (#41–59) and some binding to mixed α2–3/α2–6 branched sialosides (#60–64) (Fig. 4 and Supplementary Table 2). By using x-ray crystallography, the structure of swMO HA was determined to 2.9-Å resolution (Table 3). The overall structure of swMO HA is similar to other reported HA structures with a globular head containing the receptor binding site (RBS) and vestigial esterase domain, and a membrane proximal domain with its distinctive, central helical stalk and HA1/HA2 cleavage site (Fig. 5A). Although eight asparagine-linked glycosylation sites are predicted in the swMO HA monomer, interpretable electron density was observed at only five sites, Asn11, Asn23, Asn164, Asn285 in HA1 and Asn154 in HA2. At each of these sites, only one asparagine-linked N-acetyl glucosamine could be interpreted. During viral replication, HA is synthesized as a single chain precursor (HA0) and cleaved by specific host proteases into the infectious HA1/HA2 form. In baculovirus expression systems, low pathogenic virus HAs, such as SwMO, possess a monobasic cleavage site and are normally expressed as the HA0 form. While the HA2 fusion peptide for the infectious HA1/HA2 form inserts into the HA2 trimerization cavity, the loop in the HA0 is more exposed. Three HA0 structures, with different loop conformations have been described: H3 (PDB ID 1HA0), H1 (PDB ID 1RD8) and H16 (PDB ID 4F23). Like the H3 and H1 HA0 cleavage sites, swMO H2 HA0 also forms a loop-like structure, but is positioned further away from the glycoprotein surface. As for the H16 HA0 cleavage site, it forms a short α-helix structure.
Fig. 4.
Glycan microarray analysis of swMO recombinant HA. Colored bars highlight glycans that contain α2–3 SA (blue) and α2–6 SA (red), α2–6/α2–3-mixed SA (purple), N-glycolyl SA (green), α2–8 SA (brown), β2–6 and 9-O-acetyl SA, and non-SA (gray). Error bars reflect the standard deviation in the signal for six independent replicates on the array. Structures of each of the numbered glycans are found in Supplementary Table 2.
Table 3.
Data collection and refinement statistics.
| Data collection | swMO |
|---|---|
| Space group | P212121 |
| Cell dimensions | 70.29 Å, 160.03 Å, 170.60 Å 90°, 90°, 90° |
| Resolution (Å) | 50–2.9 (3.00–2.90)a |
| Rsym | 9.0 (78.7) |
| I/σ | 20.4 (2.9) |
| Completeness (%) | 99.8 (100) |
| Redundancy | 7.2 (7.3) |
| Refinement | |
| Resolution (Å) | 50–2.9 (2.98–2.90) |
| No. of reflections (total) | 41,095 |
| No. of reflections (test) | 2170 |
| Rwork/Rfree | 23.3/26.2 |
| No. of atoms | 11,826 |
| r.m.s.d.—bond length (Å) | 0.011 |
| r.m.s.d.—bond angle (°) | 1.416 |
| MolProbityb scores | |
| Favored (%) | 92.6 |
| Allowed (%) | 99.4 |
| Outliers (%) (No. of residues) | 0.6(9/1459) |
Numbers in parentheses refer to the highest resolution shell.
Ref. Davis et al. (2007).
Fig. 5.
Structural overview of swMO HA. (A) One swMO monomer is shown with the HA1 chain colored in green and the HA2 chain in cyan. The locations of the glycosylation sites are shown in red sticks. (B) Expanded view of swMO HA RBS with its three structural elements, the 130-loop, 190-helix, and 220-loop, colored in purple, red and yellow, respectively. Mutations found in the nasal washes of ferrets inoculated with ckPA (colored in orange and red sticks) and mallMD (colored in orange and blue sticks). (C) Amino acid differences in HA region that contains receptor binding amino acids (HA1 positions 130 to 230). All structural Figs were generated with MacPyMol (DeLano, 2002).
The RBS, at the membrane distal end of the HA monomer, is composed of three conserved structural elements: a 190-helix (residues 188–194), a 220-loop (residues 221–228), and a 130-loop (residues 134–138). In addition, highly conserved residues (Tyr98, Trp153, His183, and Tyr195) form the base of the pocket (Fig. 5B). Previously, mutations in the HA RBS of H1N1 (Glu190Asp/Gly225Asp) and H2N2/H3N2 (Gln226Leu and Gly228Ser) subtypes were found to be responsible for adaptation of these viruses from avian strain to human strain (Connor et al., 1994; Rogers and Paulson, 1983). Studies on H2 HA structures showed that the 226 and 228 substitutions result in a slight elongation of the RBS in Human H2 HA (Xu et al., 2010). SwMO RBS has Leu226 and Gly228 which is an intermediate state between avian and human strains. Because of the Leu226 substitution, the subtle movement of the 220-loop increased the α2–6-linked sialosides binding, and the glycan results presented here are consistent with previous studies with a 1957 human H2 HA (Xu et al., 2010).
