Abstract
In utero exposure to environmental endocrine disruptors can cause transgenerational effects in the males of subsequent generations. DNA methylation (5 mC) was suggested, but being challenged as the molecular carrier of such epigenetic information. In a recent study, Schuster et al. show a changed small RNA profile changed in the sperm of F3 generation after F0 in utero vinclozolin exposure, suggesting additional transgenerational epigenetic carriers for endocrine disruptor effects, other than DNA methylation.
Gestational exposure to environmental endocrine disruptors at the critical time window of germline development can result in epigenetic changes (i.e. epimutations) in the germline, and the epimutations and their associated phenotypes can be memorized and transmitted to subsequent generations [1]. It was first reported in 2005 [2], that phenotypes (impaired spermatogenesis and subfertility) induced by gestational exposure to an endocrine disruptor vinclozolin could be inherited up to F4 subsequent generation through the male germline, without initial toxin exposures in rats. However, the exact molecular carriers in the sperm responsible for this transgenerational relay of epigenetic information remain elusive.
Previous studies have focused on DNA methylation (5 mC) as a potential epigenetic mark that underlies the vinclozolin-induced transgenerational effects [3], because documented evidences have indicated that DNA methylation at certain loci can escape the epigenetic reprogramming and pass through germline [4]. However, data on altered DNA methylation profiles between vinclozolin exposed and control sperm remain inconclusive, and recent evidence has reported an inconsistent DNA methylation profiles between F1 and F2 mouse sperm after their mother or grandmother’s exposure to vinclozolin [5], suggesting that even if the DNA methylation in the fetal germ cells are affected, it could be corrected in the subsequent generation [5], thus casting doubt on the role of DNA methylation as the epigenetic carrier for the transgenerational inheritance of phenotypes. Similar cases of corrected DNA methylation profiles during the subsequent germline reprogramming were also reported in gestational nutritional stress [6] and intracytoplasmic sperm injection (ICSI), a type of assisted reproductive technologies [7]. These studies have raised questions over the role of DNA methylation in transgenerational epigenetic inheritance, at least as a persistent heritable mark across generations. Nevertheless, it should be noted that in the cases of vinclozolin exposures, different groups usually used different species, different way of drug delivery and different duration of treatment, which might cause variations in results. Moreover, different methods for DNA methylation profiling may also cause inconsistent results, and no groups have used the whole-genome shotgun bisulfite sequencing (WGSBS) approach [8], which has the highest coverage and could be used to settle the arguments. For example, WGSBS have been recently used to assess the effects of different diets (high-fat diet, low-protein diet and normal diet) on DNA methylation in mouse sperm [9]. The authors did not find consistent changes in sperm methylome of exposed males among the three groups, which led to a conclusion that DNA methylation may not be the epigenetic carrier in the sperm of exposed males [9], suggesting the involvement of other epigenetic players. Interestingly, using these paternal nutritional stress models (high-fat diet, low-protein diet), recent emerging evidences have shown that a subtype of sperm non-coding RNAs: tsRNAs (tRNA-derived small RNAs), is enriched in sperm [10], and their levels are sensitive to the paternal high-fat diet [11] and low-protein diet [12], in addition to the changes in sperm miRNAs [11–13] and RNA modifications in sperm tsRNAs-enriched RNA fractions [11]. Further functional analyses have demonstrated that injections of sperm tsRNAs or miRNAs could change embryonic transcriptome and induce offspring phenotypes that recapitulate their paternal nutritional exposures [11–13], suggesting RNAs and RNA modifications as active epigenetic carriers in sperm that can transmit acquired phenotypes to the offspring.
Under these backgrounds, Schuster et al. [14] now provided fresh insights into the potential molecular carrier of vinclozolin-induced transgenerational effects in a rat model as previously used [2], by identifying altered small RNA profiles in the sperm of F3 rats, whose great grandmothers were intraperitoneal injected with vinclozolin during gestation. Schuster et al. found in their study that a majority of tsRNAs were notably dysregulated in the sperm of F3 rats after ancestral vinclozolin exposure, in addition to the alterations of sperm miRNAs, and mitosRNAs (mitochondrial genome–encoded small RNAs), which was recently discovered in testicular cells [15] with their function remains undefined. Schuster et al. [14] further provided a correlation analysis between the altered sperm miRNAs/tsRNAs and the vinclozolin-induced sperm differential DNA methylation regions (DMRs) in the males of F3 generation. They found that sequence targets of tsRNAs, but not miRNAs, showed a correlation with DMRs of male PGCs and sperm. The discovery of the transgenerationally altered sperm RNA profiles and the association with DMRs is interesting, which implicated a scenario that the altered sperm RNA profiles, particularly tsRNAs, might facilitate the formation of certain DMRs, and meanwhile generate the same altered RNA profiles in subsequent generations. Such a scenario remains highly speculative given the limited data provided, but warrants further investigations; particularly a further examination of F1 and F2 sperm RNAs is needed.
