Abstract
Epigenetic mechanisms may be important for a native species’ response to rapid environmental change. Red Imported Fire Ants (Solenopsis invicta Santschi, 1916) were recently introduced to areas occupied by the Eastern Fence Lizard (Sceloporus undulatus Bosc & Daudin, 1801). Behavioral, morphological and physiological phenotypes of the Eastern Fence Lizard have changed following invasion, creating a natural biological system to investigate environmentally induced epigenetic changes. We tested for variation in DNA methylation patterns in Eastern Fence Lizard populations associated with different histories of invasion by Red Imported Fire Ants. At methylation sensitive amplified fragment length polymorphism loci, we detected a higher diversity of methylation in Eastern Fence Lizard populations from Fire Ant uninvaded versus invaded sites, and uninvaded sites had higher methylation. Our results suggest that invasive species may alter methylation frequencies and the pattern of methylation among native individuals. While our data indicate a high level of intrinsic variability in DNA methylation, DNA methylation at some genomic loci may underlie observed phenotypic changes in Eastern Fence Lizard populations in response to invasion of Red Imported Fire Ants. This process may be important in facilitating adaptation of native species to novel pressures imposed by a rapidly changing environment.
Keywords: ecological epigenetics, MS-AFLP, invasive species, rapid adaptation, Eastern Fence Lizard, red imported fire ant
Introduction
Environmental changes, such as those associated with invasive species, are occurring at increasing rates due to anthropogenic activities [1–3]. Understanding the mechanisms underlying an organism’s response to these changes is critical to assessing environmental impacts on natural populations and predicting the potential of populations to respond to future change. While invasive species affect the ecology of native populations, we understand little about the processes by which native species adapt to rapid environmental change and the long-term implications of these adaptations [4].
Molecular epigenetic mechanisms may be important for a native species’ response to environmental changes, including the presence of invasive species. Epigenetic mechanisms encompass changes in gene expression not caused by changes in DNA sequence [5], and these changes can be induced within the lifetime of an individual [6]. DNA methylation, the most commonly studied epigenetic mechanism [7], can vary among individuals and populations and in response to environmental stressors [6, 8–10]. Furthermore, variation in DNA methylation patterns can result in phenotypic changes that can be faithfully transmitted to future generations [10–14]. As a result, epigenetic modification of gene expression may enable organisms to respond quickly to a changing environment [15] and produce offspring pre-adapted to the environment experienced by the parents [16].
The environmental pressures imposed by invasive species can provide natural experiments with which to examine mechanisms driving rapid phenotypic responses, including changes in epigenetic variation [17]. The invasion of the Eastern Fence Lizard’s (Sceloporus undulatus Bosc & Daudin, 1801) range by the venomous Red Imported Fire Ant (Solenopsis invicta Santschi, 1916; hereafter Fire Ant) represents an excellent system to study the adaptive potential of environmentally induced epigenetic changes. The Fire Ant was introduced into the southeastern United States in the 1930’s and quickly spread to 14 states [18, 19]. This venomous ant is able to prey upon organisms much larger than itself, and can envenomate animals that eat it, creating strong novel selective pressures [18, 20–22].
Across almost half its range, the Eastern Fence Lizard is exposed to Fire Ants, and the Fire Ant-invaded populations have responded phenotypically in patterns that suggest both evolutionary (cross-generational) and plastic (within lifetime) responses (T. R. Robbins et al. unpublished data; [23–26]). These species use similar habitat [23, 27] and frequently encounter one another in nature [24]. Fire Ants will attack Eastern Fence Lizards, with as few as 12 ants able to kill an adult lizard in 1 min [27], and will envenomate Fence Lizards that eat them [20]. Multiple phenotypes of the Eastern Fence Lizard are altered within areas invaded by Fire Ants. In Fire Ant-invaded areas, Eastern Fence Lizards exhibit behavior that promotes escape from Fire Ant attack: twitching to remove Fire Ants and fleeing from the source of attack [24, 27]. Lizards from Fire Ant-invaded sites also have longer hind limbs, a heritable trait that increases the efficacy of this anti-predator behavior [27]. Furthermore, Fence Lizards from invaded populations innately avoid eating Fire Ants [25], have an elevated physiological response to stress (plasma concentrations of the stress-relevant hormone, corticosterone) in the presence of this invasive ant [27], and altered behavioral responses to increased levels of corticosterone if their habitat has been invaded [29].