Presence of virus subpopulations in ferret tissues of the respiratory tract and nasal washes
HA1 is important for recognition of sialic acid receptors and efficient transmission in mammals (Pappas et al., 2010; Van Hoeven et al., 2009). We first performed sequence analysis on tissue specimens from inoculated ferrets to look for the presence of genetically distinct viral subpopulations. The amino acid sequences of the HA1 region corresponding to the nucleotides encoding the receptor binding amino acids (positions 130 through 230, H3 numbering) were compared to those of the original viral stocks. Although no mutations were present in the ckPA virus stock, we found evidence of virus subpopulations containing amino acid changes at positions 156 and 200 among samples (day 3 p.i.) from nasal turbinates and lung homogenates of 3 of 3 ckPA-inoculated ferrets (Table 4). No virus RNA could be extracted from the trachea supernatants of ferrets infected with ckPA. Examination of mallMD-infected ferrets revealed that all HA1 sequences from viruses in the nasal turbinates, trachea, and lungs of 3/3 animals contained amino acid variability at position 212 (Arg and/or Met). This variation was also detected in the stock virus used to inoculate ferrets (Table 4). In swMO virus infected tissues, the amino acid switch Leu226Gln crucial for change of receptor binding preference from α2–6 to α2–3 SA was observed in the tracheas of 2/3 ferrets but not in nasal turbinates, lung tissues or stock virus.
Table 4.
Determination of HA1 mutations in viruses extracted from ferret tissues.
| ckPA | mallMD | swMO | ||
|---|---|---|---|---|
|
|
|
|||
| 156 | 200 | 212 | 226 | |
| Virus stock | E | T | M/R | L |
| Nasal turbinates | E/K (2/3) | T | M/R (3/3) | L (3/3) |
| Trachea | ND | ND | R (2/2) | Q (2/3) |
| Lungs | E (3/3) | T/P (pooled)a | R (1/3) M/R (2/3) | L (3/3) |
ND not possible to obtain enough DNA for sequencing analysis.
Amino acid separated by “/” means the presence of mixed populations containing either amino acid co-replicating in the tissue sample.
RNAs from the three ferrets’ tissue homogenates were pooled together before RT-PCR preparation for sequencing.
To determine if there were any HA1 mutations during H2 virus transmission, sequencing was performed on the nasal washes of ferrets used in the DC or RD transmission experiments. For the swMO transmission experiments, one HA1 amino acid change (Leu226Gln) was observed from nasal washes, and that occurred in only 1 of 6 ferrets used in DC or RD transmission experiments (Table 5). The virus sequence from nasal washes of the other swMO exposed ferrets (1/3 from RD and 3/3 from DC transmission) remained unchanged (sample from day 5 p.i.c./p.d.c.). The HA1 sequences of ckPA viruses that were collected during the DC transmission experiment revealed that the virus in both inoculated and contact ferrets (pairs #1 and #3) underwent two amino acid changes at positions 190 (Glu190Gly pair #1 and Glu190Asp pair #3) and 216 (Glu216Asp) in both ferret pairs. For inoculated ferret #2 that did not transmit ckPA virus to its cage mate, changes were not present at those positions but only at amino acid 206 (Thr/Pro206). Interestingly, the mallMD virus from one inoculated and one unrelated DC contact ferrets had subpopulations with the key RBS amino acid substitution at position 226 (Gln226Leu). Additional amino acid changes at positions Glu190Val, Gly205Val, and Met/Arg212Arg or Met in different combinations were also found depending on the ferret pair of both RD and DC transmission experiments (Table 5).
Table 5.
Determination of HA mutations in viruses extracted from ferret nasal washes during DC and RD transmission.
| DC swMOa | RD swMOa | DC mallMDb | RD mallMDb | DC ckPAc | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
||||||||||||||
| Amino acid positionsd | 226 | 226 | 190 | 205 | 212 | 226 | 228 | 190 | 205 | 212 | 226 | 228 | 190 | 206 | 216 | |
| Virus stock | L | L | E | G | M/R | Q | G | E | G | M | Q | G | E | T | E | |
| Ferrets #1 | Inoculated | – | – | – | V | R | – | – | – | V | R | – | – | G | – | D |
| Contact | L/Q | – | V | R | – | – | G | – | D | |||||||
| Ferrets #2 | Inoculated | – | – | – | – | R | – | – | – | V | R | – | – | – | T/P | – |
| Contact | – | – | – | – | R | Q/L | – | – | – | |||||||
| Ferrets #3 | Inoculated | – | – | – | V | R | Q/L | – | – | V | R | – | D | – | D | |
| Contact | – | e | V | – | M | – | – | D | – | D | ||||||
DC direct contact transmission.
RD respiratory droplet transmission.
–No amino acid change.
Blank: no virus or RNA detected by either egg titration or RT-PCR, respectively.
Amino acid separated by “/” means the presence of mixed populations containing either amino acid co-replicating in the tissue sample.
Nasal wash samples collected on days 3 p.i. and 5 p.d.c/p.i.c.
Nasal wash samples collected on days 5 p.i. and 7 p.d.c/p.i.c.
