Abstract
Epigenetics is a gene regulation mechanism that does not depend on genomic DNA sequences but depends on chemical modification of genomic DNA and histone proteins around which DNA is wrapped. The failure of epigenetic mechanisms is known to cause various congenital disorders. It is also known that the failures of epigenetic mechanisms causes various acquired disorders since epigenetic modifications of the genome (i.e., “epigenome”) are more vulnerable to environmental stress, such as malnutrition, environmental chemicals, and mental stress, than the “genome,” especially during the early period of life. However, the epigenome has a reversible property since it is based on removable residues on genomic DNA. Thus, environmentally induced epigenomic alterations can be potentially restored. In fact, some medicines, especially for psychiatric diseases, are known to restore an altered epigenome, resulting in the correction of gene expression. Several lines of evidence suggest that environmentally induced epigenomic alterations are not erased completely during gametogenesis, but are transmitted to subsequent generations with disease phenotypes. In accordance with these understandings, I would like to propose the development of epigenomic-based preemptive medicine that consists of the early detection of the developmental origins of diseases using epigenomic signatures and the early intervention that take advantages of the use of epigenomic reversibility.
Keywords: neurodevelopmental disorder, metabolic disorder, environmental stress, epigenetics: epigenome, preemptive medicine
Introduction
Diabetes mellitus (DM) comprises a group of heterogeneous metabolic disorders that share an increase in the concentration of blood glucose. Both environmental and genetic factors are thought to contribute to the occurrence of DM. A number of studies have demonstrated that various environmental factors including overeating, passive smoking in those who are not themselves active smokers, and ambient air pollution such as by PM 2.5 induce systemic insulin resistance as a predisposition to type 2 DM (T2DM) [ 1 , 2 ].
As for genetic factors associated with DM, including maturity onset diabetes of the young (MODY), several causative genes have been identified and >75 genetic variants. However, mutations in genes associated with MODY are rare and whether the identified common variants are causal and how these genetic variants exert their effect on the pathogenesis of T2DM is largely unknown [ 3 , 4 ].
Several lines of evidence suggest that epigenetic alterations induced by environmental factors (e.g. nutritional factors and mental stress) during the fetal and neonatal periods are underlying mechanisms of the predisposition to T2DM [ 5–9 ].
A number of environmental factors are also known to be involved in the pathogenesis of neurodevelopmental disorders (NDs), such as inappropriate child rearing (e.g. child abuse and malnutrition by the parents with mental problems) [ 10–13 ], viral infections with rubella and cytomegalovirus, which induce immunological reactions in the brain [ 14–19 ], and environmental chemicals such as endocrine-disrupting chemicals e.g. tobacco, air pollutants, solvents, metals, pesticides, flame retardants, non-stick chemicals, phthalates, and bisphenol A (BPA) [ 20 ].
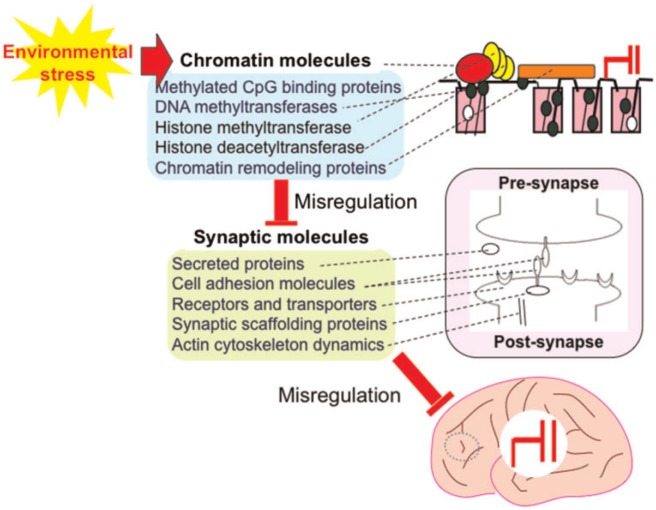
As for genetic factors associated with NDs, causative genes associated with brain function have been identified. Mutations in genes encoding secreted proteins (e.g. RELN ), cell adhesion molecules (e.g. NLGN3 and NLGN4 ), receptors and transporters (e.g. GRIN2A ), synaptic scaffolding proteins (e.g. SHANK3 and LIN7B ), and actin cytoskeleton dynamics (e.g. TSC1 and TSC2 ) [ 21–27 ]. These findings suggest that NDs can be recognized as “synaptic disorders” [ 21 , 28 ] ( Fig. 1 ).
Figure 1:
Mutations in the genes encoding proteins associated with synaptic function are found in a subset of patients with NDs. Mutations in the genes encoding chromatin-associated proteins are found in a subset of patients with NDs via misregulation of synapse-associated genes. Various types of environmental stress affect the function of chromatin-associated genes. All of these genetic alterations and epigenetic misregulation by environmental stresses lead to the misregulation of brain function
Unexpectedly, mutations have also been identified in chromatin-remodeling factors that are apparently not involved directly in brain function. These include methylated CpG-binding proteins [e.g. MEPC2 associated with Rett syndrome (RTT)], DNA methyltransferases (e.g. DNMT3A associated with intellectual disability with overgrowth), histone methyltransferase (e.g. EHMT1 associated with Kleefstra syndrome), and chromatin remodeling proteins (chromodomain helicases) (e.g. CHD8 associated with an autistic disorder) [ 29–33 ]. These findings suggest that NDs can also be recognized as “chromatin (a unit of DNA and histone proteins that are chemically modified) disorders” [ 24–35 ] ( Fig. 1 ).
Recent studies revealed that ASC2 (a protein involved in cortical neuronal migration and neurogenesis), which is associated with a subtype of NDs binds to Polycomb repressive complex 1 (a chromatin remodeling protein) and controls the expression of genes (e.g. neurocan) involved in axon guidance in the developing forebrain [ 36–40 ], suggesting that a close interaction between neuronal molecules and chromatin molecules is essential for brain development and failure of this interaction leads to misregulation of brain development-associated genes, resulting in NDs.
In this review, I introduce various congenital disorders caused by epigenetic misregulation and disorders caused by acquired epigenetic misregulation and discuss epigenomic-based preemptive medicine taking advantage of the use of the epigenetic reversibility for patients with metabolic and NDs and for future generations in terms of transgenerational epigenetic inheritance.
Congenital Disorders Caused by Epigenetic Misregulation
RTT is a representative ND characterized by repetitive and stereotypic hand movements, seizures, gait ataxia, and autistic features, which is caused by mutations in the gene encoding methyl-CpG-binding protein 2 (MeCP2) [ 29 ]. Since RTT is an X-linked dominant disorder, the affected patients are females. In males, the disorder is thought be embryonic lethal, although in rare cases, male RTT patients have been reported [ 41 ].
The MeCP2 protein binds methylated DNA regions and controls the expression of a number of genes including synapse-associated genes such as BDNF , DLX5 , ID, CRH, IGFBP3 , CDKL1, PCDHB1 , and PCDH7 by interacting with histone deacetylases in neuronal cells and brain tissue [ 42–52 ], resulting in the regulation of excitatory synaptic strength [ 53 ] ( Fig. 1 ).