The objectives of the present study were to determine if DNA methylation patterns differed between Eastern Fence Lizard populations from Fire Ant-invaded versus uninvaded areas and to determine if DNA methylation was correlated with individual phenotypes. We show that variation in DNA methylation patterns may play an important role in driving the phenotypic responses of Eastern Fence Lizards to Fire Ant invasion, providing insight into molecular mechanisms that may underlie adaptation.
Results
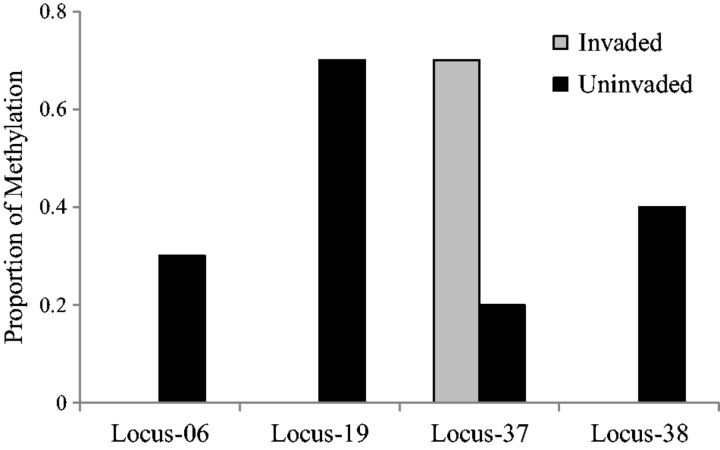
We analysed 35 variable loci of 45 total loci scored at methylation sensitive amplified fragment length polymorphism (MS-AFLP). Loci removed from the analysis were either not variable or had <10% observed methylation. We detected a significantly higher proportion of methylation in uninvaded sites (101 of 350 possible methylated sites) compared with invaded sites (65 of 350 possible methylated sites; z-score = 2.92, P = 0.002). A posthoc test identified Locus-19 (P = 0.04) and Locus-37 (P = 0.04) as contributing to this difference (Fig. 1). However, we did not detect any significant relationships with mass or hind limb length standardized by snout vent length (RHL/SVL), an adaptive phenotype of interest.
Figure 1.
proportion of methylation between invaded (gray) and uninvaded (black) sites at the four loci with the largest contribution
Epi-h ranged from 0.23 to 0.39 (Table 1). Epi-h was higher in uninvaded sites compared with invaded sites (Table 1). There was significant epigenetic differentiation among all loci (ΦST = 0.07, P = 0.03), which was driven by pairwise differences between the uninvaded site SF and the invaded sites SD (ΦST = 0.15, P = 0.02) and BWR (ΦST = 0.12, P = 0.03). However, these comparisons were not significant after Bonferroni correction. Locus-by-locus AMOVA identified four loci that had the highest contribution to differences in methylation frequency among sites (Locus-06, ΦST = 0.50, P = 0.03; Locus-19, ΦST = 0.64, P = 0.006; Locus-37, ΦST = 0.36, P = 0.06; Locus-38, ΦST = 0.38, P = 0.04; Fig. 1).
Table 1.
sites from which Eastern Fence Lizard samples were obtained, the status of invasion by the Red Imported Fire Ant at each site (status), number of lizards sampled (n), and epigenetic diversity (epi-h) at MS-AFLP binary multi-locus data and for the genetic haplotype diversity (h) at MspI for geographic samples
| Site (Abbreviation) | Status | N | epi-h | h |
|---|---|---|---|---|
| Solon Dixon Forestry Education Center (SD) | Invaded | 5 | 0.30 | 0.13 |
| Blackwater River State Forest (BWR) | Invaded | 5 | 0.23 | 0.22 |
| Edgar Evins State Park (EE) | Uninvaded | 5 | 0.39 | 0.15 |
| St. Francis National Forest (SF) | Uninvaded | 5 | 0.39 | 0.33 |
| Total | 20 | 0.33 | 0.21 |
When comparing genetic variation using only MspI, we observed 34 variable loci of the 35 used for MS-AFLP analysis. Haplotype diversity (h) ranged from 0.13 to 0.33. There was significant genetic differentiation among all loci (ΦST = 0.12, P = 0.01). One pairwise comparison among sites (SD vs SF; ΦST = 0.21, P = 0.03) was significant, yet not significant after Bonferroni correction. We failed to detect significant differences in epigenetic differentiation among sites at all loci when redundancy analysis (RDA) was used to investigate epigenetic variation while controlling for genetic variation.