Nasal wash samples collected on days 3 p.i. and 7 p.d.c/p.i.c.
H3 numbering.
No transmission occurred but ferret seroconverted.
Mixed populations during the course of mallMD infection and transmission
The appearance of Leu226 amino acid in the nasal washes of mallMD infected ferrets suggested a probable adaptation for this virus to replicate in mammalian tissues. We next performed a more in depth analysis of the nasal washes collected during the entire course of mallMD virus infection. RNA was extracted from each nasal wash sample of inoculated and contact ferrets during DC transmission experiments. Overall, viruses containing amino acid changes appeared early among inoculated ferrets (days 1–5 p. i.) in the course of infection (Table 6). Amino acids mutations that were consistently observed were Glu190Val, Gly205Val, Met/Arg212Arg or Met, Gln226Leu, and 228 (silent mutation). Viruses containing Leu226 were not detected in the nasal washes of the ferret pair#1, but mutant viruses containing Gly205Val and Met/Arg212Met or Arg substitutions were present, appearing in inoculated ferrets on day 5 p.i. and spreading to the contact ferret on day 7 p.d.c. In ferret pair #2, changes involving four amino acids at positions 205, 212, 226 and 228 took place during infection among inoculated and contact ferrets. Virus was detected in the contact ferret on day 1 p.d.c.; mixed populations containing either Gln226 or Leu226 were present in the nasal washes of the inoculated ferrets on day 7 p.i. and in its contact on days 5 and 7 p.d.c. (Table 6). The most consistent mutations that were present in the viruses collected from both ferrets were the Met/Arg212Met or Arg substitutions. Of note, a silent nucleotide mutation at residue 228 was also detected in most NW viruses carrying the Leu266 mutation. In ferret pair #3, amino acid changes occurred in the inoculated ferret starting with one HA mutation (Met/Arg212Arg) on day 1 p.i. and progressed to up to 3 mutations containing the substitutions, Gly205Val or Val/Gly, Met/Arg212Arg, Gln226Leu, including the silent mutation at position 228. In the contact ferret, the virus was first detected on day 7 p.d.c. but contained only one mutation at position Glu190Val.
Table 6.
Time course analysis of amino acid variation in mallMD virus collected from ferret nasal washes during DC transmission experiment.
| Days p.i./p.c. | Virus stock | Amino acid positions in hemagglutinina | |||||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| 190 | 205 | 212 | 226 | 228 | |||
| E | G | M/R | Q | G | |||
| Ferret pair #1 | 1 | Inoculated | – | – | M | – | – |
| Contact | |||||||
| 3 | Inoculated | – | – | M | – | – | |
| Contact | |||||||
| 5 | Inoculated | – | V | R | – | – | |
| Contact | |||||||
| 7 | Inoculated | – | V | R | – | – | |
| Contact | – | V | R | – | – | ||
| 9 | Inoculated | ||||||
| Contact | – | V | R | – | – | ||
| 11 | Inoculated | ||||||
| Contact | – | V | R | – | – | ||
| Ferret pair #2 | 1 | Inoculated | – | – | M | – | – |
| Contact | – | – | R | – | – | ||
| 3 | Inoculated | – | V/G | R | – | – | |
| Contact | – | – | R | – | – | ||
| 5 | Inoculated | – | – | R | – | Gsilb | |
| Contact | – | – | R | Q/L | – | ||
| 7 | Inoculated | – | G/V | R | L/Q | Gsil | |
| Contact | – | – | R | Q/L | – | ||
| 9 | Inoculated | ||||||
| Contact | – | – | R | – | Gsil | ||
| Ferret pair #3 | 1 | Inoculated | – | – | R | – | – |
| Contact | |||||||
| 3 | Inoculated | – | – | R | – | – | |
| Contact | |||||||
| 5 | Inoculated | – | V | R | – | Gsil | |
| Contact | |||||||
| 7 | Inoculated | – | V/G | R | Q/L | Gsil | |
| Contact | V | – | M | – | Gsil | ||
| 9 | Inoculated | ||||||
| Contact | V | – | M | – | Gsil | ||
| 11 | Inoculated | ||||||
| Contact | V | – | M | – | Gsil | ||
– no amino acid mutations were present.
Blank: no virus or RNA detected by either egg titration or RT-PCR, respectively.
Amino acid separated by “/” means the presence of mixed populations containing either amino acid co-replicating in the tissue sample.
H3 numbering.
No amino acid change, silent mutation from GGA to GGG codon.
It was also of interest to investigate whether these HA mutations occurred during the mallMD RD transmission experiment. Thus, nasal washes collected from the three mallMD inoculated ferrets were also sequenced. The sequencing results revealed that mutations in HA1 were confined to the same positions and amino acids that were found in the nasal washes of the DC transmission experiment; Gly205Val, Met/Arg212Arg, 228 silent mutation (data not shown). However interestingly, no evidence of viruses containing the Gln226Leu amino acid substitutions was present. Taken together, these findings identify significant amino acid mutations that occurred during H2 virus transmission, including Gln226Leu substitution that has been associated with α2,6 sialic acid (human-like) binding preference.