Instead of studying the inaccessible brain cells of RTT patients during development, neural cells can be generated by using induced pluripotent stem cell (iPSC) technology. Several iPSC studies demonstrated that RTT neurons have abnormalities in maturation [ 54–56 ] and differentiation, in which a subset of glia cell-specific genes, such as GFAP , are aberrantly expressed due to de-suppression because of a deficiency of MeCP2 [ 57 ] ( Fig. 2 ).
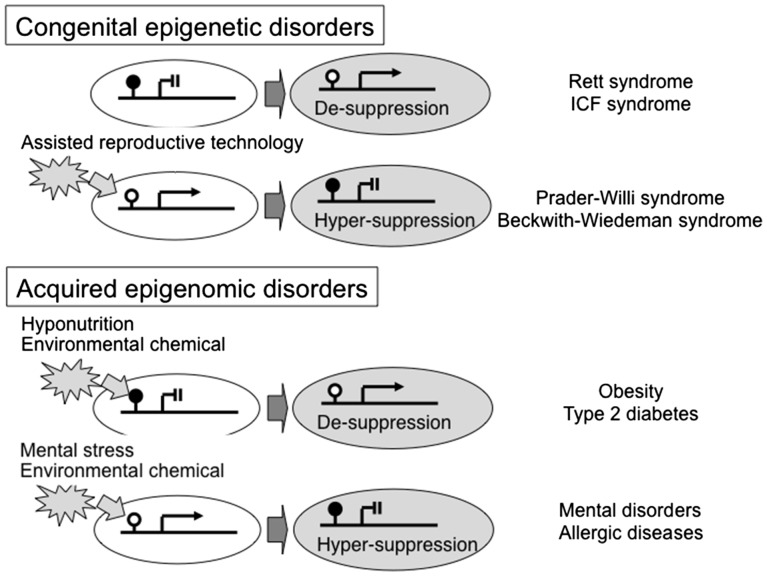
Figure 2:
Epigenetic misregulation in congenital epigenetic disorders and acquired epigenetic disorders. ( A ) Abnormalities in a methylated CpG binding protein or a DNA methyltransferase result in Rett syndrome or ICF syndrome, respectively. An aberrant genomic pattern due to failure of genomic imprinting results in Prader-Willi syndrome. ( B ) Environmental factors alter epigenomic status, resulting in predispositions to various common disorders
The brain is a gene-dosage-sensitive organ in which either under-expression due to mutation or deletion of a gene or over-expression due to duplication of the same gene results in neurological abnormalities. The effects of aberrant gene expression are exemplified by conditions such as Pelizaeus–Merzbacher disease, a severe congenital myelination disorder associated with deletion, mutation, or duplication of PLP1 [ 58 ], lissencephaly, a severe congenital neural migration disorder associated with either deletion or duplication of LIS1 ( PAFAH1B1 ) [ 59 , 60 ], Charcot–Marie–Tooth disease, an adult-onset neuromuscular disease associated with mutation or duplication of PMP22 [ 61 ], and Parkinson’s disease associated with either mutation or multiplication of SNCA [ 62 ].
Similarly, not only mutations in MECP2 but also duplication of MECP2 cause severe mental retardation especially in males [ 63 ] and cognitive impairment with learning difficulties and speech delay in females [ 64 ]. The over-dosage effect of Mecp2 has been found in transgenic mice and monkeys that show motor coordination deficits, heightened anxiety, and impairments of learning and memory. These animals also exhibit various behavioral problems such as a higher frequency of repetitive circular locomotion, increased stress responses, stereotypic cognitive behaviors, and reduced interactions with other animals [ 65 , 66 ].
These findings results from genetic and epigenetic studies suggest that the brain is extremely sensitive to the dosage of proteins associated with synaptic or neuronal function, such as BDNF and LIS1, and is also sensitive to the dosage of proteins associated with chromatin structure or epigenetic gene regulation such as MeCP2, and further indicate that the brain is an organ that requires strict gene control to maintain the corrected levels of proteins associated with brain function.
ICF syndrome, which is characterized by Immunodeficiency (e.g., IgG and IgA), Centromere instability (breakage of the pericentric heterochromatic regions of chromosomes 1, 9, and 16 due to abnormally low levels of DNA methylation), and Facial anomalies, is a congenital autosomal recessive disorder caused by mutations in the DNA methyltransferase gene, DNMT3B, which lead to de-suppression due to hypomethylation of genes [ 67 , 68 ] ( Fig. 2 ). A recent study demonstrated a hypomethylation pattern in mesenchymal stem cells differentiated from iPSCs of ICF patients, which is potentially associated with immunological pathogenesis [ 69 ].
Prader–Willi syndrome (PWS) is a congenital epigenetic disorder characterized by muscle hypotonia during infancy, cryptorchidism (in boys), short stature, small hands and feet, hyperphagia starting from childhood and subsequent obesity and T2DM in adulthood, and various neurodevelopmental features such as obsessive–compulsive disorder. PWS is not caused by abnormalities in a single epigenetic molecule, but is caused by an abnormal epigenomic pattern in which the expressed paternal genes, located on chromosome 15q12, are missing physically or functionally due to paternal deletion or uniparental maternal disomy, respectively [ 70–73 ]; PWS patients have only maternally inherited genes on chromosome 15q12 that are imprinted (methylated) and are thus not expressed ( Fig. 2 ).
Angelman syndrome, characterized by severe intellectual disability and intractable epilepsy with puppet-like ataxic movements and paroxysms of laughter, is another epigenetic disorder caused by an abnormal epigenomic pattern in which the expressed maternal allele of UBE3A , located on chromosome 15q12, is missing physically or functionally due to maternal deletion or uniparental paternal disomy, respectively [ 74 ]. Conversely, over-dosage of the expressed genes located on the maternal chromosome due to chromosomal duplication or triplication causes autistic-like features [ 75 ]. Is interesting to note that assisted reproductive technologies (e.g. in vitro fertilization and intracytoplasmic sperm injection), which are used widely due to increases in age at the time of conception of the first child, alter DNA methylation status at loci and are potentially involved in an increased risk of Beckwith–Wiedemann syndrome, a congenital epigenetic disorder characterized by overgrowth in the fetal period with an increased risk of childhood cancer [ 76 , 77 ].
Acquired Disorders Caused by Epigenetic Misregulation
In the concept of gene–environment interactions for common diseases including metabolic and NDs, the combination of heritability (G: genetic factor such as a single nucleotide polymorphism) and experience (E: environmental factor), that is, the “G × E” model, has been used where G and E contribute independently to disease occurrence. I would like to propose an “E × G” model, in which E changes G dynamically, where G is not a genomic DNA sequence (i.e. the genome) but genomic DNA and histone protein modifications (i.e. the epigenome) [ 78 ]. I would like to show examples that underlie this new model, in which environmental factors alter the epigenome of individuals, changing their health status.
