Abstract
Transposons are playing an important role in the evolution of eukaryotic genomes. These endogenous virus-like elements often amplify within their host genomes in a species specific manner. Today we have limited understanding when and how these amplification events happens. What we do know is that cells have evolved multiple line of defenses to keep these potentially invasive elements under control, often involving epigenetic mechanisms such as DNA-methylation and histone modifications. Emerging evidence shows a strong link between transposon activity and human aging and diseases, as well as a role for transposons in normal brain development. Controlling transposon activity may therefore uphold the fine balance between health and disease. In this article we investigate this balance, and sets it in relation to allostatic load, which conceptualize the link between stress and the “wear and tear” of the organism that leads to aging and disease. We hypothesize that stress-induced retrotransposon reactivation in humans may be used to estimate allostatic load, and may be a possible mechanism in which transposons amplify within species genomes.
Keywords: transposons, retrotransposons, stress, allostatic load, allostasis, epigenetic silencing
Introduction
Ever since the stress concept was first seriously exploited by Hans Selye in the 1950s it has been disputed. The classic example of one of these disputes is how our bodies reacts to a rollercoaster ride: you experience fears or trills, your blood pressure goes up, heart rate increases, and your HPA-axis releases cortisol into your bloodstream. Typical symptoms of a stress response, but was this a stressful experience? Was it bad for you? Selye coined the words eustress and distress, simply meaning positive and negative stress [ 1 ], in an attempt to resolve this problem.
Today, to avoid misconceptions, stress is often described in more neutral terms, as a processes in which external or internal factors, positive or negative, may cause a threat to homeostasis where the organism is forced to change its internal physiological parameters to maintain stability (allostasis) [ 2 ]. But maintaining stability through allostasis comes with a cost, the allostatic load, which causes accumulated wear and tear on the organism, and eventually leads to aging and disease [ 3 , 4 ]. In nature, allostatic load is directly related to a less fit organism, thus the better an organism is genetically adapted to its current environment, the lower the allostatic load [ 5 ]. Allostatic load may become very high if an organism is unable to cope with its environment, say that it is unable to escape climate change or human exploitation [ 6 ]. In such scenarios, there are strong selective pressures for populations to change and genetically adapt. In this paper we attempt to explore an emerging mechanism that we recently stumbled across when we studied chronic cortisol exposure (the main human stress hormone) in preschool children [ 7 ]: mammalian stressed-induced retrotransposon (re)activation. This mechanism has the potential to help us better understand the concepts of eustress and distress, and the role of allostatic load in disease and evolution.
Presenting the transposable element
The basic principles about transposons have recently been reviewed elsewhere [ 8 ]. Transposons are virus-like elements that are native to all eukaryotic genomes. Barbra McClintock who discovered them in maize called them “jumping genes” because of their ability to move within their host’s genome [ 9 ]. There are two types: Class 1 retrotransposons that move through a copy and paste mechanism, and Class 2 DNA-transposons that move through a cut and paste mechanism. In most mammalian genomes, which will be the main focus here, retrotransposons are the most abundant [ 10 ]. Retrotransposons can be further divided into long terminal repeats (LTR) such as many endogenous retroviruses, or non-LTRs such as long and short interspersed elements (LINE and SINE). LTR retrotransposons and some non-LTR retrotransposons such as LINEs are autonomous, meaning they carry their own reverse transcriptase that they use after transcription (copy) to reintegrate themselves (paste) into a novel location of their host’s genome. This makes them closely related to retroviruses, which is very clear in some species where they can become exogenous, viral, and infect other individuals, such as the porcine endogenous retrovirus in pigs or gypsy in Drosophila [ 11 , 12 ]. Other non-LTR retrotransposons are non-autonomous, such as SINEs, meaning they do not carry their own reverse transcriptase and are therefore dependent on autonomous retrotransposons for their proliferation, thus are sometimes called “a parasite’s parasite” [ 13 , 14 ]. In most mammal species non-autonomous retrotransposons such as Alu, SVA and B1 often hijack the reverse transcriptase of the widespread LINE1 (L1) elements.
The good side: transposons and evolution
While previously often described as “junk”, “selfish” or “parasitic” DNA, transposons are today known to play an important role in gene evolution, not only by contributing with raw genetic material, but also for donating functional elements such as promoters and enhancers, and as mutagenic drivers for gene knockouts, homologous recombination, and novel gene isoforms and splice variants [ 15 , 16 ]; modifications which sometimes affect fitness [ 8 ]. In Drosophila , insertions of transposons have been estimated to account for about half of all spontaneous phenotype-inducing mutations [ 17 ].
Broadening the perspective, it may be argued that the diversity of transposable elements at least quantitatively marks species differences better than do the much more genetically conserved genes. In fact, between 40-50% of the human genome originates from retrotransposons (much less DNA-transposons), which may be compared to the 1.5% that codes for proteins [ 14 , 18 ]. This abundance is a result of recent or ongoing genomic amplification events, where certain types of transposons become reactivated in the germline of certain classes, families, species or even strains of species. This is best described with some examples.