Discussion
The natural emergence of another influenza pandemic virus is considered likely, if not inevitable. Public health concerns have mostly centered on the potential of contemporary avian influenza subtype viruses to mutate or reassort with other influenza subtypes to a form that could become transmissible among humans. As a result, multiple studies have been performed to assess the molecular determinants of avian H5, H7 and H9 virus adaptation, pathogenicity and transmissibility in mammalian hosts (Maines et al., 2006; Salomon et al., 2006; Chen et al., 2012; Russell et al., 2012; Herfst et al., 2012; Sorrell et al., 2009). However, relatively less attention has been given to H2 subtype viruses largely due to their sporadic isolation and low pathogenicity in both avian and mammalian species (Pappas et al., 2010; Jones et al., 2014). The isolation of H2N3 influenza viruses from swine in 2006 focused some awareness and attention to this subtype, and since then, a number of new studies have been published that have examined the transmission, pathogenesis and vaccine development for this subtype (Nabel et al., 2011; Chen et al., 2010; Killian et al., 2011; Pappas et al., 2010; Jones et al., 2014; Driskell et al., 2010). In the present study, we examined the pathogenicity and transmissibility of four genetically distinct H2 viruses. All four H2 viruses replicated efficiently and transmitted in the direct contact ferret model. As efficient replication at the lower temperature of 33 °C is thought to be important for virus adaptation to humans (Pappas et al., 2010; Van Hoeven et al., 2009), we determined the optimal replication temperature of each virus. The H2 viruses that transmitted readily by respiratory droplets also demonstrated an ability to replicate efficiently at 33 °C—the lower temperature found in the environment of the mammalian upper airway. Notably, this included the swine H2N3 (swMO) virus that was able to replicate efficiently at 33 °C under in vitro conditions and spread to naïve ferrets by respiratory droplets. These results prompted us to characterize the swMO virus in more detail by analyzing the receptor binding specificity on a glycan array platform and report the crystal structure of swMO HA. In addition, we attempted to gain insight into the possible origin of virus diversity by sequencing nasal wash samples collected during transmission experiments. The detection of viral subpopulations isolated from H2-infected ferrets containing key mutations, such as the Gln226Leu change (associated with increased human-like receptor binding preference) is important for identifying genetic changes of H2 viruses that can potentially increase its transmissibility.
The genetic and antigenic surveillance of avian species in the wild, in farms and in live markets has been critical for detection of newly emerging influenza viruses and for assessment of potential public health risks (World Health Organization, 2013). This includes H2 subtype viruses that are isolated from live bird markets and wild birds, often on an annual basis (Piaggio et al., 2012; Makarova et al., 1999; Liu et al., 2004; Kishida et al., 2008; Ferro et al., 2010; Ramey et al., 2010). The continuous isolation of avian H2 viruses increases the risk of human exposure to a novel influenza virus and underscores the need to better understand the potential of this subtype to acquire properties that would confer efficient RD transmission between mammals. Although there are multiple molecular determinants that affect influenza virus host adaptation, it is generally believed that the HA of avian virus origin must acquire human influenza virus-like receptor binding (α2,6 SA) properties to become transmissible in mammals (Pappas et al., 2010; Maines et al., 2011; Pearce et al., 2012; Belser et al., 2013; Tumpey et al., 2007). The avian H2 viruses (ckPA and mallMD) tested in this study exhibited classical avian α2,3 SA binding and failed to transmit by respiratory droplets. Moreover, both ckPA and mallMD viruses replicated poorly at 33 °C, further supporting a lack of adaptation of these avian influenza viruses to mammalian hosts. Conversely, the human-adapted H2 (Eng67) virus, which preferentially binds to SA in α2–6 linkage and replicated efficiently at 33 °C, transmitted efficiently to naïve ferrets by respiratory droplets. Although the avian H2 viruses failed to spread in the RD model, the efficient DC transmission results are distinguishable from other avian viruses (e.g. H5N1, H7N7) that have shown inefficient transmission in the DC ferret model (Maines et al., 2006). Avian influenza viruses that are at least able to spread under close contact conditions could possess an advantage over viruses that transmit poorly or not at all. Thus, an avian influenza virus that jumps the species barrier to infect an intermediate host, such as pigs and subsequently spreads by close contact may result in the generation of a mutant virus with adaptive mutations that confers RD transmission. Experimentally, natural virus variants with minimal molecular changes have been isolated from exposed ferrets following transmission events, such as have been described for H2, H5 and H9 subtype viruses (Pappas et al., 2010; Herfst et al., 2012; Sorrell et al., 2009).
