Abstract
There is increasing evidence, particularly from plants, that epigenetic mechanisms can contribute to environmental adaptation and evolution. The present article provides an overview on this topic for animals and highlights the special suitability of clonal, invasive, hybrid, polyploid, and domesticated species for environmental and evolutionary epigenetics. Laboratory and field studies with asexually reproducing animals have shown that epigenetically diverse phenotypes can be produced from the same genome either by developmental stochasticity or environmental induction. The analysis of invasions revealed that epigenetic phenotype variation may help to overcome genetic barriers typically associated with invasions such as bottlenecks and inbreeding. Research with hybrids and polyploids established that epigenetic mechanisms are involved in consolidation of speciation by contributing to reproductive isolation and restructuring of the genome in the neo-species. Epigenetic mechanisms may even have the potential to trigger speciation but evidence is still meager. The comparison of domesticated animals and their wild ancestors demonstrated heritability and selectability of phenotype modulating DNA methylation patterns. Hypotheses, model predictions, and empirical results are presented to explain how epigenetic phenotype variation could facilitate adaptation and speciation. Clonal laboratory lineages, monoclonal invaders, and adaptive radiations of different evolutionary age seem particularly suitable to empirically test the proposed ideas. A respective research agenda is presented.
Keywords: epigenetic variation, adaptation, general-purpose genotype, speciation, genome reconfiguration, monoclonal invaders
Introduction
The generation of phenotypic variation by genetic mechanisms and natural selection of the best suited phenotypes is the central tenet of the prevailing neo-Darwinian theory of evolution [1]. However, phenotypic diversity can also result from the production of epigenetic variants from the same genome either via developmental stochasticity or environmental induction [2–6]. Earlier, genetic variation was regarded as the sole type of variation that could produce an evolutionary response. Epigenetic sources of phenotype variation were seen as a one-generation phenomenon without evolutionary impact because they were considered non-heritable.
The importance of the environment for shaping of the phenotype and evolution has been recognized since Darwin’s times and has been reinforced in recent discussions concerning phenotypic plasticity [7–12]. Meanwhile, there is sound evidence that developmental and environmental influences on phenotype are mediated by epigenetic mechanisms such as DNA methylation, histone modifications and microRNAs [2, 13–15]. The involvement of epigenetic mechanisms in the production of phenotypic diversity is much better investigated in plants than in animals [16–19]. However, if the role of epigenetic phenotype variation is to be understood in ecology and evolution, its significance must also be demonstrated for animals. Although research on environmental and evolutionary epigenetics is still in an early stage there have already been attempts to integrate it into evolutionary theory [3, 8, 12].
Most animal populations are obligatory sexual and recombine the phenotype-determining genes in each generation, which makes research on the role of epigenetics in animal ecology and evolution difficult. Genetically identical or genetically depauperate populations may help to overcome this constraint, as will be discussed below. The article starts with a comparison of stochastic developmental and environmentally induced phenotype variation. It then reviews the contribution of asexually reproducing, invasive, domesticated, hybrid, and polyploid animals to the understanding of the role of epigenetic mechanisms in ecological adaptation, consolidation of speciation, and potential triggering of speciation. The last section elaborates on the special suitability of monoclonal invaders for research on environmental and evolutionary epigenetics and proposes a research agenda.
Stochastic Developmental versus Environmentally Induced Phenotype Variation
Phenotypic variation can be subdivided into genetic, stochastic developmental, and environmental components [4, 20, 21]. The two epigenetic proportions are called ‘stochastic developmental phenotype variation’ (SPV) and ‘environmentally induced phenotype variation’ (EPV) in the following. Both epigenetic phenomena are closely intermingled but represent different mechanisms to generate phenotypic diversity [3–5, 13, 22–24].