The human genome has three known active retrotransposon families: the evolutionary wide-spread L1 with more than 500 000 copies, the primate specific Alu (a SINE) with more than 1 million copies, and the SVA with about 3000 copies that is only found in the great apes [ 14 ]. In other species other transposons are active and contribute to genome amplification. The mouse genome is for instance marked by recent activities of L1, B1, IAP and MusD/ETn retrotransposons. It has been shown that 15% of all spontaneous germ-line mutations in laboratory mice originate from IAP and MusD/Etn transpositions [ 19 ], and that as much as 60% of the IAP and 25% of MusD/ETn are strain-specific when comparing closely related mouse strains [ 20 ]. A similar phenomena is observed in humans where one-third of the human-specific L1 insertions differ between unrelated individuals, and where a novel L1 or Alu germline insertion is estimated to occur in about every 100:th [ 21 , 22 ] and 20:th [ 23 ] birth, respectively. So active retrotransposons are highly polymorphic, which means that they shape mammalian genomes as we speak, but the impact of transposon invasion is probably best illustrated in an invertebrate. In two Drosophila species, D. melanogaster [ 24 ] and D. simulans [ 25 ] the P element (a cut and paste DNA-transposon) have independently invaded their genomes within a few decades; during the time we have studied them in the lab.
Furthermore, the LINE class of autonomous retrotransposons is particularly interesting in mammalian evolution since it is highly active in most mammals but not in bats, in which it has become extinct [ 26 ]. The reason for this is poorly understood, but bats are also genetically unique since they are the only mammalian order showing active DNA-transposons, of which some are uniquely found in bats and insects (but not other mammals) [ 27 , 28 ]. This proves the amazing phenomena of horizontal gene transfer between phylum, maybe between a predator and its prey. In other vertebrates, similar gene transfers have been observed, such as the AviRTE retrotransposon between bird species and human pathogenic endoparasitic nematodes [ 29 ], and in the case of the BovB retrotransposon, between diverse groups of vertebrates and ticks that parasite them [ 30 ].
The examples above illustrates only a few different aspects of how transposons contribute to germline evolution by generating genetic diversity and amplification within species, and sometimes horizontal transfers between species. While all these aspects, as long as they are kept under some control, seem beneficial for species evolution, as will be discussed below, there is a tradeoff between evolvability in populations and the risk of disease in individuals.
The bad side: Controlling retrotransposons to avoid disease
“Jumping” transposons can also be a threat to the cell. The risk of inducing maladaptive genomic changes, such as knocking out important genes or their regulatory regions, are inevitable when transposons are active. At least 50 human diseases are known to be immediately caused by retrotransposon insertions into important regions of the germline genome [ 15 , 31 ]. When transposition gets too high it leads to genome instability, where the integrity of the genome becomes compromised, resulting in higher degree of aneuploidy, chromosomal rearrangements, and DNA damage [ 32 , 33 ]. When genome instability occurs in the germline it often leads to infertility [ 34 ], but it is also frequently observed in somatic tumor cells [ 32 ], or cell lines derived from cancerous tissue [ 35 ].
To keep genomes intact, healthy cells keep very tight control over the expression of transposons. In fact, the many possible routes to keep transposable elements silenced may be a consequence of the risks involved in losing control. Firstly, there are post-transcriptional mechanisms that may inhibit transposition either immediately after transcription, through micro-RNA related pathways [ 36 ] or so called PIWI-interacting RNA (piRNA), as well as through proteins that target and repress retrotransposon RNA and/or transposon specific enzymes, such as the reverse transcriptase [ 37–42 ]. Secondly, epigenetic silencing mechanisms, such as DNA methylation, repressive histone modifications, and ATP-dependent chromatin remodeling, play critical roles in controlling transposable elements [ 34 ]. To exemplify how cells control transposon activity we will focus on two mechanism: [A] quickly evolving Krüppel-associated box (KRAB) zinc finger proteins (KZNF), and [B] piRNAs.
KZNFs is a very large group of transcriptional repressors containing more than 400 members in humans, often located in genomic clusters, of which about one third are primate specific [ 43 ]. When a KZNF binds DNA it recruits Krüppel associated protein 1 (KAP1; also known as TRIM28), which in turn form complexes with H3K9 methyltransferase SETDB1, the histone deacetylase-containing NuRD complex, and heterochromatin protein 1 (HP1), resulting in epigenetically silenced chromatin. KAP1 has been shown to co-localize with active types of retrotransposons such as L1 and SVA in humans [ 44–46 ], and proteins from the same family have been shown to do so in mice [ 47 , 48 ]. This suggests a conserved relationship between KZNFs, the TRIM protein family, and retrotransposons. More figuratively speaking, KZNFs may be seen as gatekeepers, keeping retrotransposons epigenetically imprisoned, always on alert to sound the alarm trough TRIM-proteins if finding open chromatin in the vicinity of a retrotransposon. The importance of KZNFs as gatekeepers is illustrated by what seems to be an evolutionary arms race between these transcription factors and retrotransposons; when a new active variant of retrotransposon evolves, this is soon followed by a KZNF able to bind and epigenetically silence it, which in turn is followed by retrotransposon mutants with changes in the binding site for the novel KZNF [ 46 ].