The swine swMO virus was of particular interest because it has been previously shown to replicate well in the respiratory tracts of mammals and transmit efficiently in the DC ferret model (Chen et al., 2010; Ma et al., 2007; Richt et al., 2012; Jones et al., 2014). Similarly, we found that swMO virus transmitted efficiently in the DC model; however we also observed transmission via respiratory droplets in 2/3 exposed ferrets. It is of concern that, in addition to its ability to transmit by respiratory droplets, swMO-inoculated ferrets exhibited more severe clinical signs (weight loss, lethargy, nasal discharge) and higher virus titers in respiratory tract tissues than those in Eng67-inoculated ferrets. In particular, the swine H2N3 virus demonstrated a tropism for the lower respiratory tract; swMO virus replicated in the ferret lung (Fig. 3) and caused gross pathological lung lesions not observed in Eng67-inoculated ferrets (not shown). These results are consistent with the finding of increased swMO virus replication and lesions in lung tissues of macaques compared to that observed among human H2N2-infected animals (Chen et al., 2010; Ma et al., 2007; Richt et al., 2012; Jones et al., 2014). This work also examined the molecular determinants that might influence virulence and transmission of the swine swMO H2N3 virus. Amino acid sequence comparison of swMO and other influenza viruses revealed that in addition to the presence of 226Leu in HA1, swMO virus contains the triple-reassortant internal gene (TRIG) cassette similar to H3N2v and A (H1N1)pdm09 viruses, both of which are capable of RD transmission (Pearce et al., 2012; Maines et al., 2009). Additional genetic markers for swMO that have been shown to confer viral replication advantages, including its ability to replicate at 33 °C, is the presence of the Ser590/Arg591 polymorphism and Ala271in PB2. The presence of these aminoacid sequences has been shown as critical elements for replication and adaptation of triple reassortant viruses in mammals (Mehle and Doudna, 2009; Liu et al., 2012; Bussey et al., 2010).
The ferret model was also used to measure the impact of within host ‘quasispecies’ diversity. In particular, the HA, which is the major determinant of virus antigenicity and receptor binding specificity can mutate during replication and adaptation in mammals (Pappas et al., 2010). By examining the HA1 amino acid sequences in samples taken from the ferret respiratory tracts on day 3 p.i., it was determined that the swine (swMO) and avian H2 viruses (ckPA, mallMD) contained mutations at distinct amino acid positions. In ckPA-infected ferret tissues (nasal turbinates or lungs), HA mutations were located at positions 156 or 200. In mallMD-infected tissue samples, virus mutants with changes at amino acid position 212 (Met/Arg) were present and swMO-infected trachea samples in 2/3 ferrets contained viruses with 226Gln mutations. Two substitutions, Glu190Gly/Asp and Glu216Asp, in the HA of ckPA virus were present in all nasal wash samples collected from the inoculated and contact ferret pairs where transmission occurred. Interestingly, the virus isolated from the inoculated ferret that did not transmit to its cage mate and possessed no mutations at 190 or 216. Instead, a mixed virus subpopulation containing Thr206Pro was present. These results suggest that mutations at amino acids 190 and 216 may be involved in the transmission of ckPA virus in ferrets via DC, but more investigation is necessary to confirm this observation. In addition to the Met/Arg212Arg or 212Met mutations found in the mallMD ferrets, Glu190Val, Gly205Val, and Gln226Leu changes were detected in the nasal washes of both inoculated and contact ferrets, but not all were present in the same virus. The appearance of mutant viruses containing Leu226 in one inoculated and one contact ferret suggested this virus was acquiring adaptive changes during its replication in the upper respiratory tract of the ferrets. A more detailed sequencing analysis was performed on the viruses obtained from all of the nasal washes collected from the 3 ferrets during the course of virus infection. The dynamic changes that mallMD virus underwent during the course of infection in the ferrets’ upper respiratory tracts were monitored. Amino acid changes were restricted to Glu190Val, Gly205Val, Met/Arg212Arg or 212Met and Gln226Leu in all 9 ferrets tested, including the inoculated ferrets used in RD transmission experiments (data not shown). It appears that the Leu226 amino acid mutation is not an essential contribution to a specific combination of mutations that are necessary for DC transmission to occur. Conversely, virus subpopulations containing the Leu226 amino acid may or may not require additional amino acid changes at positions Val205 and/or Arg212. Furthermore, the appearance of Leu226 in the contact ferret did not necessarily mean that it was acquired from the inoculated ferret; but rather, it could have occurred by virus adaptation. To understand the possible role of the amino acid mutations of mallMD and ckPA in the overall HA structure and/or binding to receptors, we mapped their positions on the globular head of H2 HA. Changes in residues located at position 190 (Glu/Val/Gly, in ckPA) and 226 (Gln/Leu, in mallMD) are in the RBS and involved in the binding to the ligands, while positions 216 (Glu/Asp, in ckPA) and 212 (Met/Arg, in mallMD) are further away from the RBS, but close to the HA trimer interface and may affect the local monomer-monomer interaction (Fig. 5B).
The present work supported previous reports showing that avian H2 viruses have not fully adapted to mammals, but can acquire mutations enabling α2,6 SA recognition that permit transmission between mammals. The presence of mutations in the two avian viruses during replication in the ferret model can be attributed to the incompatibility of avian polymerase to the new host and/or to the natural mechanism these viruses have to generate thermostable virus subpopulations as a mechanism of virus fitness (Negovetich and Webster, 2010). The fact that swMO did not present mutations other than Gln226 may indicate significant stability of the HA1 molecule, which is probably a result of the HA structural framework as well as the high efficiency of polymerase replication at lower temperature.