Epidemiological studies of populations affected by famines in the Netherlands and China demonstrated that the generation with a lower birth weight than normal had an increased risk for obesity, DM, and mental disorders [ 79 , 80 ]. These studies suggested that the number of patients with DM and an ND might increased in Japan because the rate of low birth weight infants has increased over the past 30 years due to the increase of young women who diet even while pregnant and the directions of obstetricians to minimize pregnancy weight gain to avoid a hard labor and reduce the risk of gestational DM [ 79 ]. Furthermore, recent epigenetic studies demonstrated that malnutrition with insufficient folic acid intake during pregnancy induced lower DNA methylation in genes (e.g. PPAR α) in the liver of rat offspring [ 5 , 6 ]. Similar low levels of DNA methylation were observed in the peripheral blood of individuals who lived through the Dutch famine [ 81 ]. Conversely, periconceptional micronutrient supplementation, including folate, zinc, and vitamins A, B, C, and D, increased DNA methylation levels in human offspring [ 82 ]. This kind of scientific study to clarify the mechanism of the occurrence of “adult diseases” during the early period of life for early intervention is referred to as “Developmental Origins of Heath and Disease” [ 83 ].
In addition to malnutrition, a number of environmental chemicals have been shown to alter the epigenome. For example, prenatal exposure to BPA, a chemical with reproductive toxicity that induces growth alterations and immune dysregulation, alters DNA methylation in fetal brain and in mast cells and liver of offspring [ 84–86 ]; prenatal exposure to polybrominated diphenyl ethers, which are used as flame retardants, decreases DNA methylation of TNFα and increases TNFα (a proinflammatory molecule) expression in cord blood [ 87 ]; prenatal exposure to tobacco smoking alters DNA methylation of AHRR , MYO1G , CYP1A1 , and CNTNAP2 in cord blood; and these altered DNA methylation patterns were observed in the peripheral blood of their children born to smoking mothers at the age of 17 years and may be associated with diseases, such as bronchial asthma [ 88 ]. These findings suggest that DNA methylation is changeable during the early periods of life, and these changes can persist for a long period after birth and can be associated with disease phenotypes.
Similar to patients with DM, the number of children with NDs has increased in England [ 89 ] (prevalence from 1/2,500 to 1/86) and other countries over the last 50 years. The rate of affected children has reached 100 (range, 34–264) per 10 000 children [ 90–94 ]. For these increases, twin studies have implicated the influence of environmental factors in the development of NDs [ 95–97 ].
The epigenome, characterized by epigenetic mechanisms, acts as a “physical receptor” for environmental stresses. In fact, epigenomic differences are more markedly different in older monozygotic twins than in younger monozygotic twins [ 98 ], and differential epigenomic patterns have been observed between discordant monozygotic twins with RTT, a representative ND as mentioned above, in which abnormal epigenomic patterns that lead to aberrant synaptic gene expression were observed in the RTT twin with the more severe phenotype [ 99 ]. A previous rat study demonstrated that exposure to short-term postnatal stress by separating offspring from their mother induced hypermethylation within the promoter region of NR3C1 , which encodes a glucocorticoid receptor hormone associated with resilience, in the hippocampal region of the offspring, which leads to life-long abnormal behavior [ 7 ]. Furthermore, recent human studies also demonstrated that ice storm stress in 1998 in Quebec during pregnancy altered DNA methylation of immunological genes in the peripheral blood of the offspring [ 100 ], and severe maternal stress that causes depression during pregnancy alters DNA methylation in imprinted IGF2 and GNASXL in cord blood [ 101 ] and in NR3C1 and BDNF in the buccal mucosa of 2-month-old infants [ 102 , 103 ].
All of these findings indicate that the epigenome is vulnerable to environmental stress during the early period of life, environmental stress-induced epigenomic alterations can alter or modify phenotypes, and the recent increase of ND patients may be caused by epigenomic alterations induced by environmental and social stress to children and/or mothers.
Epigenomic-Based Preemptive Medicine
For adult-onset diseases, physicians make a diagnosis based on the guidelines established for each disease. In these cases, diagnosis is made at a later stage of development after the patient fulfills the criteria for each adult disease (e.g. blood sugar and HbA1c levels for T2DM). Furthermore, gold standard therapeutic protocols are strictly determined in the guidelines, regardless of a patient’s individual genetic background, which may influence the effectiveness of the administered drugs. In this context, “personalized medicine” has been proposed as the application of treatments that take into consideration each patient’s genetic background.
“Preemptive medicine” is a type of personalized medicine that is based on the individual and is thus different from population-based preventive medicine that started as a means to prevent the spread of infectious diseases (it is now moving toward the prevention of adults diseases). In preemptive medicine, a practical approach is to detect high-risk individuals by screening with a blood biomarker, which includes genetic and epigenetic information, and to intervene in high-risk individuals at the preclinical stage to prevent serious events, such as T2DM, Alzheimer’s disease, osteoporosis, and coronary heart disease [ 104 ].
Such a preemptive approach based on an epigenomic marker has already been started for PWS, one of the congenital epigenetic disorders mentioned above. High-risk individuals (i.e. PWS patients) are identified by an abnormal pattern of SNRPN promoter methylation in peripheral blood during infancy, and a variety of physical and drug treatments are provided to the individuals to prevent future symptoms (e.g. obesity and T2DM).
For example, a program of a well-balanced low-calorie diet and regular exercise is applied to prevent weight gain by hyperphagia and the subsequent development of obesity, which begins at 2–4 years of age [ 105 ]. Physical therapy for patients younger than 3 years of age improves muscle strength and encourages the achievement of developmental milestones, and daily muscle training increases physical activity and lean body mass in older patients [ 106 ]. Growth hormone treatment normalizes height, increases lean body mass, decreases fat mass, and increases mobility, which are beneficial to weight management [ 107 ]; a longitudinal study demonstrated that growth hormone treatment normalized stature and improved weight and body composition in PWS patients compared to non-growth hormone-treated PWS patients [ 108 , 109 ]. Furthermore, a recent randomized controlled trial revealed that physical training combined with growth hormone further improved muscle thickness, which was matched by an increase in muscle strength and motor development in infants with PWS [ 110 ]. Growth hormone treatment further improves language skills in infancy, cognitive skills in childhood, and mental speed, mental flexibility, motor performance and ability to adapt to society in adulthood [ 111–115 ]. Therefore, epigenomic-based preemptive medicine is effective for patients with PWS although the number of PWS patients is limited.
In order to establish a preemptive program for acquired disorders similar to that for PWS, it will be necessary to identify “epigenomic signatures” that are modified by environmental factors and can be detected in peripheral blood. In fact, mental stress-induced hypermethylation of the brain-derived neurotrophic factor ( Bdnf ) gene was demonstrated in the hippocampal region of a mouse model of depression [ 116 ]. Subsequently, abnormal DNA methylation of BDNF was proposed as an epigenomic blood marker for the individuals with major depression [ 117 ]. Abnormal DNA methylation of FHL2 , ZNF518B , GNPNAT1 , and HLTF was also proposed as an epigenomic blood marker for individuals with T2DM [ 118 ]. Furthermore, it was demonstrated recently that exposure to BPA during the prenatal period induced lasting DNA methylation changes in Bdnf in the hippocampus and blood in mice, and that exposure to high levels of BPA in utero induced DNA methylation changes in human cord blood [ 119 ]. These findings suggest that methylation in the blood may be used as a predictor of methylation in the brain and indicate that DNA methylation in the peripheral blood can be a useful biomarker for the detection of psychopathology.