As with the KZNFs, piRNAs appear in genomic clusters, but are transcribed by RNA polymerase II as a single-stranded precursor unit [ 49 ]. These precursors of hundreds of nucleotides are processed and transported back to the nucleus where they become mature piRNAs of 26–31 nucleotides. Similar to KRAB zinc finger proteins, piRNAs are used as guides to find transposon specific sequences, and by interacting with PIWI-like family proteins, such as MILI, MIWI and MIWI2, they may silence these transposable elements [ 50 , 51 ] by inducing DNA methylation [ 52 ] or post-transcriptional gene silencing [ 53 , 54 ]. Initial findings suggested that piRNA exclusively controlled transposable elements in the male germline as a way to control transposon activity during germline epigenetic reprogramming, when epigenetic silencing is nearly completely abolished [ 55 , 56 ]. More recent findings have revealed potential roles also in somatic cell types such as neurons [ 57–59 ].
The complicated side: Transposons in somatic diseases and normal development
Transposons may not only be a problem in the germline, or in somatic tumors, but also in other somatic cells. One interesting example are recent findings in several autoimmune diseases, such as Aicardi-Goutières syndrome, chilblain lupus and systemic lupus erythematosus, where mutations in the TREX1 gene may cause impairments in a mechanism of the innate immune system that senses exogenous reverse transcription, resulting in false detection of endogenous retrotransposons as if they were exogenous retroviruses [ 40 , 60 ]. If retrotransposons play roles in other autoimmune diseases remain to be investigated. Nevertheless, these autoimmune diseases illustrate an interesting trade-off in eukaryotic evolution; between being able to tolerate endogenous retrotransposons and detecting invading retroviruses.
Furthermore, in the “Loss of heterochromatin model of aging” first proposed in 1997, it is suggested that silent heterochromatin formed during embryogenesis decays during the aging process, resulting in reactivation of epigenetically silenced genomic regions [ 61 ]. Since then, global loss of heterochromatin particularly at gene-poor transposon-rich regions has repeatedly been associated with aging in several tissues of many eukaryotic species [ 62–65 ], as well as human diseases that result in premature aging such as Hutchinson-Gilford Progeria Syndrome [ 66 ] or Werner syndrome [ 67 ]. Furthermore, heterochromatin protein 1 (HP1) plays an important role in maintaining several repressive histone methylations, such as H3K9me3, that are highly associated with heterochromatin and retrotransposons in humans [ 68 ]. Overexpressing HP1 in Drosophila , increases longevity, while repressing HP1 decreases longevity [ 69 ] compared to wild type flies, which illustrates a direct relationship between aging and loss of repressive chromatin. Finally, there are some indications that heterochromatin loss is more related to functional decline (frailty) than actual biological age [ 69 , 70 ], which is in line with the allostatic load concept; that at least some aspects of aging is a consequence of accumulated wear and tear, and not the biological clock. Exactly what role retrotransposons play in aging, if they are directly involved or just passive markers, is still poorly understood. Nonetheless, it has recently been shown that the loss of epigenetic silencing in aging reactivates functional retrotransposons in several animal species including humans [ 71–74 ], which may contribute to increased genomic instability and decreased viability of the aging cell. Further proof for this is seen in yeast where meta-analysis shows a strong overlap between genes associated with phenotypes that have increased lifespan and genes involved in transposition of mobile elements [ 74 ]
While aging can be seen as an increase in frailty with an associated general increase in the risk of many diseases, there are also examples of specific somatic diseases associated with retrotransposon reactivation. For example, it was recently shown that L1 copy number was elevated in the prefrontal cortex of deceased patients with schizophrenia, bipolar disorder and major depression [ 75 ]. This is particularly interesting since actively dividing neural precursor cells in brain regions with high levels of neuronal plasticity (such as prefrontal cortex, hippocampus and caudate nucleus) have shown higher activity of retrotransposons compared to many other somatic tissues [ 76 , 77 ]. This has given rise to the idea that genetic mosaicism driven by retrotransposon reactivation may play a role in creating neural diversity in these regions, which may be critical for learning and memory.
A parallel may again be seen in Drosophila , where small RNA pathways controlling the expression of transposons are repressed in mushroom body neurons known to be involved in learning and memory, but not in other neural cell types, resulting in increased expression of transposons in this type of neuron [ 78 ]. To relate back to the aging discussion, the same pathways are further repressed in the aging fly [ 73 ]. This indicates a conserved role for transposable elements in age-dependent neurodevelopment and plasticity that may be controlled by small RNAs. The novel findings of functional piRNA in the mammalian brain [ 57–59 ] and loss of retrotransposon silencing in age-dependent neurodegenerative disorders such as Alzheimer's [ 79 ] supports this, but it also adds another layer of complexity to the roles played by retrotransposons in the brain. On the one hand the activity of transposable elements are needed for normal brain development, and on the other hand too much activity are associated with aging and disease.
Stress-induced transposon reactivation
So what does stress, or more specifically, allostatic load –the accumulated consequences of living in a non-optimal physical, physiological or psychological environment– have to do with retrotransposons? Stress-related diseases are thought to arise when the allostatic load is too high or has accumulated over time [ 3 ]. This is supported by the many human diseases that are affected by chronic and/or severe types of stress, functioning either as a direct trigger (e.g. post-traumatic stress disorder, ischemic heart disease), or increasing the risk and severity for the disorder (e.g. diabetes, obesity, auto-immune disease, heart diseases, drug addiction, major depression, schizophrenia) [ 80 ]. As outlined above, the control of transposable elements are partly lost in many of these complex diseases. Similarly, severe chronic stress, especially during early life, has been suggested to increase the rate of biological senescence [ 4 ]. Again, as we pointed out above, retrotransposons are reactivated as we get older. So stress, just as transposons, has an intimate relationship with both aging and disease. The question is: If stress-induced retrotransposon reactivation in humans is a reality, can we use it as an estimator of allostatic load? Furthermore, could stress-induced retrotransposon reactivation play a more active role in stress-induced disease and aging?