Influenza H2 viruses represent a potential pandemic subtype should they acquire a mammalian transmissible phenotype. In particular, contemporary H2N3 influenza virus appears to be already partially adapted to mammals (Ma et al., 2007; Killian et al., 2011). Identification of the molecular determinants that confer viral transmission and receptor-binding properties of H2 viruses are needed to understand the factors that may lead to the emergence of future pandemic influenza viruses. While H2 viruses are isolated at a relatively low number each year, the potential for these viruses to acquire a transmissible phenotype underscores their pandemic potential.
Materials and methods
Viruses
The two avian viruses, A/chicken/Pennsylvania/298101-4/2004 (ckPA, H2N2) and A/mallard/Maryland/235/2001 (mallMD, H2N3) were kindly provided by David Swayne (U.S. Dept. of Agriculture, Southeastern Poultry Research Laboratory) and A/swine/Missouri/2124514/2006 virus (swMO, H2N3) was provided by the University of Minnesota. Stocks of the two avian viruses were prepared using 10 day-old embryonated hen’s eggs that were incubated at 37 °C for 48 h. The human, A/England/10/1967 (Eng/67, H2N2), and swine viruses were propagated using Madin–Darby canine kidney cells (MDCK) as previously described (Maines et al., 2006). The genome of each virus stock was confirmed by full sequencing to ensure no inadvertent mutations were introduced during propagation of virus. All experiments performed using these viruses were conducted in biosafety level 3 level containment laboratories with enhancements (BSL-3E) (Richmond, 2007).
Ferret pathogenesis and transmission experiments
Male Fitch ferrets, 8–12 months of age (Triple F Farms, Sayre, PA), serologically negative for currently circulating seasonal influenza viruses, were housed in cages placed inside a Duo-Flo Bioclean mobile clean unit (Lab Products, Seaford, DE). Pathotyping, direct contact (DC) and respiratory droplet (RD) virus transmission experiments were performed as described (Maines et al., 2005). Briefly, ferrets were intranasally (i.n.) inoculated with 106 EID50 of virus in a total volume of 1 mL (0.5 mL/nostril). Nasal wash samples were collected from ferrets every other day post-virus inoculation (p.i.), or post-ferret direct contact (p.d.c.)/indirect contact p.i.c. ). For determination of virus replication, tissue specimens were collected from the ferret’s nasal turbinates, trachea and lungs on day 3 p.i. Virus transmission was verified by the presence of H2 virus specific antibodies in convalescent serum. Sera were collected on day 21 p.i. for determination of seroconversion by hemagglutination inhibition (HI) assay. All ferret experiments were conducted under the guidance of the Centers for Disease Control and Prevention’s Institutional Animal Care and Use Committee (IACUC) in facilities accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Viral protein sequence analysis
Viral RNA was extracted from virus stocks, ferrets’ nasal washes (collected on days 3 or 5 p.i. and days 5 or 7 p.d.c./p.i.c. ) or tissue homogenate supernatants (day 3 p.i.) using QIAamp Viral RNA mini kit (Qiagen, Valencia, CA). Virus gene segments were amplified by reverse transcription-PCR using Access Quick (Promega, Madison, WI), followed by gel purification of the fragments for sequencing reaction using Big Dye V3.1 Terminator kit and analysis using a 3730xl DNA sequencer (Applied Biosystems, Foster City, CA). Oligonucleotide primer sequences can be provided upon request. Sequence analysis was performed using SeqMan, MegAlign (Lasergene) and BioEdit.
Recombinant swMO HA cloning and expression
The cDNA for the ectodomain of the HA from A/swine/Missouri/2124514/2006 was codon optimized and synthesized by GeneArt® (Life Technologies) and subcloned into the pAcGP67-B, baculovirus transfer vector (BD Biosciences). The recombinant HA protein contained a C-terminal thrombin site followed by a trimerizing sequence (foldon) from the bacteriophage T4 fibritin for generating functional trimers, and a His-Tag to aid purification (Yang et al., 2010b). Secreted protein was recovered from the culture supernatant and purified by metal affinity chromatography and size exclusion chromatography (SEC). For structural analyses, the recombinant protein was subjected to thrombin cleavage to remove the foldon and His-Tag and purified by SEC. Thrombin-treated protein was buffer exchanged into 10 mM Tris–HCl and 50 mM NaCl (pH 8.0) and the HAs were concentrated to 14 mg/ml for crystallization trials.
Crystallization and data collection
Initial crystallization trials were set up using a Topaz™ Free Interface Diffusion (FID) Crystallizer system (Fluidigm Corporation, San Francisco, CA). Following optimization, diffraction quality crystals for HA were obtained at 20 °C using sitting drop by vapor diffusion against a reservoir solution containing 20% PEG 3000, 0.1 M Tris–HCl pH 7.6. Crystals were flash-cooled at 100 K. Data were collected at Advanced Photon Source (APS) beamline 22-ID at 100 K and processed with the DENZO-SACLEPACK suite (Otwinowski and Minor, 1997). Statistics for data collection are presented in Table 3.