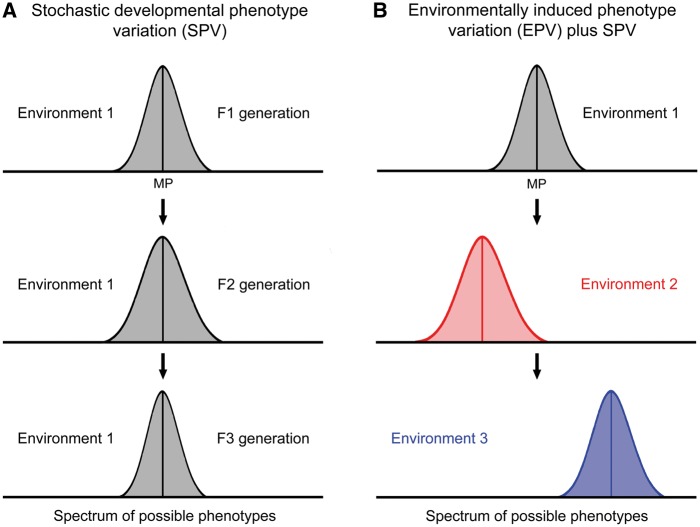
The difference between SPV and EPV can be best illustrated by raising genetically identical populations either in the same or different environments [24]. SPV (sometimes called ‘developmental noise’) generates a range of epigenetically mediated phenotypes around a mean phenotype (MP) that is determined by the interaction of the genome and the prevailing environment (Fig. 1A). The range of SPV can randomly vary between identically raised populations or between subsequent generations grown in the same environment but the MP is thought to remain constant in both cases (Fig. 1A). This a priori production of a range of epigenetically diverse phenotypes from the same genotype (DNA sequence and its copies) without knowing the future conditions serves for risk-spreading that enhances the chance to stay in the game of life when the environmental conditions change. It is thought to contribute to evolutionary bet-hedging [3–5]. SPV seems to be particularly important for asexual species and lineages but is probably also effective in sexually reproducing species, which use genetic recombination as a major generator of phenotypic diversity.
Figure 1.
Stochastic developmental and environmentally induced phenotype variation exemplified by genetically identical populations. (A) In a given environment, developmental stochasticity produces a range of epigenetically diverse phenotypes around a mean phenotype (MP). The MP is thought to express an optimal epigenetic profile resulting from the interaction of the genome and the prevailing environment. The range around the MP can be symmetric or asymmetric. In a stable environment the MP holds its position on the scale of possible phenotypes throughout the following generations but the stochastically produced ranges of phenotypes around it may vary to some degree. (B) Different environments induce shifts of the MP on the scale of possible phenotypes. Environmental cues may also have an influence on the range of SPV around the MP
EPV is synonymous with phenotypic plasticity sensu strictu, the adaptive response of populations to environmental signals [3, 5, 8, 25, 26]. Researchers on phenotypic plasticity have often subsumed stochastic developmental and environmental influences on phenotype under this term [9]. For reasons of clarity, I use EPV instead of phenotypic plasticity in the following. Cues from different environments can induce different epigenetically mediated phenotypes from the same genotype (Fig. 1B). Snell-Rood [27] defined two forms of phenotypic plasticity: developmental and activational. Developmental plasticity refers to the capacity of a genotype to adopt different developmental trajectories in different environments (not to be confused with SPV). Activational plasticity refers to differential activation of an underlying network in different environments such that an individual expresses various phenotypes throughout its lifetime. In contrast to SPV, EPV is thought to be directional shifting the MP to different positions on the scale of possible phenotypes (Fig. 1B). The MPs of all environments together form the reaction norm curve for phenotypic traits [28]. SPV and EPV are closely intermingled because the environment determines the position of the MP on the scale of possible phenotypes and sets the frame in which SPV can vary (Fig. 1B).
A special case of environmentally induced phenotype induction is polyphenism, the switch between alternative phenotypes in response to environmental signals [29–31]. A good example is the queens and workers of honey bee, which originate from the same genome but differ in morphology, social behavior, and life span due to different feeding of their larval stages (royal jelly for the designated queens, pollen for the workers). These phenotypic differences are mediated by epigenetic mechanisms. Lyko et al. [31] established that DNA methylation in brains of queens and workers of bee differs in more than 550 genes, including genes involved in metabolism, brain development, and neural functions. Herb et al. [32] revealed that DNA methylation is even involved in the reversible switch of honey bee workers between nursing and foraging periods.
All traits in animals seem to vary due to SPV but at different degrees, depending on species and condition [4]. As a rule of thumb, morphological traits show the lowest degree of SPV whereas behavioral traits show the highest degree [4]. Probably, only few of these variations have an adaptive potential, among them traits involved in feeding, camouflage, or temperature resistance. Examples of traits without adaptive potential are the fingerprints of primates, which are highly variable among genetically identical twins [4]. Similarly, the majority of environmental cues do not induce EPV. Many result in a short-term physiological response and many in no response at all. Most examples on EPV published so far for animals concern environmental toxicants [33, 34]. Food shortage, predator pressure, and harsh environmental conditions are thought to be the main elicitors of non-pathological EPV [3, 26, 35].
Animals seem to have sensitive windows for the susceptibility to SPV, which depends much on trait and life history of the species considered [4]. As a rule of thumb, embryonic development seems to be of prime importance in determinately growing mammals and insects, whereas the adult life period appears equally important in indeterminately growing crustaceans, molluscs, and fish [4]. For example, differences in adult body weight among identically raised littermates of inbred rat were shown to have their roots in stochastic events in the zygote and first cleavage stages [36], whereas in isogenic batch-mates of marbled crayfish such differences were more attributable to the adult life [24]. SPV-related differences in ageing and diseases of humans can have their origin in both early development and the later life [37, 38]. Similar sensitive windows also exist for EPV [3, 33, 39]. The environmental exposure at a critical window can alter the epigenetic programming and subsequently change gene expression [33].