Discussion
Currently, environmental epigenetic studies focus primarily on plants [7]; however, there is a growing body of work that shows DNA methylation has an important role in the ecology of vertebrates [30, 31–34]. Our results reveal that epigenetic variation in DNA methylation patterns can vary with the presence of an invasive species. Some loci had among-site variation in methylation frequency that did not map with Fire Ant invasion status; however, there was more DNA methylation in uninvaded than in Red Imported Fire Ant-invaded sites (Fig. 1). These findings suggest that invasion of Red Imported Fire Ants may disrupt patterns of DNA methylation.
Our results detected greater epigenetic diversity than genetic diversity at each site with a relatively low level of signal in DNA methylation among sites. There was a pattern of greater methylation in uninvaded sites, and an uninvaded site had different frequency of DNA methylation compared with two invaded sites. Contrasting epigenetic variation with genetic variation identified a comparison between an invaded and uninvaded site that was epigenetically different yet not genetically different. Yet, there was a comparison between an invaded and uninvaded site that differed both epigenetically and genetically. This suggests that ecologically relevant differences in both epigenetic and genetic data may exist among Eastern Fence Lizards whose habitat has been invaded by Red Imported Fire Ants. When comparing all markers, the RDA failed to detect significant differences in epigenetic variation after controlling for genetic variation. We believe this test was inhibited by a general lack of power based on small observed effect sizes, not that it indicates no epigenetic signal exists. This position is supported by the presence of differences in epigenetic data not mirrored in the genetic data, and that there was an overall pattern of greater methylation in uninvaded sites.
While there were different patterns of DNA methylation between invaded and uninvaded sites, the data suggest that there is a high level of intrinsic variability in DNA methylation variation. It is possible that the relatively high level of intrinsic variability coupled with low power of our study created noise and low level of signal in our data. This noise is likely exacerbated by the anonymity of the genomic elements being screened.
An important next step will be to directly determine if different DNA methylation states are associated with the phenotypic differences exhibited by Eastern Fence Lizard populations associated with the invasion of Red Imported Fire Ants. Next-generation sequencing-based techniques will be important in addressing this question and in identifying what parts of the genome are differentially methylated and associated with differences in phenotype [7]. Previous studies indicate that the behavioral, morphological and physiological adaptations of Eastern Fence Lizards to Fire Ant invasion are influenced by both cross-generational and within lifetime factors [23, 25, 27]. Change in epigenetic marks and epigenetic variation may provide an important process allowing rapid responses to novel environmental conditions. Common garden studies and developmental experimental manipulation [6, 36, 37] will allow more targeted investigation of environmentally induced DNA methylation changes and their role in promoting rapid adaptive phenotypic plasticity.
Methods
To determine if DNA methylation of Eastern Fence Lizards differed with the Fire Ant invasion status of a site, we compared methylation frequencies among Eastern Fence Lizards from invaded and uninvaded areas. We collected tissue samples (toe clips) from Eastern Fence Lizards captured from two sites invaded by Fire Ants (57–62 years ago; [35]): Solon Dixon Forestry Education Center, Escambia County, AL (SD, N = 5) and Blackwater River State Forest, Okaloosa County, FL (BWR, N = 6); and two sites not yet invaded by fire ants; Edgar Evins State Park, DeKalb County, TN (EE, N = 5) and St. Francis National Forest, Lee County, AR (SF, N = 6) (Table 1). We characterized individual morphology by body mass (g), snout vent length (SVL; cm), and hind limb length (RHL; cm). For comparisons with methylation among individuals we standardized RHL with SVL (as RHL/SVL) to focus on allometrically corrected variation in limb length. All samples were stored in 95% ethanol until DNA extraction with the Qiagen DNeasy Animal Tissue Kit (Qiagen, Valencia, CA).