Structure determination and refinement
The swMO HA structure was determined by molecular replacement with Phaser (McCoy et al., 2005) using the HA structure from A/Japan/305/1957(H2N2) (H3N2), PDB: 3KU5 (HA1, 91% identity; HA2, 96% identity) as a search model. Three hemagglutinin monomers making one non-crystallographic trimer occupy the asymmetric unit with an estimated solvent content of 60% based on a Matthews’ coefficient (Vm) of 3/Da. The model was then “mutated” to the correct sequence and rebuilt by Coot (Emsley and Cowtan, 2004), and then the protein structures were refined with REFMAC (CCP4, 1994) using TLS refinement (Winn et al., 2001) and Phenix refine (Adams et al., 2010). The final model was assessed using MolProbity (Davis et al., 2007) and statistics for data processing and refinement are presented in Table 2.
Glycan binding analyses
Glycan microarray printing and recombinant HA analyses have been described previously (Yang et al., 2010b; Stevens et al., 2004, 2006a, 2006b; Yang et al., 2010a, 2012). Supplementary Table 2 lists glycans used in these experiments.
Hemagglutination assays
Hemagglutination assays using resialyated turkey red blood cells were performed as previously described with minor modifications (Glaser et al., 2006). Turkey red blood cells were enzymatically desialyated using Vibrio cholera Sialidase (Roche-Applied Science, Indianapolis, IN), followed by resialylation using either α2–6-(N)-sialyltransferase or α2–3-(N)-sialyltransferase (Sigma-Aldrich, St. Louis, MO). The HI assay was performed using homologous viruses along with 1% turkey red blood cells.
PDB accession code
The atomic coordinates and structure factors of swMO HA are available from the RCSB Protein Data Bank (PDB) under accession code 4W8N.
Supplementary Material
Acknowledgments
This work was funded by the Centers for Disease Control and Prevention. Glycan microarray slides were produced under contract for the Centers for Disease Control and Prevention using a glycan library generously provided by the Consortium for Functional Glycomics (CFG) (www.functionalglycomics.org) funded by National Institute of General Medical Sciences Grant GM62116. We thank the staff of SER-CAT sector 22 for their help with data collection. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Appendix A. Supplementary information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/
References
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013;501(7468):556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J Virol. 2010;84(9):4395–4406. doi: 10.1128/JVI.02642-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen GL, et al. Evaluation of replication and cross-reactive antibody responses of H2 subtype influenza viruses in mice and ferrets. J Virol. 2010;84(15):7695–7702. doi: 10.1128/JVI.00511-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, et al. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology. 2012;422(1):105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conenello GM, Zamarin D, Perrone LA, Tumpey T, Palese P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007;3(10):1414–1421. doi: 10.1371/journal.ppat.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205(1):17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35(Web Server issue):W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMol Molecular Graphics Systems. 2002 〈 www.pymol.org〉.
- Dowdle WR. Influenza pandemic periodicity, virus recycling, and the art of risk assessment. Emerg Infect Dis. 2006;12(1):34–39. doi: 10.3201/eid1201.051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driskell EA, Jones CA, Stallknecht DE, Howerth EW, Tompkins SM. Avian influenza virus isolates from wild birds replicate and cause disease in a mouse model of infection. Virology. 2010;399(2):280–289. doi: 10.1016/j.virol.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferro PJ, et al. Multiyear surveillance for avian influenza virus in waterfowl from wintering grounds, Texas coast, USA. Emerg Infect Dis. 2010;16(8):1224–1230. doi: 10.3201/eid1608.091864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser L, et al. Sequence analysis and receptor specificity of the hemagglutinin of a recent influenza H2N2 virus isolated from chicken in North America. Glycoconj J. 2006;23(1–2):93–99. doi: 10.1007/s10719-006-5441-0. [DOI] [PubMed] [Google Scholar]
- Herfst S, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336(6088):1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci USA. 2008;105(11):4381–4386. doi: 10.1073/pnas.0800482105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JC, et al. Risk assessment of H2N2 influenza viruses from the avian reservoir. J Virol. 2014;88(2):1175–1188. doi: 10.1128/JVI.02526-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian ML, Zhang Y, Panigrahy B, Trampel D, Yoon KJ. Identification and characterization of H2N3 avian influenza virus from backyard poultry and comparison with novel H2N3 swine influenza virus. Avian Dis. 2011;55(4):611–619. doi: 10.1637/9749-040111-Reg.1. [DOI] [PubMed] [Google Scholar]
- Kishida N, et al. H2N5 influenza virus isolates from terns in Australia: genetic reassortants between those of the Eurasian and American lineages. Virus Genes. 2008;37(1):16–21. doi: 10.1007/s11262-008-0235-z. [DOI] [PubMed] [Google Scholar]
- Liu J, et al. Structures of receptor complexes formed by hemagglutinins from the Asian Influenza pandemic of 1957. Proc Natl Acad Sci USA. 2009a;106(40):17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, et al. Interregional transmission of the internal protein genes of H2 influenza virus in migratory ducks from North America to Eurasia. Virus Genes. 2004;29(1):81–86. doi: 10.1023/B:VIRU.0000032791.26573.f1. [DOI] [PubMed] [Google Scholar]