It was reported recently that epigenomic restoration of histone acetylation can be achieved by the administration of psychotropic drugs, such as valproic acid and imipramine [ 116 , 120 , 121 ]. A recent epidemiological study further demonstrated that supplementation of folic acid during pregnancy, which is an important nutrient for DNA methylation, reduced the risk of NDs in the offspring [ 122 ]. Studies using RTT or Mecp2 -duplication mouse models have demonstrated that genetic supplementation of MeCP2, bone marrow transplantation, antisense oligonucleotides, or deep brain electrical stimulation after birth successfully attenuates neurological symptoms [ 123–126 ]. These findings support the notion that the phenotypes of NDs caused by epigenetic dysregulation are reversible and thus treatable. In this context, drugs are under development by many pharmacological companies taking advantage of the use of epigenomic reversibility.
Studies using mouse models also suggest that the clinical features of a congenital epigenomic disorder can be ameliorated not only by the administration of “ultimately designed” epigenomic drugs but also by providing a better nurturing environment. For example, environmental enrichment with larger-sized home cages containing a variety of objects including running wheels, improved motor coordination with a slight increase in BDNF protein levels in the cerebellum, rescued memory deficits in the Morris water maze, and decreased anxiety-related behavior in RTT model (heterozygous Mecp2+/- ) female mice [ 127 , 128 ]. Similarly, environmental enrichment improved locomotor activity with reduced ventricular volume and decreased the expression of synaptic markers, such as synaptophysin and PSD95 in the hypothalamus and syntaxin 1a and synaptotagmin [ 129 , 130 ]. The precise molecular mechanism underlying the environmental enrichment effect remains to be elucidated, but it is intriguing to think that the epigenomic status of MeCP2-target synaptic genes may be restored by environmental enrichment.
Conclusions
In this article, I introduced congenital disorders with epigenetic abnormalities caused by genetic alterations such as RTT and PWS and acquired disorders with environmentally induced epigenomic abnormalities. Furthermore, I discussed the concept of epigenomic-based preemptive medicine, taking advantage of the use of epigenomic reversibility.
Several lines of evidence suggest that environmental stress that alters a phenotype affect not only the exposed individual but also subsequent progeny for successive generations. In other words, ancestral experiences could influence subsequent generations, the concept of which is termed “transgenerational inheritance.” Furthermore, environmental factors such as endocrine-disrupting chemicals and nutrition do not promote genetic mutations but instead promote epigenetic changes; the permanent programming of an altered epigenome in the germline can allow for the transmission of transgenerational epigenetic phenotypic variations and subsequent disease risk [ 131 , 132 ].
The evidence supports the theory of Lamarckian inheritance in which an organism can pass on phenotypes that it acquired during its lifetime to its offspring. More precisely, a hypothesis has emerged that environmental stress results in epigenetic changes at some loci in the genome and these can escape the epigenetic reprogramming that normally occurs between generations [ 133 , 134 ].
Short-term postnatal mental stress by separating offspring from their mother alters DNA methylation not only in the brain but also in the sperm of male offspring, and then, the environmentally induced epigenetic and expression alterations of Crfr2 are transmitted up to the third generation (F1 sperm and F2 brain) along with behavioral abnormalities [ 135 ]. Furthermore, exposure to prenatal stress induces changes in DNA methylation and micro-RNA expression in the placenta and brain, which leads to an increase in risk for NDs, schizophrenia, and anxiety- or depression-related disorders later in life [ 136 ].
Exposure to an environmental chemical (e.g. vinclozolin) during embryonic gonadal sex determination can alter male germ-line epigenetics, and the alteration of DNA methylation in the germ line appears to result in the transmission of transgenerational adult-onset diseases, such as spermatogenic defects, prostate and kidney diseases, and cancer [ 137 ]. A recent study demonstrated that exposure to BPA in early life induces glucose intolerance and β-cell dysfunction, with hypermethylation and associated decreased expression of Igf2 in the islets of male F2 offspring; this suggests that exposure to BPA during early life can result in the generational transmission of glucose intolerance and β-cell dysfunction through the male germ line by an epigenetic mechanism [ 138 ].
However, evidence that such effects persist in subsequent generations has been inconclusive [ 133 , 139 , 140 ]. These effects must be observed in the F3 generation to be considered transgenerational because the in utero nature of the ancestral perturbation affects not only the somatic and germ cells of the developing F1 fetus but also the germ cells of the F2 generation [ 132 ]. In this context, a recent study demonstrated that !treatment of pregnant mice with methoxychlor altered the methylation of all genes examined in the F1 offspring, but these effects disappeared gradually from F1 to F3 [ 141 ]. This suggests that transgenerational epigenetic inheritance is not “solid (complete)” genetic inheritance but “soft (incomplete)” epigenomic inheritance [ 142 , 143 ]. Nevertheless, epigenomic-based preemptive medicine will be important not only for the exposed first generation but also for subsequent successive generations in terms of disrupting the vertical transmission of epigenomic disorders.
Acknowledgments
The authors would like to thank the Ministry of Education, Science, Sports and Culture (MEXT), Grants-in-Aid (KAKENHI) for Scientific Research (B) (#26293245) and Exploratory Research (#15K15388), and the Japan Agency for Medical Research and Development (AMED) for funds for the development of core technologies for innovative drug development based upon IT. T.K. wrote the article.
Conflict of interest statement . None declared.