Facilitation of Ecological Adaptation by Epigenetic Phenotype Variation Exemplified by Invasions
Investigation of natural animal populations revealed that epigenetic diversity is usually higher than genetic diversity. This was shown for species diverse as sandhoppers, fish, and bats [40–42]. These findings suggest that wild animals might use epigenetic phenotype variation in addition to genetic variation to adapt to different environments. In sexually reproducing species, the generation of different phenotypes via genetic recombination and selection of the best suited phenotypes is regarded as the prime mechanism of ecological adaptation. However, this mode of adaptation to new environments is not applicable for asexually reproducing populations and genetically uniform invasive groups. The generation of a range of epigenetically different phenotypes from the same genome is probably an alternative in these organisms.
Epimutations are apparently much more frequent than genetic mutations, e.g. 3 × 10−4 methylation gains or losses per CpG and generation compared with 7 × 10−9 base substitutions per site and generation in the model plant Arabidopsis thaliana [43, 44]. One may conclude from this fact that the generation of phenotypic diversity by epigenetic mechanisms occurs with much higher speed when compared with random mutation, which is the only genetic mechanism to generate phenotypic diversity in asexual lineages. However, it is largely unknown how many single nucleotide polymorphisms (SNPs) or single base methylation polymorphisms (SMPs) are required to change phenotypic traits. The relationship between genetic, epigenetic, and phenotypic variation is presently investigated in animals, plants, and humans by genome-wide association studies [45–48]. Individual SMPs seem to have no direct phenotypic effect [49] but many of them together produce differentially methylated regions (DMRs) and epialleles, which have been shown to underlie phenotypic changes [48–52]. Epialleles are often considered to have a low stability as will be discussed in detail later on, and some theoretical models suggest that epigenetic variation may be less responsive to natural selection than genetic variation, even in cases where levels of heritability appear similar [53]. These argument seem to speak against a significant role of epigenetic phenotype variation in adaptation.
Arguments for a supportive role of epigenetic phenotype variation in environmental adaptation of animals come from biological invasions [54]. Adaptation is the process by which species become fitted to their environment. It is the result of the action of natural selection upon heritable variation. Invaders can show two different types of adaptation, plastic and genetic [26, 55]. Plastic adaptation is based on EPV plus SPV and is mediated by epigenetic mechanisms. Genetic adaptation is based on genetic variation and selection. The relationship between the two is complex and poorly understood [26]. Interestingly, invasive groups can be ecologically and evolutionarily very successful even when genetically impoverished. This phenomenon is well known as ‘invasion paradox’ but the underlying mechanisms are largely unknown [56–59]. Particularly the first steps of invasion, the survival of the invading group and the establishment of a founder population, may depend much on epigenetic variation.
An animal example of the involvement of epigenetic mechanisms in adaptation to new environments is the invasion of house sparrow, Passer domesticus, in Kenya [60]. House sparrows were introduced to Mombasa in the 1950s and since evolved significant phenotypic variation. Analysis of DNA methylation sensitive-amplified fragment length polymorphisms (MS-AFLP) and microsatellite loci revealed that Kenyan house sparrow populations have a low genetic diversity but a high epigenetic diversity. There was a significant negative correlation between epigenetic and genetic diversity (Fig. 2) and a positive correlation between epigenetic diversity and the degree of inbreeding, suggesting that DNA methylation may help to overcome genetic barriers typically associated with invasions such as genetic bottlenecks and inbreeding.
Figure 2.