To resolve these issues we need to study direct relationships between stress and retrotransposon reactivation. In plants this relationship has been clear ever since Barbra McClintock discovered her “jumping genes” in maize [ 9 , 81 ], but in mammals, and more specifically humans, it has hardly been studied at all. Not until recently. We used next generation sequencing to study how chronically high levels of cortisol (measured in hair) in healthy 5-year old children affected whole-genome DNA-methylation, and found that it led to a general loss of DNA-methylation in blood cell retrotransposons [ 7 ]. This finding is particularly important since it is prospectively unbiased. There were no age, disease or even stress (measured by exposure to serious life events) differences that could have affected the results, only an association between chronically increased levels of systemic cortisol, caused by either eustress or distress, and loss of epigenetic silencing in retrotransposons. Furthermore, regions where high cortisol children had lost DNA-methylation, were enriched for the binding site of a “gatekeeper” KZNF (ZNF263), which points at a possible mechanism for the loss [ 7 ].
To fully understand the potential for retrotransposon reactivation as an estimator and/or a mediator of allostatic load, we need to distinguish between the acute and chronic effects of stress. Many transcription factors involved in the stress-response, such as the glucocorticoid receptor, bind promoters of epigenetic modifying enzymes and regulate their transcription, like DNA-methyltransferases [ 82 ] or histone methyltransferases such as Suv39h2 [ 83 ]. Bruce McEwen, that together with Eliot Stellar coined the word allostatic load [ 3 ], has together with Donald Pfaff’s lab showed that in rat brains, acute restraint stress increases Suv39h2 expression, followed by a global increase of repressive H3K9me3 histone marks, resulting in downregulation of B1 and IAP retrotransposons [ 83 , 84 ]. This would indicate that stress could lead to more epigenetic silencing and less retrotransposon activity, thus it may not be so relevant for allostatic load. On the contrary, other repressive histone methylations, such as H3K9me2, are lower after acute stress, and when exposing animals to chronic stress also H3K9me3 to some extent decreases [ 85 ]. So a global loss of epigenetic silencing could be a specific effect of chronic stress which would be interesting for the allostatic load concept. In brains of mice susceptible to chronic social defeat a global loss of the repressive H3K9me2 histone marks following the defeat confirms this relationship [ 86 ].
An interesting parallel to acute and chronic stress is observed in the brains of mice given acute and chronic injections of drugs of abuse. Both cocaine and morphine, when administered chronically in rodents, result in a global loss of repressive histone marks in mobile elements, sometimes followed by higher activity of retrotransposons [ 87–89 ]. As with acute stress exposure, H3K9me3 shows a strong opposite relationship after an acute cocaine injection [ 89 ]. Thus, while H3K9me3 are sensitive to pervious exposure, other repressive histone marks, such as H3K9me2, seem to always experience a global loss after stress or drug exposure, independent of previous exposure.
Because of the temporal complexity in retrotransposon regulation, it is critical for future research to focus on the long term consequences of stress by estimating the number of successful transpositions following chronic exposure to internal or external constraints (e.g. restraint stress, social defeat, drug exposure). This has been done in animal prenatal and postnatal stress models that induce schizophrenia like symptoms in the offspring. Here, pregnant mice were injected with polyriboinosinic-polyribocytidilic acid or neonatal macaques were chronically exposed to epidermal growth factor, in both cases resulting in more copy numbers of brain L1 retrotransposons [ 75 ].