References
- 1. Wei XEM, Yu S. A meta-analysis of passive smoking and risk of developing Type 2 Diabetes Mellitus . Diabetes Res Clin Pract 2015. ; 107 : 9 – 14 . [DOI] [PubMed] [Google Scholar]
- 2. Haberzettl P, O'Toole TE, Bhatnagar A , et al. . Exposure to fine particulate air pollution causes vascular insulin resistance by inducing pulmonary oxidative stress . Environ Health Perspect 2016. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amed S, Oram R. Maturity-Onset Diabetes of the Young (MODY): making the right diagnosis to optimize treatment . Can J Diabetes 2016. : S1499 – S2671 . [DOI] [PubMed] [Google Scholar]
- 4. Kwak SH, Park KS. Recent progress in genetic and epigenetic research on type 2 diabetes . Exp. Mol. Med 2016. ; 48 : e220.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lillycrop KA, Phillips ES, Jackson AA , et al. . Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring . J Nutr 2005. ; 135 : 1382 – 6 . [DOI] [PubMed] [Google Scholar]
- 6. Lillycrop KA, Phillips ES, Torrens C , et al. . Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring . Br. J. Nutr 2008. ; 100 : 278 – 28 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Weaver IC, Cervoni N, Champagne FA , et al. . Epigenetic programming by maternal behavior . Nat. Neurosci 2004. ; 7 : 847 – 54 . [DOI] [PubMed] [Google Scholar]
- 8. Ehara T, Kamei Y, Yuan X , et al. . Ligand-activated PPARα-dependent DNA demethylation regulates the fatty acid β-oxidation genes in the postnatal liver . Diabetes 2015. ; 64 : 775 – 84 . [DOI] [PubMed] [Google Scholar]
- 9. Sterns JD, Smith CB, Steele JR , et al. . Epigenetics and type II diabetes mellitus: underlying mechanisms of prenatal predisposition . Front Cell Dev. Biol 2014. ; 2 : 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Larsson HJ, Eaton WW, Madsen KM , et al. . Risk factors for autism: perinatal factors, parental psychiatric history, and socioeconomic status . Am. J. Epidemiol 2005. ; 161 : 916 – 25 . [DOI] [PubMed] [Google Scholar]
- 11. Jokiranta E, Brown A, Heinimaa M , et al. . Parental psychiatric disorders and autism spectrum disorders . Psychiatry Res 2013. ; 207 : 203 – 11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roberts AL, Lyall K, Rich-Edwards JW , et al. . Association of maternal exposure to childhood abuse with elevated risk for autism in offspring . JAMA Psychiatry 2013. ; 70 : 508 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Belluscio LM, Berardino BG, Ferroni NM , et al. . Early protein malnutrition negatively impacts physical growth and neurological reflexes and evokes anxiety and depressive-like behaviors . Physiol. Behav 2014. ; 129 : 237 – 54 . [DOI] [PubMed] [Google Scholar]
- 14. Berger BE, Navar-Boggan AM, Omer SB. Congenital rubella syndrome and autism spectrum disorder prevented by rubella vaccination–United States.; 2001-2010 . BMC Public Health 2011. ; 11 : 340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakamoto A, Moriuchi H, Matsuzaki J , et al. . Retrospective diagnosis of congenital cytomegalovirus infection in children with autism spectrum disorder but no other major neurologic deficit . Brain Dev 2015. ; 37 : 200 – 5 . [DOI] [PubMed] [Google Scholar]
- 16. Heilbrun LP, Palmer RF, Jaen CR , et al. . Maternal Chemical and Drug Intolerances: Potential Risk Factors for Autism and Attention Deficit Hyperactivity Disorder (ADHD) . J Am. Board Fam Med 2015. ; 28 : 461 – 70 . [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez JI, Kern JK. Evidence of microglial activation in autism and its possible role in brain underconnectivity . Neuron Glia Biol 2011. ; 7 : 205 – 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Theoharides TC, Asadi S, Patel AB. Focal brain inflammation and autism . J. Neuroinflammation 2013. ; 10 : 46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDougle CJ. Toward an immune-mediated subtype of autism spectrum disorder . Brain Res 2015. ; 1617 : 72 – 92 . [DOI] [PubMed] [Google Scholar]
- 20. Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence . Curr Probl Pediatr Adolesc Health Care 2014. ; 44 : 277 – 318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues . Trends Neurosci 2006. ; 29 : 349 – 58 . [DOI] [PubMed] [Google Scholar]
- 22. Corradi A, Fadda M, Piton A , et al. . SYN2 is an autism predisposing gene: loss-of-function mutations alter synaptic vesicle cycling and axon outgrowth . Hum Mol Genet 2014. ; 23 : 90 – 103 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamilton PJ, Campbell NG, Sharma S , et al. . De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder . Mol Psychiatry 2013. ; 18 : 1315 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hill-Yardin EL, Argyropoulos A, Hosie S , et al. . Reduced susceptibility to induced seizures in the Neuroligin-3(R451C) mouse model of autism . Neurosci Lett 2015. ; 589 : 57 – 61 . [DOI] [PubMed] [Google Scholar]
- 25. Sala C, Vicidomini C, Bigi I , et al. . Shank synaptic scaffold proteins: keys to understanding the pathogenesis of autism and other synaptic disorders . J. Neurochem 2015. ; 135 : 849 – 58 . [DOI] [PubMed] [Google Scholar]
- 26. Leblond CS, Nava C, Polge A , et al. . Meta-analysis of SHANK Mutations in Autism Spectrum Disorders: a gradient of severity in cognitive impairments . PLoS Genet 2014. ; 10 : e1004580.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizuno M, Matsumoto A, Hamada N , et al. . Role of an adaptor protein Lin-7B in brain development: possible involvement in autism spectrum disorders . J Neurochem 2015. ; 132 : 61 – 9 . [DOI] [PubMed] [Google Scholar]
- 28. Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science 2003. ; 302 : 826 – 30 . [DOI] [PubMed] [Google Scholar]
- 29. Amir RE, Van den Veyver IB, Wan M , et al. . Rett syndrome is caused by mutations in X-linked MECP2.; encoding methyl-CpG-binding protein 2 . Nat Genet 1999. ; 23 : 185 – 8 . [DOI] [PubMed] [Google Scholar]
- 30. Tatton-Brown K, Seal S, Ruark E , et al. . Childhood Overgrowth Consortium . Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability . Nat Genet 2015. ; 46 : 385 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kleefstra T, Brunner HG, Amiel J , et al. . Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome . Am J Hum Genet 2006. ; 79 : 370 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balan S, Iwayama Y, Maekawa M , et al. . Exon resequencing of H3K9 methyltransferase complex genes.; EHMT1.; EHTM2 and WIZ.; in Japanese autism subjects . Mol Autism 2014. ; 5 : 49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barnard RA, Pomaville MB, O'Roak BJ. Mutations and modeling of the chromatin remodeler CHD8 define an emerging autism etiology . Front Neurosci 2015. ; 9 : 477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Rubeis S, He X, Goldberg AP , et al. . DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium . Synaptic, transcriptional and chromatin genes disrupted in autism . Nature 2014. ; 515 : 209 – 15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder . Nat Rev Neurosci 2015. ; 16 : 551 – 63 . [DOI] [PubMed] [Google Scholar]
- 36. Hori K, Nagai T, Shan W , et al. . Cytoskeletal regulation by AUTS2 in neuronal migration and neuritogenesis . Cell Rep 2014. ; 9 : 2166 – 79 . [DOI] [PubMed] [Google Scholar]