We performed MS-AFLP following [36] We modified the typical AFLP protocol by replacing MseI with MspI and HpaII, methylation sensitive isoschizomeric enzymes that cut a CCGG restriction site but have different sensitivities to cytosine methylation. Together, four different types of variation can be scored see [38] Thus, if the MS-AFLP protocol is performed independently for each enzyme for each individual, the resulting banding pattern indicates the methylation state of a particular restriction site.
We digested ∼200 ng of DNA with 10U of restriction enzymes (MspI plus EcoRI and HpaII plus EcoRI) in a 20 μl reaction incubated at 37°C for 3 h (all enzymes New England Biolabs Ipswich, MA). We then ligated double stranded adaptors to the digested fragments with T4 DNA ligase (New England Biolabs Ipswich, MA). We conducted preselective polymerase chain reaction (PCR) with primers designed for the adaptors (MspI/HpaII: ATCATGAGTCCTGCTCGG; EcoRI: TACTGCGTACCAATTCA) at a final volume of 25 μl. We then performed selective PCR using primers with additional bases (MspI/HpaII: ATCATGAGT CCTG CTCGGTCAT, EcoRI: 6-FAM-TACTGCGTACCAATTCAGC and HEX-TACTGCGTACCAATTCACG). We sent PCR products to the Iowa State University DNA facility for fragment analysis. We used PEAKSCANNER (Applied Biosystems Foster City, CA) to analyse gel images and we scored band presence or absence manually.
We scored all individuals at each enzyme combination and identified the methylation state for each restriction site (locus). We ran the protocol twice for two or three individuals to determine which restriction sites were reliably detected (we only present reliable loci here). We adopted a conservative approach to scoring the gel images as AFLP-type reactions can generate variable banding among and within individuals. For a scored position to be considered reliable, the bands had to be identical and clearly distinguishable in each replicate of a given sample. Also, if subsequent reactions on additional samples generated inconsistent or unclear bands, or bands occurred at highly variable intensities at a locus, that locus was dropped from the analysis. For epigenetic analysis, we generated data as two categories; methylated loci (MS-AFLP Type II or Type III) or not methylated loci (MS-AFLP Type I or Type IV) and constructed binary epi-haplotypes. For genetic analysis, we used the results from the MspI/EcoRI reactions.
To determine if presence of Fire Ants affected patterns of DNA methylation in Eastern Fence Lizards, we used a z-test to determine if the proportion of methylation differed between invaded and uninvaded sites; we then used a GLM in a posthoc test to identify the loci that had the highest contribution to any significant difference observed. We compared methylation state at each locus with invasion status, mass, and RHL/SVL using generalized linear models for binary distributions in R with package BRGLM for bias reduction due to separation of data (i.e. when treatments are near to or are one hundred percent different).
We assessed epi-haplotype (epi-h) diversity of the binary methylated/not methylated data to estimate diversity of DNA methylation using GENALEX-6 [39]. We then estimated differentiation of DNA methylation among samples by AMOVA using GENALEX-6 to calculate ΦST over all loci, pairwise among sites, and independently for each locus to identify outlier loci that had different methylation patterns among sites. We used 9999 permutations to estimate statistical significance for all AMOVA runs.
We assessed genetic variation among individuals using the MspI/EcoRI restriction enzyme data. We estimated haplotype diversity (h) for each site, conducted AMOVA over all and pairwise among sites, and calculated genetic distances as the number of pairwise differences using GENALEX-6. We used RDA to determine if significant differences observed in epigenetic differentiation remained after controlling for the observed genetic differentiation among sites using the online MASAME RDA app for R (available at http://mb3is.megx.net/gustame/constrained-analyses/rda last accessed 4/23/2016).