Conclusion and future directions
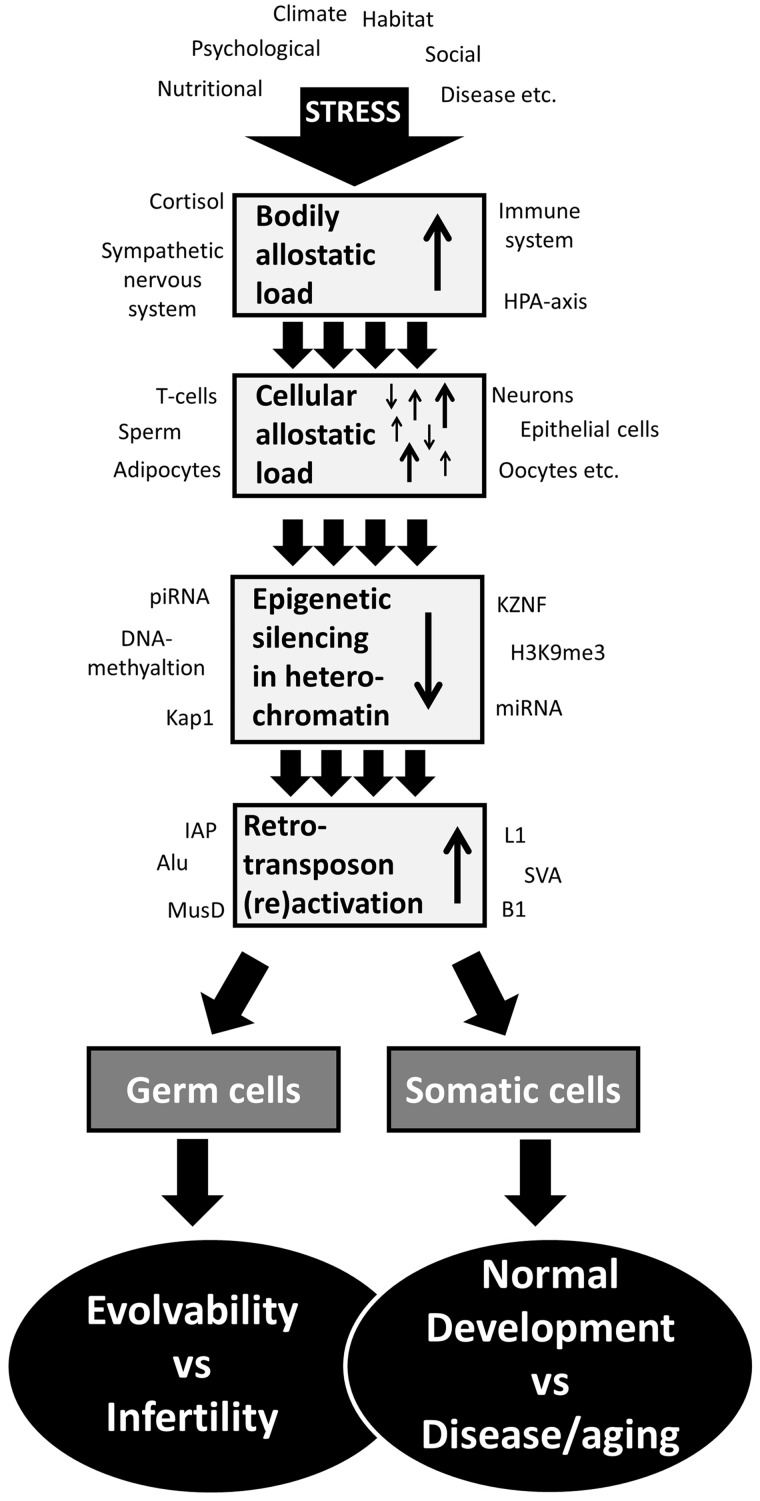
Integrating stress-induced retrotransposon reactivation into the allostatic load concept holds many promises ( Fig. 1 ). Still it is evident that there are several aspects of stress-induced retrotransposon reactivation that needs to be further investigated before we can do that. Firstly, transposons are challenging to study because of their repetitive nature, both locally (they are often flanked by other genomic repeats which makes them sometimes difficult to sequence), and globally (they are often found in multiple copy numbers across a genome which sometimes makes it difficult to pinpoint their exact location). Thus many common genome analysis pipelines disregard retrotransposon sequences, simply because they are difficult to interpret, which probably have led to an underestimation of the impact of retrotransposons in the current literature [ 90 ]. Furthermore, we still know very little of how chronic eustress may affect retrotransposon reactivation. Some studies have shown that voluntary and forces exercise may both increase retrotransposon reactivation [ 91–93 ]. Another problem in associating the allostatic load concept with retrotransposon reactivation is the limitations of the current tools to estimate organismal “wear and tear”, which needs to be considered from a whole body perspective. During the last decade promising attempts have been made to combine neuroendocrine (e.g. blood cortisol, dopamine and catecholamine release), immune (e.g. blood Interleukin-6 and Insulin-like growth factor release), metabolic (e.g. blood cholesterol, glucose and insulin release), cardiovascular and respiratory (e.g. blood pressures, heart rate and peak expiratory flow), as well as anthropometric (e.g. waist-to-hip ratio and body mass index) health biomarkers into indexes of allostatic load (reviewed by Juster et. al. [ 94 ]). Still none of these indexes has been used in relation to retrotransposon activity. It becomes even more complicated if we presume that different types of environmental challenges may increase allostatic load in some cells, while not affecting or even decreasing it in others, maybe in an activity dependent manner. This is for example seen in rats exposed to acute restraint stress, which increases retrotransposon activity in the cerebellum but decreases it in the hippocampus [ 83 ]. Nonetheless, the differences in retrotransposon activity between cell types of different tissues may help us understand why some stressors increase disease risks in some tissues and not in others. The potential to find a cell type that reliably measures average allostatic load of the body is off course very intriguing. Monitoring transposon activity and epigenetic control mechanisms in such a cell type could be an estimator of the current allostatic state, while counting retrotransposon copy numbers would indicate the “wear and tear” of the body accumulated over a lifetime. Furthermore, monitoring transposon activity and copy number in germ cells, will not only give us estimates of how fit an individual may be to its current living habitat, but may also reveal an evolutionary mechanism: that stress-induced transposon reactivation plays an important role in initiating transposon amplification events. Hypothetically, this mechanism would be adaptive in its own right, since it signals the germline to start changing if the allostatic load gets too high, ultimately meaning that you are poorly adapted to your current living habitat.