- 37. Kalscheuer VM, FitzPatrick D, Tommerup N , et al. . Mutations in autism susceptibility candidate 2 (AUTS2) in patients with mental retardation . Hum Genet 2007. ; 121 : 501 – 9 . [DOI] [PubMed] [Google Scholar]
- 38. Beunders G, Voorhoeve E, Golzio C , et al. . Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus . Am J Hum Genet 2013. ; 92 : 210 – 20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao Z, Lee P, Stafford J , et al. . An AUTS2-Polycomb complex activates gene expression in the CNS . Nature 2014. ; 516 : 349 – 54 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oksenberg N, Haliburton GD, Eckalbar WL , et al. . Genome-wide distribution of Auts2 binding localizes with active neurodevelopmental genes . Transl Psychiatry 2014. ; 4 : e431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reichow B, George-Puskar A, Lutz T , et al. . Brief report: systematic review of Rett syndrome in males . J Autism Dev Disord 2015. ; 45 : 3377 – 83 . [DOI] [PubMed] [Google Scholar]
- 42. Ballestar E, Yusufzai TM, Wolffe AP. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA . Biochemistry 2000. ; 39 : 7100 – 6 . [DOI] [PubMed] [Google Scholar]
- 43. Wolffe AP. Histone deacetylase: a regulator of transcription . Science 1996. ; 272 : 371 – 2 . [DOI] [PubMed] [Google Scholar]
- 43. Jones PL, Veenstra GJ, Wade PA , et al. . Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription . Nat Genet 1998. ; 19 : 187 – 91 . [DOI] [PubMed] [Google Scholar]
- 45. Nan X, Bird A. The biological functions of the methyl-CpG-binding protein MeCP2 and its implication in Rett syndrome . Brain Dev 2001. ; 23 : S32 – 7 . [DOI] [PubMed] [Google Scholar]
- 46. Chen WG, Chang Q, Lin Y , et al. . Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2 . Science 2003. ; 302 : 885 – 9 . [DOI] [PubMed] [Google Scholar]
- 47. Martinowich K, Hattori D, Wu H , et al. . DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation . Science 2003. ; 302 : 890 – 3 . [DOI] [PubMed] [Google Scholar]
- 48. Horike S, Cai S, Miyano M , et al. . Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome . Nat Genet 2005. ; 37 : 31 – 40 . [DOI] [PubMed] [Google Scholar]
- 49. Peddada S, Yasui DH, LaSalle JM. Inhibitors of differentiation (ID1.; ID2.; ID3 and ID4) genes are neuronal targets of MeCP2 that are elevated in Rett syndrome . Hum Mol Genet 2006. ; 15 : 2003 – 14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Itoh M, Ide S, Takashima S , et al. . Methyl CpG-binding protein 2 (a mutation of which causes Rett syndrome) directly regulates insulin-like growth factor binding protein 3 in mouse and human brains . J Neuropathol Exp Neurol 2007. ; 66 : 117 – 23 . [DOI] [PubMed] [Google Scholar]
- 51. Carouge D, Host L, Aunis D , et al. . CDKL5 is a brain MeCP2 target gene regulated by DNA methylation . Neurobiol Dis 2010. ; 38 : 414 – 24 . [DOI] [PubMed] [Google Scholar]
- 52. Miyake K, Hirasawa T, Soutome M , et al. . The protocadherins.; PCDHB1 and PCDH7.; are regulated by MeCP2 in neuronal cells and brain tissues: implication for pathogenesis of Rett syndrome . BMC Neurosci 2011. ; 12 : 81.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chao HT, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number . Neuron 2007. ; 56 : 58 – 65 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Marchetto MC, Carromeu C, Acab A , et al. . A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells . Cell 2010. ; 143 : 527 – 39 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Farra N, Zhang WB, Pasceri P , et al. . Rett syndrome induced pluripotent stem cell-derived neurons reveal novel neurophysiological alterations . Mol Psychiatry 2012. ; 17 : 1261 – 71 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Djuric U, Cheung AY, Zhang W , et al. . MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells . Neurobiol Dis 2015. ; 76 : 37 – 45 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Andoh-Noda T, Akamatsu W, Miyake K , et al. . Differentiation of multipotent neural stem cells derived from Rett syndrome patients is biased toward the astrocytic lineage . Mol Brain 2015. ; 8 : 31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Inoue K, Kanai M, Tanabe Y , et al. . Prenatal interphase FISH diagnosis of PLP1 duplication associated with Pelizaeus-Merzbacher disease . Prenat Diagn 2001. ; 21 : 1133 – 6 . [DOI] [PubMed] [Google Scholar]
- 59. Reiner O, Carrozzo R, Shen Y , et al. . Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats . Nature 1993. ; 364 : 717 – 21 . [DOI] [PubMed] [Google Scholar]
- 60. Bi W, Sapir T, Shchelochkov OA , et al. . Increased LIS1 expression affects human and mouse brain development . Nat. Genet 2009. ; 41 : 168 – 77 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Online Mendelian Inheritance of Men (OMIM) #118220. http://omim.org/entry/118220 . (30 July 2016, date last accessed).
- 62. Obi T, Nishioka K, Ross OA , et al. . Clinicopathologic study of a SNCA gene duplication patient with Parkinson disease and dementia . Neurology 2008. ; 70 : 238 – 41 . [DOI] [PubMed] [Google Scholar]
- 63. Van Esch H, Bauters M, Ignatius J , et al. . Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males . Am J Hum Genet 2005. ; 77 : 442 – 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Novara F, Simonati A, Sicca F , et al. . MECP2 duplication phenotype in symptomatic females: report of three further cases . Mol Cytogenet 2014. ; 7 : 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Liu Z, Li X, Zhang JT , et al. . Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2 . Nature 2016. ; 530 : 98 – 102 . [DOI] [PubMed] [Google Scholar]
- 66. Cyranoski D. Monkeys genetically modified to show autism symptoms . Nature 2016. ; 529 : 449.. [DOI] [PubMed] [Google Scholar]
- 67. Okano M, Bell DW, Haber DA , et al. . DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development . Cell 1999. ; 99 : 247 – 57 . [DOI] [PubMed] [Google Scholar]
- 68. Kubota T, Furuumi H, Kamoda T , et al. . ICF syndrome in a girl with DNA hypomethylation but without detectable DNMT3B mutation . Am J Med Genet A 2004. ; 129A : 290 – 3 . [DOI] [PubMed] [Google Scholar]
- 69. Huang K, Wu Z, Liu Z , et al. . Selective demethylation and altered gene expression are associated with ICF syndrome in human-induced pluripotent stem cells and mesenchymal stem cells . Hum Mol Genet 2014. ; 23 : 6448 – 57 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kubota T, Das S, Christian SL , et al. . Methylation-specific PCR simplifies imprinting analysis . Nat Genet 1997. ; 16 : 16 – 7 . [DOI] [PubMed] [Google Scholar]
- 71. Schaaf CP, Gonzalez-Garay ML, Xia F , et al. . Truncating mutations of MAGEL2 cause Prader-Willi phenotypes and autism . Nat Genet 2013. ; 45 : 1405 – 8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stelzer Y, Sagi I, Yanuka O , et al. . The noncoding RNA IPW regulates the imprinted DLK1-DIO3 locus in an induced pluripotent stem cell model of Prader-Willi syndrome . Nat Genet 2014. ; 46 : 551 – 7 . [DOI] [PubMed] [Google Scholar]
- 73. Online Mendelian Inheritance of Men (OMIM) #176270. http://omim.org/entry/176270 (checked on 9 May 2016).