Acknowledgments
We thank S. Graham and J. Newman for help with lizard collection, G. Dewitt, D. Fricken, M. Goldy-Brown, A. Hollowell, M. Hook, C. Norjen, M. S. McGinley, L. Horne, A. Pianovich, A. Jacobs and M. O’Brien for lizard care. We thank the Landsdale family for access to their land and lizards and personnel at St. Francis National Forest, Edgar Evins State Park, Blackwater River State Forest, Geneva State Forest, Conecuh National Forest, and especially the Solon Dixon Forestry Education Center for logistical support. We thank the Armstrong State University STEP program, Department of Biology, and College of Science and Technology and the National Science Foundation (DEB0949483 to TL) for funding. This research adheres to the Guidelines for the Use of Animals in Research and the Institutional Guidelines of Penn State University (IACUC #35780), and animal collection was permitted by the respective States.
Data available at: DRYAD. doi:10.5061/dryad.91c0t
Conflict of interest statement. None declared.
References
- 1. Gibbons JW, Scott DE, Ryan TJ. et al. The global decline of reptiles, déjà vu amphibians. Bioscience 2000;50:653–66. [Google Scholar]
- 2. Goldewijk KK. Estimating global land use change over the past 300 years: the HYDE Database. Global Biogeochem Cycles 2001;15:417–33. [Google Scholar]
- 3. Rosenzweig C, Karoly D, Vicarelli M. et al. Attributing physical and biological impacts to anthropogenic climate change. Nature 2008;453:353–7. [DOI] [PubMed] [Google Scholar]
- 4. Strauss SY, Lau JA, Carroll SP. Evolutionary responses of natives to introduced species: what do introductions tell us about natural communities? Ecol Lett 2006;9:357–74. [DOI] [PubMed] [Google Scholar]
- 5. Holliday R. Epigenetics: a historical overview. Epigenetics 2006;1:76–80. [DOI] [PubMed] [Google Scholar]
- 6. Herrera CM, Pozo MI, Bazaga P. Jack of all nectars, master of most: DNA methylation and the epigenetic basis of niche width in a flower living yeast. Mol Ecol 2012;21:2602–16. [DOI] [PubMed] [Google Scholar]
- 7. Schrey A, Alvarez M, Foust CM. et al. Ecological epigenetics: beyond MS-AFLP. Integr Comp Bio 2013;53:340–50. [DOI] [PubMed] [Google Scholar]
- 8. Herrera CM, Bazaga P. Epigenetic differentiation and relationship to adaptive genetic divergence in discrete populations of the violet Viola cazorlensis. New Phytol 2010;187:867–876. [DOI] [PubMed] [Google Scholar]
- 9. Herrera CM, Bazaga P. Untangling individual variation in natural populations: ecological, genetic and epigenetic correlates of long-term inequality in herbivory. Mol Ecol 2011;20:1675–88. [DOI] [PubMed] [Google Scholar]
- 10. Verhoeven KJF, Jansen JJ, van Dijk PJ. et al. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 2010;185:1108–18. [DOI] [PubMed] [Google Scholar]
- 11. Nätt D, Rubin CJ, Wright D. et al. Heritable genome-wide variation of gene expression and promoter methylation between wild and domesticated chickens. BMC Genomics 2012;13:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jablonka E, Raz G. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Q Rev Biol 2009;84:131–76. [DOI] [PubMed] [Google Scholar]
- 13. Johannes F, Porcher E, Teixeira FK. et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genetics 2009;5:e1000530.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cropley JE, Dang THY, Martin DIK. et al. The penetrance of an epigenetic trait in mice is progressively yet reversibly increased by selection and environment. Proc Biol Sci 2012;279:2347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rando O, Verstrepen K. Timescales of genetic and epigenetic inheritance. Cell 2007;128:655–68. [DOI] [PubMed] [Google Scholar]
- 16. Sheriff MJ, Love OP. Determining the adaptive potential of maternal stress. Ecol Lett 2013;16:271–80. [DOI] [PubMed] [Google Scholar]
- 17. Gao L, Geng Y, Li B. et al. Genome-wide DNA methylation alterations of Alternathera philoxeroides in natural and manipulated habitats: implications for epigenetic regulation of rapid responses to environmental fluctuation and phenotypic variation. Plant Cell Environ 2010;33:1820–1827. [DOI] [PubMed] [Google Scholar]
- 18. Allen CR, Epperson DM, Garmestani AS. Red imported fire ant impacts on wildlife: a decade of research. Am Mid Nat 2004;152:88–103. [Google Scholar]
- 19. Code of Federal Regulations. 2015. Title 7. Agriculture. Part 301.81. Subpart: imported fire ant. https://www.law.cornell.edu/cfr/text/7/301.81-11 (23 April 2015, date last accessed: 4/23/2016).