Figure 1:
An integrated model of stress-induced retrotransposon (re)activation and the allostatic load concept. Exogenous and endogenous challenges increase the allostatic load on the body, which represents an average allostatic load experienced by all cells in the organism. A consequence of this is that not all cells will experience higher allostatic load following stress; some might even experience less allostatic load. High cellular allostatic load leads to loss of epigenetic silencing in heterochromatic genomic regions, which is followed by (re)activation of retrotransposons. Losing control of retrotransposons has different consequences and tradeoffs depending on cell type. In somatic cells, it may help an individual animal to generate cellular diversity important for normal development, but may also cause genome instability which may increase the risk of disease. In germ cells, retrotransposon (re)activation may cause infertility, but also contribute to species evolvability by increasing genetic diversity. Hypothetically, a challenging environment could, by increasing the allostatic load, induce the genome-wide transposon amplification events observed in most eukaryotes, thereby representing a tool for a population/species to adapt to the challenging environment.
Conflict of interest statement . None declared.
References
- 1. Selye H. Confusion and controversy in the stress field. J Human Stress 1975. ; 1 : 37 – 44 . [DOI] [PubMed] [Google Scholar]
- 2. Sterling P, Eyer J. Allostasis: A new paradigm to explain arousal pathology. 1988. .
- 3. McEwen BS, Stellar E. Stress and the individual: mechanisms leading to disease. Arch Intern Med 1993. ; 153 : 2093 – 101 . [PubMed] [Google Scholar]
- 4. Epel ES, Lithgow GJ. Stress biology and aging mechanisms: toward understanding the deep connection between adaptation to stress and longevity. J Gerontol Series A: Biol Sci Med Sci 2014. ; 69 : S10 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Korte SM, Koolhaas JM, Wingfield JC. et al. . The Darwinian concept of stress: benefits of allostasis and costs of allostatic load and the trade-offs in health and disease. Neurosci Biobehav Rev 2005. ; 29 : 3 – 38 . [DOI] [PubMed] [Google Scholar]
- 6. Bijlsma R, Loeschcke V. Environmental stress, adaptation and evolution: an overview. J Evol Biol 2005. ; 18 : 744 – 9 . [DOI] [PubMed] [Google Scholar]
- 7. Nätt D, Johansson I, Faresjö T. et al. . High cortisol in 5-year-old children causes loss of DNA methylation in SINE retrotransposons: a possible role for ZNF263 in stress-related diseases. Clin Epigenet 2015. ; 7 : 1 – 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Warren IA, Naville M, Chalopin D , et al. . Evolutionary impact of transposable elements on genomic diversity and lineage-specific innovation in vertebrates. Chromosome Res 2015. ; 23 : 505 – 31 . [DOI] [PubMed] [Google Scholar]
- 9. McClintock B. The significance of responses of the genome to challenge. Science 1984. ; 226 : 792 – 801 . [DOI] [PubMed] [Google Scholar]
- 10. Chalopin D, Naville M, Plard F. et al. . Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol 2015. ; 7 : 567 – 80 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim A, Terzian C, Santamaria P. et al. . Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc Natl Acad Sci USA 1994. ; 91 : 1285 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Song SU, Gerasimova T, Kurkulos M. et al. . An env-like protein encoded by a Drosophila retroelement: evidence that gypsy is an infectious retrovirus. Genes Dev 1994. ; 8 : 2046 – 57 . [DOI] [PubMed] [Google Scholar]
- 13. Weiner AM. SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol 2002. ; 14 : 343 – 50 . [DOI] [PubMed] [Google Scholar]
- 14. Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet 2009. ; 10 : 691 – 703 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodier JL, Kazazian HH., Jr , Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 2008. ; 135 : 23 – 35 . [DOI] [PubMed] [Google Scholar]
- 16. Feschotte C. The contribution of transposable elements to the evolution of regulatory networks. Nat Rev Genet 2008. ; 9 : 397 – 405 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Finnegan DJ. Transposable elements. Curr Opin Genet Dev 1992. ; 2 : 861 – 7 . [DOI] [PubMed] [Google Scholar]
- 18. Groenen MAM, Archibald AL, Uenishi H. et al. . Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012. ; 491 : 393 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Initial sequencing and comparative analysis of the mouse genome. Nature 2002. ; 420 : 520 – 62 . [DOI] [PubMed] [Google Scholar]
- 20. Zhang Y, Maksakova IA, Gagnier L. et al. . Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet 2008. ; 4 : e1000007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang CRL, Schneider AM, Lu Y. et al. . Mobile interspersed repeats are major structural variants in the human genome. Cell 2010. ; 141 : 1171 – 82 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ewing AD, Kazazian HH. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res 2011. ; 21 : 985 – 90 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xing J, Zhang Y, Han K. et al. . Mobile elements create structural variation: Analysis of a complete human genome. Genome Res 2009. ; 19 : 1516 – 26 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Daniels SB, Peterson KR, Strausbaugh LD. et al. . Evidence for horizontal transmission of the P transposable element between Drosophila species. Genetics 1990. ; 124 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hill T, Schlötterer C, Betancourt AJ. Hybrid dysgenesis in Drosophila simulans associated with a rapid invasion of the P-element. PLoS Genet 2016. ; 12 : e1005920.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cantrell MA, Scott L, Brown CJ. et al. . Loss of LINE-1 activity in the megabats. Genetics 2008. ; 178 : 393 – 404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas J, Sorourian M, Ray D . et al. . The limited distribution of Helitrons to vesper bats supports horizontal transfer. Gene 2011. ; 474 : 52 – 8 . [DOI] [PubMed] [Google Scholar]