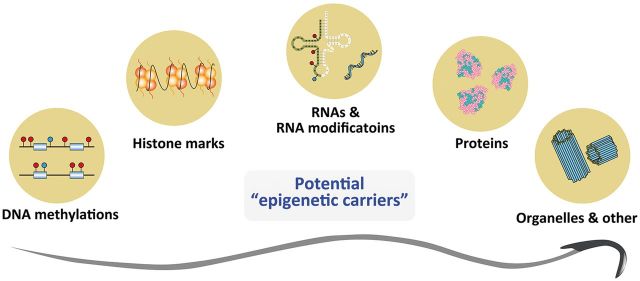
While increasing evidence has demonstrated the phenomenon of acquired epigenetic inheritance, the identity of epigenetic carriers in the germline remain elusive, although recent data have provided a direct causal effect of sperm RNAs [11–13, 16] Since RNAs are less stable than DNAs, it remains unknown whether the sperm-borne RNAs act only transiently after entering oocytes or should they persist their effects through early embryo development. In either case, mechanisms such as RNA modifications to maintain, or regulatory loops to renew or relay the effect of initial RNAs may facilitate their effects in transgenerational epigenetic inheritance. Also, to identify the interplay between sperm RNAs and DNA modifications [both 5 mC and the newly identified N6-mA (N6-methyladenine) [17] as well as histone modifications [18] or long-range genome interactions would represent a future direction of research. Moreover, it should be noticed that in addition to the epigenetic players mentioned above, sperm can also deliver other unexpected cargos into oocytes, such as functional proteins, chemicals and remnant organelles (Fig. 1). Indeed, recent studies have shown that centrioles contributed by sperm showed exceptional persistence during the embryo development in C. elegans [19]; could they provide unidentified information to the developing embryos, in a similar or distinct manner of the contribution of maternally derived mitochondria? The significance of these sperm-borne molecules/components in the context of acquired inheritance might represent new realms of future research.
Figure 1:
A versatile of potential molecular carriers in the rat sperm that have the potential to mediate epigenetic inheritance of acquired traits from environmental exposures.
“The absence of evidence is not the evidence of absence”-Carl Sagan (1934-1996, astronomer)
Given the mystery of acquired inheritance remains, and the potential molecular carriers underlying these epigenetic phenomena now expanding, the same patience might be required to equal the efforts of intensive scientific explorations in this developing field.
Acknowledgement
Research in Qi Chen lab is currently supported by startup funds from University of Nevada School of Medicine.
Conflict of interest statement. None declared.
References
- 1. Skinner MK. Endocrine disruptors in 2015: Epigenetic transgenerational inheritance. Nat Rev Endocrinol 2016;12:68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anway MD, Cupp AS, Uzumcu M. et al. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 2005;308:1466–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guerrero-Bosagna C, Settles M, Lucker B. et al. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One 2010;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 2012;13:153–62. [DOI] [PubMed] [Google Scholar]
- 5. Iqbal K, Tran DA, Li AX. et al. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol 2015;16:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Radford EJ, Ito M, Shi H. et al. In utero effects. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 2014;345:1255903.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Waal E, Yamazaki Y, Ingale P. et al. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci USA 2012;109:4163–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010;11:191–203. [DOI] [PubMed] [Google Scholar]
- 9. Shea JM, Serra RW, Carone BR. et al. Genetic and Epigenetic Variation, but Not Diet, Shape the Sperm Methylome. Dev Cell 2015;35:750–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng H, Shi J, Zhang Y. et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 2012;22:1609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen Q, Yan M, Cao Z. et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016;351:397–400. [DOI] [PubMed] [Google Scholar]
- 12. Sharma U, Conine CC, Shea JM. et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 2016;351:391–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grandjean V, Fourré S, De Abreu DA. et al. RNA-mediated paternal heredity of diet-induced obesity and metabolic disorders. Sci Rep 2015;5: 18193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuster A, Skinner MK, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2016;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ro S, Ma HY, Park C. et al. The mitochondrial genome encodes abundant small noncoding RNAs. Cell Res 2013;23:759–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gapp K, Jawaid A, Sarkies P. et al. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 2014;17:667–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu TP, Wang T, Seetin MG. et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016;532:329–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siklenka K, Erkek S, Godmann M. et al. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015;350:6261. [DOI] [PubMed] [Google Scholar]
- 19. Balestra FR, von Tobel L, Gonczy P. Paternally contributed centrioles exhibit exceptional persistence in C. elegans embryos. Cell Res 2015;25:642–4. [DOI] [PMC free article] [PubMed] [Google Scholar]