- Liu Q, et al. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J Virol. 2012;86(2):1233–1237. doi: 10.1128/JVI.05699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, et al. Panorama phylogenetic diversity and distribution of Type A influenza virus. PloS One. 2009b;4(3):e5022. doi: 10.1371/journal.pone.0005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, et al. Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci USA. 2007;104(52):20949–20954. doi: 10.1073/pnas.0710286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79(18):11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci USA. 2006;103(32):12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, et al. Transmission and pathogenesis of swine-origin 2009A (H1N1) influenza viruses in ferrets and mice. Science. 2009;325(5939):484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413(1):139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova NV, Kaverin NV, Krauss S, Senne D, Webster RG. Transmission of Eurasian avian H2 influenza virus to shorebirds in North America. J Gen Virol. 1999;80(Pt 12):3167–3171. doi: 10.1099/0022-1317-80-12-3167. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- Mehle A, Doudna JA. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc Natl Acad Sci USA. 2009;106(50):21312–21316. doi: 10.1073/pnas.0911915106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel GJ, Wei CJ, Ledgerwood JE. Vaccinate for the next H2N2 pandemic now. Nature. 2011;471(7337):157–158. doi: 10.1038/471157a. [DOI] [PubMed] [Google Scholar]
- Negovetich NJ, Webster RG. Thermostability of subpopulations of H2N3 influenza virus isolates from mallard ducks. J Virol. 2010;84(18):9369–9376. doi: 10.1128/JVI.01170-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Scalepack Manual. 1997. [Google Scholar]
- Pappas C, et al. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PloS One. 2010;5(6):e11158. doi: 10.1371/journal.pone.0011158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MB, et al. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc Natl Acad Sci USA. 2012;109(10):3944–3949. doi: 10.1073/pnas.1119945109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piaggio AJ, et al. Molecular surveillance of low pathogenic avian influenza viruses in wild birds across the United States: inferences from the hemagglutinin gene. PloS One. 2012;7(12):e50834. doi: 10.1371/journal.pone.0050834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyhala R. Antibody status to influenza A/Singapore/1/57(H2N2) in Finland during a period of outbreaks caused by H3N2 and H1N1 subtype viruses. J Hyg. 1985;95(2):437–445. doi: 10.1017/s0022172400062860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramey AM, et al. Transmission and reassortment of avian influenza viruses at the Asian-North American interface. Virology. 2010;406(2):352–359. doi: 10.1016/j.virol.2010.07.031. [DOI] [PubMed] [Google Scholar]
- Richmond JY, McKinney RW. Biosafety in Microbiological and Biomedical Laboratories. 5. Centers for Disease Control and Prevention; Atlanta, GA: 2007. [Google Scholar]
- Richt JA, et al. Recently emerged swine influenza A virus (H2N3) causes severe pneumonia in Cynomolgus macaques. PloS One. 2012;7(7):e39990. doi: 10.1371/journal.pone.0039990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127(2):361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- Rogers GN, et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- Russell CA, et al. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Science. 2012;336(6088):1541–1547. doi: 10.1126/science.1222526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med. 2006;203(3):689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer JR, et al. Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology. 1993;194(2):781–788. doi: 10.1006/viro.1993.1319. [DOI] [PubMed] [Google Scholar]
- Simonsen L, et al. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998;178(1):53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci USA. 2009;106(18):7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, et al. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science. 2004;303(5665):1866–1870. doi: 10.1126/science.1093373. [DOI] [PubMed] [Google Scholar]
- Stevens J, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006a;355(5):1143–1155. doi: 10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Stevens J, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006b;312(5772):404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315(5812):655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- Van Hoeven N, et al. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc Natl Acad Sci USA. 2009;106(9):3366–3371. doi: 10.1073/pnas.0813172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan K, et al. Determinants of glycan receptor specificity of H2N2 influenza A virus hemagglutinin. PloS One. 2010;5(10):e13768. doi: 10.1371/journal.pone.0013768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG. Predictions for future human influenza pandemics. J Infect Dis. 1997;176(Suppl 1):S14–S19. doi: 10.1086/514168. [DOI] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57(Pt 1):122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Influenza at the Human–Animal Interface (HAI) 2013. [Google Scholar]
- Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol. 2010;84(4):1715–1721. doi: 10.1128/JVI.02162-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010a;6(9):e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney P, Stevens J. Structure and Receptor binding properties of a pandemic H1N1 virus hemagglutinin. PLoS Curr. 2010b:2. doi: 10.1371/currents.RRN1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Donis RO, Stevens J. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol. 2012;86(16):8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.