- 74. Rangasamy S, D'Mello SR, Narayanan V. Epigenetics.; autism spectrum.; and neurodevelopmental disorders . Neurotherapeutics 2013. ; 10 : 742 – 56 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Scoles HA, Urraca N, Chadwick SW , et al. . Increased copy number for methylated maternal 15q duplications leads to changes in gene and protein expression in human cortical samples . Mol. Autism 2011. ; 2 : 19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lim D, Bowdin SC, Tee L. Clinical and molecular genetic features of Beckwith-Wiedemann syndrome associated with assisted reproductive technologies . Hum Reprod 2009. ; 24 : 741 – 7 . [DOI] [PubMed] [Google Scholar]
- 77. Bliek J, Alders M, Maas SM , et al. . Lessons from BWS twins: complex maternal and paternal hypomethylation and a common source of haematopoietic stem cells . Eur J Hum Genet 2009. ; 17 : 1625 – 34 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Crews D, Gillette R, Miller-Crews I , et al. . Nature, nurture and epigenetics . Mol Cell Endocrinol 2014. ; 398 : 42 – 52 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gluckman PD, Seng CY, Fukuoka H , et al. . Low birthweight and subsequent obesity in Japan . Lancet 2007. ; 369 : 1081 – 2 . [DOI] [PubMed] [Google Scholar]
- 80. Painter RC, de Rooij SR, Bossuyt PM , et al. . Early onset of coronary artery disease after prenatal exposure to the Dutch famine . Am J Clin. Nutr 2006. ; 84 : 322 – 7 . [DOI] [PubMed] [Google Scholar]
- 81. Tobi EW, Lumey LH, Talens RP , et al. . DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific . Hum Mol Genet 2009. ; 18 : 4046 – 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Cooper W, Khulan B, Owens S , et al. . DNA methylation profiling at imprinted loci after periconceptional micronutrient supplementation in humans: results of a pilot randomized controlled trial . Faseb J 2012. ; 26 : 1782 – 90 . [DOI] [PubMed] [Google Scholar]
- 83. Benyshek DC. The “early life” origins of obesity-related health disorders: new discoveries regarding the intergenerational transmission of developmentally programmed traits in the global cardiometabolic health crisis . Am J Phys Anthropol 2013. ; 152 : 79 – 93 . [DOI] [PubMed] [Google Scholar]
- 84. Yaoi T, Itoh K, Nakamura K , et al. . Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A . Biochem Biophys Res Commun 2008. ; 376 : 563 – 7 . [DOI] [PubMed] [Google Scholar]
- 85. O'Brien E, Dolinoy DC, Mancuso P. Perinatal bisphenol A exposures increase production of pro-inflammatory mediators in bone marrow-derived mast cells of adult mice . J Immunotoxicol 2014. ; 11 : 205 – 12 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Weinhouse C, Bergin IL, Harris C , et al. . Stat3 is a candidate epigenetic biomarker of perinatal Bisphenol A exposure associated with murine hepatic tumors with implications for human health . Epigenetics 2015. ; 10 : 1099 – 110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dao T, Hong X, Wang X , et al. . Aberrant 5'-CpG methylation of cord blood TNFα associated with maternal exposure to polybrominated diphenyl ethers . PLoS One 2015. ; 10 : e0138815.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Richmond RC, Simpkin AJ, Woodward G , et al. . Prenatal exposure to maternal smoking and offspring DNA methylation across the lifecourse: findings from the Avon Longitudinal Study of Parents and Children (ALSPAC) . Hum Mol Genet 2015. ; 24 : 2201 – 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Baird G, Simonoff E, Pickles A , et al. . Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) . Lancet 2006. ; 368 : 210 – 5 . [DOI] [PubMed] [Google Scholar]
- 90. Yeargin-Allsopp M, Rice C, Karapurkar T , et al. . Prevalence of Autism in a US Metropolitan Area . JAMA 2003. ; 289 : 49 – 55 . [DOI] [PubMed] [Google Scholar]
- 91. Honda H, Shimizu Y, Imai M , et al. . Cumulative incidence of childhood autism: a total population study of better accuracy and precision . Dev Med Child Neurol 2005. ; 47 : 10 – 18 . [DOI] [PubMed] [Google Scholar]
- 92. Holden C. Autism Now . Science 2009. ; 323 : 565. [Google Scholar]
- 93. Fombonne E. Epidemiology of pervasive developmental disorders . Pediatr Res 2009. ; 65 : 591 – 8 . [DOI] [PubMed] [Google Scholar]
- 94. Kim YS, Leventhal BL, Koh YJ , et al. . Prevalence of autism spectrum disorders in a total population sample . Am J Psychiatry 2011. ; 168 : 904 – 12 . [DOI] [PubMed] [Google Scholar]
- 95. Herbert MR. Contributions of the environment and environmentally vulnerable physiology to autism spectrum disorders . Curr Opin Neurol 2010. ; 23 : 103 – 10 . [DOI] [PubMed] [Google Scholar]
- 96. Hoekstra RA, Bartels M, Hudziak JJ , et al. . Genetics and environmental covariation between autistic traits and behavioral problems . Twin Res Hum Genet 2007. ; 10 : 853 – 86 . [DOI] [PubMed] [Google Scholar]
- 97. Hallmayer J, Cleveland S, Torres A , et al. . Genetic heritability and shared environmental factors among twin pairs with autism . Arch Gen Psychiatry 2011. ; 68 : 1095 – 102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fraga MF, Ballestar E, Paz MF , et al. . Epigenetic differences arise during the lifetime of monozygotic twins . Proc Natl Acad Sci U. S. A 2005. ; 102 : 10604 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Miyake K, Yang C, Minakuchi Y , et al. . Comparison of genomic and epigenomic expression in monozygotic twins discordant for Rett syndrome . PLoS ONE 2013. ; 8 : e66729.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cao-Lei L, Massart R, Suderman MJ , et al. . DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm . PLoS One 2014. ; 9 : e107653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Vangeel EB, Izzi B, Hompes T , et al. . DNA methylation in imprinted genes IGF2 and GNASXL is associated with prenatal maternal stress . Genes Brain Behav 2015. ; 14 : 573 – 82 . [DOI] [PubMed] [Google Scholar]
- 102. Braithwaite EC, Kundakovic M, Ramchandani PG , et al. . Maternal prenatal depressive symptoms predict infant NR3C1 1F and BDNF IV DNA methylation . Epigenetics 2015. ; 10 : 408 – 17 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Palma-Gudiel H, Córdova-Palomera A, Eixarch E , et al. . Maternal psychosocial stress during pregnancy alters the epigenetic signature of the glucocorticoid receptor gene promoter in their offspring: a meta-analysis . Epigenetics 2015. ; 10 : 893 – 902 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Imura H. Life course health care and preemptive approach to non-communicable diseases . Proc Jpn Acad Ser B Phys Biol Sci 2013. ; 89 : 462 – 73 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Miller JL, Lynn CH, Shuster J , et al. . A reduced-energy intake, well-balanced diet improves weight control in children with Prader-Willi syndrome . J Hum Nutr Diet 2013. ; 26 : 2 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schlumpf M, Eiholzer U, Gygax M , et al. . A daily comprehensive muscle training programme increases lean mass and spontaneous activity in children with Prader-Willi syndrome after 6 months . J Pediatr Endocrinol Metab 2006. ; 19 : 65 – 74 . [DOI] [PubMed] [Google Scholar]
- 107. Angulo MA, Castro-Magana M, Lamerson M , et al. . Final adult height in children with Prader-Willi syndrome with and without human growth hormone treatment . Am J Med Genet A 2007. ; 143A : 1456 – 61 . [DOI] [PubMed] [Google Scholar]
- 108. Butler MG, Lee J, Cox DM , et al. . Growth charts for Prader-Willi syndrome during growth hormone treatment . Clin Pediatr (Phila) 2016. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Reus L, Pillen S, Pelzer BJ , et al. . Growth hormone therapy, muscle thickness, and motor development in Prader-Willi syndrome: An RCT . Pediatrics 2014. ; 134 : e1619 – 27 . [DOI] [PubMed] [Google Scholar]
- 110. Myers SE, Whitman BY, Carrel AL , et al. . Two years of growth hormone therapy in young children with Prader-Willi syndrome: Physical and neurodevelopmental benefits . Am J Med Genet A 2007. ; 143A : 443 – 8 . [DOI] [PubMed] [Google Scholar]
- 111. Siemensma EP, Tummers-de Lind van Wijngaarden RF, Festen DA , et al. . Beneficial effects of growth hormone treatment on cognition in children with Prader-Willi syndrome: a randomized controlled trial and longitudinal study . J Clin Endocrinol Metab 2012. ; 97 : 2307 – 14 . [DOI] [PubMed] [Google Scholar]
- 112. Osório J., Growth development Growth hormone therapy improves cognition in children with Prader-Willi syndrome . Nat Rev Endocrinol 2012. ; 8 . [DOI] [PubMed] [Google Scholar]
- 113. Höybye C, Thorén M, Böhm B. Cognitive, emotional, physical and social effects of growth hormone treatment in adults with Prader-Willi syndrome . J Intellect Disabil Res 2005. ; 49 : 245 – 52 . [DOI] [PubMed] [Google Scholar]
- 114. GeneReviews ® [Internet]. Prader-Willi Syndrome Available online: http://www.ncbi.nlm.nih.gov/books/NBK1330/ . (30 July 2016, date last accessed).