- 28. Tang Z, Zhang H-H, Huang K. et al. . Repeated horizontal transfers of four DNA transposons in invertebrates and bats. Mobile DNA 2015. ; 6 : 1 – 10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suh A, Witt CC, Menger J. et al. . Ancient horizontal transfers of retrotransposons between birds and ancestors of human pathogenic nematodes. Nat Commun 2016. ; 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Walsh AM, Kortschak RD, Gardner MG. et al. . Widespread horizontal transfer of retrotransposons. Proc Natl Acad Sci 2013. ; 110 : 1012 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaer K, Speek M. Retroelements in human disease. Gene 2013. ; 518 : 231 – 41 . [DOI] [PubMed] [Google Scholar]
- 32. Schmitt MW, Prindle MJ, Loeb LA. Implications of genetic heterogeneity in cancer. Ann New York Acad Sci 2012. ; 1267 : 110 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kemp JR, Longworth MS. Crossing the LINE Toward Genomic Instability: LINE-1 Retrotransposition in Cancer. Front Chem 2015. ; 3 : 68.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang F, Wang PJ. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline . Semin Cell Dev Biol . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Criscione SW, Zhang Y, Thompson W. et al. . Transcriptional landscape of repetitive elements in normal and cancer human cells. BMC Genomics 2014. ; 15 : 1 – 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heras SR, Macias S, Plass M. et al. . The Microprocessor controls the activity of mammalian retrotransposons. Nat Struct Mol Biol 2013. ; 20 : 10.1038/nsmb.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bogerd HP, Wiegand HL, Hulme AE. et al. . Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci 2006. ; 103 : 8780 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goodier JL, Cheung LE, Kazazian HH. Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 2013. ; 41 : 7401 – 19 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Knisbacher BA, Levanon EY. DNA editing of LTR retrotransposons reveals the impact of APOBECs on vertebrate genomes. Mol Biol Evol 2016. ; 33 : 554 – 67 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stetson DB, Ko JS, Heidmann T. et al. . Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 2008. ; 134 : 587 – 98 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Moldovan JB, Moran JV. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet 2015. ; 11 : e1005121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goodier JL, Pereira GC, Cheung LE. et al. . The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet 2015. ; 11 : e1005252.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Huntley S, Baggott DM, Hamilton AT. et al. . A comprehensive catalog of human KRAB-associated zinc finger genes: Insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 2006. ; 16 : 669 – 77 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rowe HM, Jakobsson J, Mesnard D. et al. . KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010. ; 463 : 237 – 40 . [DOI] [PubMed] [Google Scholar]
- 45. Turelli P, Castro-Diaz N, Marzetta F. et al. . Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res 2014. ; 24 : 1260 – 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jacobs FMJ, Greenberg D, Nguyen N. et al. . An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 2014. ; 516 : 242 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herquel B, Ouararhni K, Martianov I. et al. . Trim24-repressed VL30 retrotransposons regulate gene expression by producing noncoding RNA. Nat Struct Mol Biol 2013. ; 20 : 339 – 46 . [DOI] [PubMed] [Google Scholar]
- 48. Isbel L, Srivastava R, Oey H. et al. . Trim33 binds and silences a class of young endogenous retroviruses in the mouse testis; a novel component of the arms race between retrotransposons and the host genome. PLoS Genet 2015. ; 11 : e1005693.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Aravin A, Gaidatzis D, Pfeffer S. et al. . A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006. ; 442 : 203 – 7 . [DOI] [PubMed] [Google Scholar]
- 50. Aravin AA, Sachidanandam R, Girard A. et al. . Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 2007. ; 316 : 744 – 7 . [DOI] [PubMed] [Google Scholar]
- 51. Betel D, Sheridan R, Marks DS. et al. . Computational analysis of mouse piRNA sequence and biogenesis. PLoS Comput Biol 2007. ; 3 : e222.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Watanabe T, Tomizawa S-i, Mitsuya K. et al. . Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science (New York, N.Y.) 2011. ; 332 : 848 – 52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reuter M, Berninger P, Chuma S. et al. . Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 2011. ; 480 : 264 – 7 . [DOI] [PubMed] [Google Scholar]
- 54. De Fazio S, Bartonicek N, Di Giacomo M. et al. . The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 2011. ; 480 : 259 – 63 . [DOI] [PubMed] [Google Scholar]
- 55. Girard A, Sachidanandam R, Hannon GJ. et al. . A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006. ; 442 : 199 – 202 . [DOI] [PubMed] [Google Scholar]
- 56. Grivna ST, Beyret E, Wang Z. et al. . A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 2006. ; 20 : 1709 – 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rajasethupathy P, Antonov I, Sheridan R. et al. . A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012. ; 149 : 693 – 707 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ghosheh Y, Seridi L, Ryu T. et al. . Characterization of piRNAs across postnatal development in mouse brain. Scientific Reports 2016. ; 6 : 25039.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vella S, Gallo A, Lo Nigro A. et al. . PIWI-interacting RNA (piRNA) signatures in human cardiac progenitor cells. Int J Biochem Cell Biol 2016. ; 76 : 1 – 11 . [DOI] [PubMed] [Google Scholar]