- 20. Epperson DM, Heise CD. Nesting and hatchling ecology of gopher tortoises (Gopherus polyphemus) in southern Mississippi. J Herpetol 2003;37:315–24. [Google Scholar]
- 21. Tschinkel WR. 2006. The Fire Ants. Cambridge, Massachusetts: Harvard University/Belknap Press [Google Scholar]
- 22. Langkilde T, Freidenfelds NA. Consequences of envenomation: red imported fire ants have delayed effects on survival but not growth of native fence lizards. Wildlife Res 2010;37:566–73. [Google Scholar]
- 23. Langkilde T. Holding ground in the face of invasion: native fence lizards (Sceloporus undulatus) do not alter their habitat use in response to introduced fire ants (Solenopsis invicta). Can J Zool 2009a;87:626–34. [Google Scholar]
- 24. Freidenfelds NA, Robbins TR, Langkilde T. Evading invaders: the effectiveness of a behavioral response acquired through lifetime exposure. Behav Ecol 2012;23:659–64. [Google Scholar]
- 25. Robbins TR, Langkilde T. The consequences of lifetime and evolutionary exposure to toxic prey: changes in avoidance behavior through ontogeny. J Evol Bio 2012;25:1937–46. [DOI] [PubMed] [Google Scholar]
- 26. Robbins T, Freidenfelds N, Langkilde T. Native predator eats invasive toxic prey: evidence for increased incidence of consumption rather than aversion-learning. Biol Invasions 2013;15:407–15. [Google Scholar]
- 27. Langkilde T. Invasive fire ants alter behavior and morphology of native lizards. Ecology 2009b;90:208–217. [DOI] [PubMed] [Google Scholar]
- 28. Graham SP, Freidenfelds NA, McCormick GL. et al. The impacts of invaders: basal and acute stress glucocorticoid profiles and immune function in native lizards threatened by invasive ants. Gen Comp Endocrinol 2012;176:400–408. [DOI] [PubMed] [Google Scholar]
- 29. Trompeter WP, Langkilde T. Invader danger: lizards faced with novel predators exhibit an altered behavioral response to stress. Horm Behav 2011;60:152–8. [DOI] [PubMed] [Google Scholar]
- 30. Massicotte R, Angers B. General-purpose genotype or how epigenetics extend the flexibility of a genotype. Gen Res Int 2012;2012:7 doi:10.1155/2012/317175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Angers B, Castonguay E, Massicotte R. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol Ecol 2010;19:1283–95. [DOI] [PubMed] [Google Scholar]
- 32. Massicotte R, Whitelaw E, Angers B. DNA methylation: a source of random variation in natural populations. Epigenetics 2011;6:421–7. [DOI] [PubMed] [Google Scholar]
- 33. Liu S, Sun K, Jiang T. et al. Natural epigenetic variation in the female great roundleaf bat (Hipposideros armiger) populations. Mol Genet Genom 2012;287:643–50. [DOI] [PubMed] [Google Scholar]
- 34. Liebl A, Schrey A, Richards C. et al. Patterns of DNA methylation throughout a range expansion of an introduced songbird. Integr Comp Biol 2013;53:351–8. [DOI] [PubMed] [Google Scholar]
- 35. Callcott AMA, Collins HL. Invasion and range expansion of imported fire ants (Hymenoptera: Formicidae) in North America from 1918–1995. Florida Entomologist 1996;79: 240–51. [Google Scholar]
- 36. Richards C, Schrey A, Pigliucci M. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol Lett 2012;15:1016–25. [DOI] [PubMed] [Google Scholar]
- 37. Nicotra AB, Segal D, Hoyle GL. et al. Adaptive plasticity and epigenetic variation in response to warming in an Alpine plant. Ecol Evol 2015;5:634–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salmon A, Clotault J, Jenczewski E. et al. Brassica oleracea displays a high level of DNA methylation polymorphism. Plant Sci 2008;174:61–70. [Google Scholar]
- 39. Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinformatics 2012;28:2537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]