- 115. Sode-Carlsen R, Farholt S, Rabben KF , et al. . One year of growth hormone treatment in adults with Prader-Willi syndrome improves body composition: Results from a randomized, placebo-controlled study . J Clin Endocrinol Metab 2010. ; 95 : 4943 – 50 . [DOI] [PubMed] [Google Scholar]
- 116. Tsankova NM, Berton O, Renthal W , et al. . Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action . Nat Neurosci 2006. ; 9 : 519 – 25 . [DOI] [PubMed] [Google Scholar]
- 117. Fuchikami M, Morinobu S, Segawa M , et al. . DNA methylation profiles of the brain-derived neurotrophic factor (BDNF) gene as a potent diagnostic biomarker in major depression . PLoS ONE 2011. ; 6 : e23881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Bacos K, Gillberg L, Volkov P , et al. . Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes . Nat Commun 2016. ; 7 : 11089.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kundakovic M, Gudsnuk K, Herbstman JB , et al. . DNA methylation of BDNF as a biomarker of early-life adversity . Proc Natl Acad Sci USA 2015. ; 112 : 6807 – 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Jessberger S, Nakashima K, Clemenson GD, Jr , et al. . Epigenetic modulation of seizure-induced neurogenesis and cognitive decline . J Neurosci 2007. ; 27 : 5967 – 75 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Nghia NA, Hirasawa T, Kasai H , et al. . Long-term imipramine treatment increases N-methyl-d-aspartate receptor activity and expression via epigenetic mechanisms . Eur J Pharmacol 2015. ; 752 : 69 – 77 . [DOI] [PubMed] [Google Scholar]
- 122. Surén P, Roth C, Bresnahan M , et al. . Association between maternal use of folic acid supplements and risk of autism spectrum disorders in children . JAMA 2013. ; 309 : 570 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Guy J, Gan J, Selfridge J , et al. . Reversal of neurological defects in a mouse model of Rett syndrome . Science 2007. ; 315 : 1143 – 7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Derecki NC, Cronk JC, Lu Z , et al. . Wild-type microglia arrest pathology in a mouse model of Rett syndrome . Nature 2012. ; 484 : 105 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sztainberg Y, Chen HM, Swann JW , et al. . Reversal of phenotypes in MECP2 duplication mice using genetic rescue or antisense oligonucleotides . Nature 2015. ; 528 : 123 – 6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Hao S, Tang B, Wu Z , et al. . Forniceal deep brain stimulation rescues hippocampal memory in Rett syndrome mice . Nature 2015. ; 526 : 430 – 4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Kondo M, Gray LJ, Pelka GJ , et al. . Environmental enrichment ameliorates a motor coordination deficit in a mouse model of Rett syndrome- Mecp2 gene dosage effects and BDNF expression . Eur J Neurosci 2008. ; 27 : 3342 – 50 . [DOI] [PubMed] [Google Scholar]
- 128. Lonetti G, Angelucci A, Morando L , et al. . Early environmental enrichment moderates the behavioral and synaptic phenotype of MeCP2 null mice . Biol Psychiatry 2010. ; 67 : 657 – 65 . [DOI] [PubMed] [Google Scholar]
- 129. Nag N, Moriuchi JM, Peitzman CG , et al. . Environmental enrichment alters locomotor behaviour and ventricular volume in Mecp2 1lox mice . Behav Brain Res 2009. ; 196 : 44 – 8 . [DOI] [PubMed] [Google Scholar]
- 130. Kerr B, Silva PA, Walz K , et al. . Unconventional transcriptional response to environmental enrichment in a mouse model of Rett syndrome . PLoS One 2010. ; 5 : e11534.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Skinner MK, Manikkam M, Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology . Trends Endocrinol. Metab 2010. ; 21 : 214 – 22 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Hanson MA, Skinner MK. Developmental origins of epigenetic transgenerational inheritance . Environ Epigenet 2016. ; 2 : 1 – 9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Whitelaw E. Disputing Lamarckian epigenetic inheritance in mammals . Genome Biol 2015. ; 16 : 60.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals . Nat Rev Genet 2012. ; 13 : 153 – 62 . [DOI] [PubMed] [Google Scholar]
- 135. Franklin TB, Russig H, Weiss IC , et al. . Epigenetic transmission of the impact of early stress across generations . Biol Psychiatry 2010. ; 68 : 408 – 15 . [DOI] [PubMed] [Google Scholar]
- 136. Babenko O, Kovalchuk I, Metz GA. Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health . Neurosci Biobehav Rev 2015. ; 48 : 70 – 91 . [DOI] [PubMed] [Google Scholar]
- 137. Anway MD, Skinner MK. Epigenetic programming of the germ line: effects of endocrine disruptors on the development of transgenerational disease . Reprod Biomed Online 2008. ; 16 : 23 – 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Mao Z, Xia W, Chang H , et al. . Paternal BPA exposure in early life alters Igf2 epigenetic status in sperm and induces pancreatic impairment in rat offspring . Toxicol Lett 2015. ; 238 : 30 – 8 . [DOI] [PubMed] [Google Scholar]
- 139. Dias BG, Ressler KJ. Experimental evidence needed to demonstrate inter- and trans-generational effects of ancestral experiences in mammals . Bioessays 2014. ; 36 : 919 – 23 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Heard E, Martienssen RA. Transgenerational epigenetic inheritance: myths and mechanisms . Cell 2014. ; 157 : 95 – 109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Stouder C, Paoloni-Giacobino A. Specific transgenerational imprinting effects of the endocrine disruptor methoxychlor on male gametes . Reproduction 2011. ; 141 : 207 – 16 . [DOI] [PubMed] [Google Scholar]
- 142. Hanson MA, Low FM, Gluckman PD. Epigenetic epidemiology: the rebirth of soft inheritance . Ann Nutr Metab 2011. ; 58 : 8 – 15 . [DOI] [PubMed] [Google Scholar]
- 143. Dickins TE, Rahman Q. The extended evolutionary synthesis and the role of soft inheritance in evolution . Proc Biol Sci 2012. ; 279 : 2913 – 21 . [DOI] [PMC free article] [PubMed] [Google Scholar]