- 60. Volkman HE, Stetson DB. The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 2014. ; 15 : 415 – 22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Villeponteau B. The heterochromatin loss model of aging. Exp Gerontol 1997. ; 32 : 383 – 94 . [DOI] [PubMed] [Google Scholar]
- 62. Haithcock E, Dayani Y, Neufeld E , et al. . Age-related changes of nuclear architecture in Caenorhabditis elegans. Proc Natl Acad Sci USA 2005. ; 102 : 16690 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Scaffidi P, Misteli T. Lamin A-dependent nuclear defects in human aging. Science 2006. ; 312 : 1059 – 63 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet 2007. ; 23 : 413 – 8 . [DOI] [PubMed] [Google Scholar]
- 65. Tsurumi A, Li W. Global heterochromatin loss: a unifying theory of aging? Epigenetics 2012. ; 7 : 680 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Shumaker DK, Dechat T, Kohlmaier A. et al. . Mutant nuclear lamin A leads to progressive alterations of epigenetic control in premature aging. Proc Natl Acad Sci USA 2006. ; 103 : 8703 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang W, Li J, Suzuki K. et al. . A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science (New York, N.Y.) 2015. ; 348 : 1160 – 3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eissenberg JC, Elgin SCR. HP1a: a structural chromosomal protein regulating transcription. Trends Genet 2014. ; 30 : 103 – 10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larson K, Yan S-J, Tsurumi A. et al. . Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 2012. ; 8 : e1002473.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bellizzi D, DÁquila P, Montesanto A. et al. . Global DNA methylation in old subjects is correlated with frailty. Age 2012. ; 34 : 169 – 79 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bollati V, Schwartz J, Wright R. et al. . Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev 2009. ; 130 : 234 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. De Cecco M, Criscione SW, Peterson AL . et al. . Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013. ; 5 : 867 – 83 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li W, Prazak L, Chatterjee N. et al. . Activation of transposable elements during aging and neuronal decline in Drosophila. Nat Neurosci 2013. ; 16 : 529 – 31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Maxwell PH. What might retrotransposons teach us about aging? Curr Genet 2016. ; 62 : 277 – 82 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bundo M, Toyoshima M, Okada Y , et al. . Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron 81 : 306 – 13 . [DOI] [PubMed] [Google Scholar]
- 76. Coufal NG, Garcia-Perez JL, Peng GE. et al. . L1 retrotransposition in human neural progenitor cells. Nature 2009. ; 460 : 1127 – 31 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Baillie JK, Barnett MW, Upton KR. et al. . Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011. ; 479 : 534 – 37 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perrat PN, DasGupta S, Wang J. et al. . Transposition driven genomic heterogeneity in the Drosophila brain. Science (New York, N.Y.) 2013. ; 340 : 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bollati V, Galimberti D, Pergoli L. et al. . DNA methylation in repetitive elements and Alzheimer disease. Brain Behav Immun 2011. ; 25 : 1078 – 83 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sapolsky RM. Why zebras don't get ulcers: The acclaimed guide to stress, stress-related diseases, and coping-now revised and updated. 2004. : Macmillan.
- 81. Ito H. Small RNAs and transposon silencing in plants. Dev Growth Differ 2012. ; 54 : 100 – 7 . [DOI] [PubMed] [Google Scholar]
- 82. Stankiewicz AM, Swiergiel AH, Lisowski P. Epigenetics of stress adaptations in the brain. Brain Res Bull 2013. ; 98 : 76 – 92 . [DOI] [PubMed] [Google Scholar]
- 83. Hunter RG, Murakami G, Dewell S. et al. . Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci USA 2012. ; 109 : 17657 – 62 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hunter RG. et al. . Stress and the dynamic genome: Steroids, epigenetics, and the transposome. Proc Natl Acad Sci 2015. ; 112 : 6828 – 33 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hunter RG, McCarthy KJ, Milne TA. et al. . Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA 2009. ; 106 : 20912 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Covington HE, Maze I, Sun H. et al. . A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 2011. ; 71 : 656 – 70 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Sun H, Maze I, Dietz DM. et al. . Morphine epigenomically regulates behavior through alterations in histone H3 lysine 9 dimethylation in the nucleus accumbens. J Neurosci 2012. ; 32 : 17454 – 64 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Maze I, Covington HE, Dietz DM. et al. . Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science (New York, N.Y.) 2010. ; 327 : 213.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Maze I, Feng J, Wilkinson MB. et al. . Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci USA 2011. ; 108 : 3035 – 40 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2012. ; 13 : 36 – 46 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Muotri AR, Zhao C, Marchetto MCN. et al. . Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus 2009. ; 19 : 1002 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Capomaccio S, Verini-Supplizi A, Galla G. et al. . Transcription of LINE-derived sequences in exercise-induced stress in horses. Animal Genet 2010. ; 41 : 23 – 7 . [DOI] [PubMed] [Google Scholar]
- 93. Frese S, Ruebner M, Suhr F. et al. . Long-term endurance exercise in humans stimulates cell fusion of myoblasts along with fusogenic endogenous retroviral genes in vivo. PLoS ONE 2015. ; 10 : e0132099.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Juster R-P, McEwen BS, Lupien SJ. Allostatic load biomarkers of chronic stress and impact on health and cognition. Neurosci Biobehav Rev 2010. ; 35 : 2 – 16 . [DOI] [PubMed] [Google Scholar]