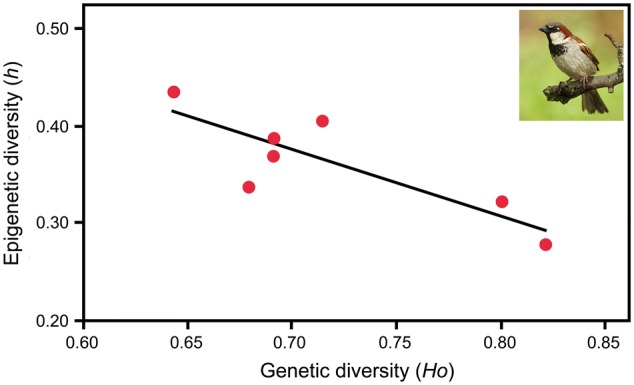
Relationship of genetic and epigenetic diversity in invasive house sparrow, Passer domesticus, from seven Kenyan cities. House sparrows were introduced to Mombasa in the 1950s and have considerably expanded their range since then. Epigenetic diversity was determined by MS-AFLP and genetic diversity by microsatellite analysis. The graph shows that epigenetic diversity is negatively correlated with genetic diversity, suggesting that epigenetic variation may compensate for low genetic variation. h, haplotype diversity; Ho, heterozygosity (redrawn after [60]; photograph by Andreas Mrowetz)
The production of epigenetically diverse phenotypes from the same genome may also explain why many clonal animals and plants are able to live in a wide range of habitats and geographical regions, despite their genetic uniformity. This phenomenon is often explained by the existence of so-called ‘general-purpose genotypes’ or ‘superclones’, which are thought to be able to cope with a broad spectrum of environmental challenges [61–63]. An interesting animal example is the gynogenetic all-female fish Chrosomus eos–neogaeus, which occurs in North America in 14 genetically identical lineages originating from different hybridization events between the redbelly dace C. eos and the fine-scale dace C. neogaeus [64]. Dating of hybridization events suggested a relatively recent origin (<50 000 years ago). Each hybrid lineage apparently originated from a single hybrid zygote and is genetically uniform with the exception of random mutations that accumulated over time. These lineages frequently display a large geographical distribution at a regional scale. The analysis of DNA methylation polymorphisms within such lineages revealed epigenetic similarity of individuals in a given lake but significant differences between lakes, suggesting that epigenetic phenotype diversification may be the mechanistic explanation of the ‘general purpose genotype’ [65].
Investigation of clonal populations has the advantage that genetic variability is usually much smaller than in sexually reproducing populations but it can still be present. Thus, the observed epigenetic variation might mimick genetic variation to some extent (see discussion on polyclonal and monoclonal invaders below).
Recent analysis of DNA methylation in C. eos–neogaeus lineages from predictable and unpredictable environments (lakes versus intermittent headwater streams) in Canada identified the relative contributions of SPV and EPV to phenotype modification [5]. Comparison of clones from the wild revealed that risk-spreading SPV prevails in unpredictable environments whereas directional EPV is predominant in predictable environments (Fig. 3). Differences in environmental effects on epigenetic variation between sympatric lineages (Fig. 3, ET3 and ET4) and lineages reared in similar experimental conditions (Fig. 3, LR1 and ET2) further showed that the epigenetic response to environmental signals is strongly influenced by the genotype. Common garden experiments revealed that the proportion of pure environmental effects can considerably change when clone members are transplanted into a new environment (Fig. 3, LR1 and ET2 under natural and experimental conditions). The example of C. eos–neogaeus demonstrates that SPV and EPV always act conjointly but have different weighting in different environments.
Figure 3.
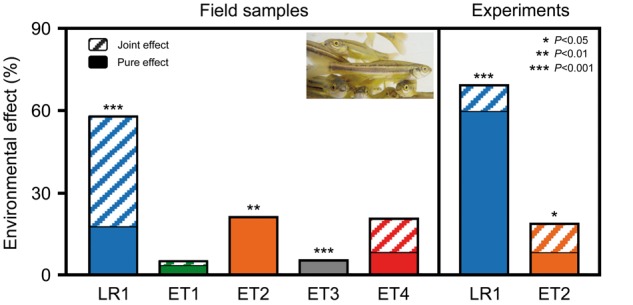
Environmental effects on epigenetic variation in asexual fish Chrosomus eos–neogaeus under natural and experimental conditions. Environmental and genetic joint effect is separated from the pure environmental effect. Epigenetic differences were determined by MS-ALFP and genetic differences by microsatellite analysis. LR1 to ET4 refer to distinct hybrid lineages from the Laurentians region (LR) and Eastern Townships (ET) of southern Quebec (Canada). LR1 lineages are from environmentally stable lakes whereas ET1–ET4 are from environmentally unstable streams. Experimental animals were sampled as larvae from the indicated sites and raised in common garden experiments for five months until adults. P-values refer to significance of the proportion of epigenetic variation explained uniquely by environment. The graph shows differences of environmental effects on epigenetic variation between predictable (LR1) and unpredictable environments (ET1–ET4) and among lineages even if they occur in sympatry (ET3 and ET4). Comparison of LR1 and ET2 in the field and laboratory suggests rapid epigenetic response to environmental change (redrawn after [5]; photograph from [5])
Facilitation of Evolution by Epigenetic Mechanisms Exemplified by Hybrids, Polyploids, Adaptive Radiations, and Domesticated Animals
Even if considered as a one-generation phenomenon only, the production of a range of epigenetically determined phenotypes from the same genetic template would have some evolutionary consequences since it helps populations, particularly clonal ones, to stay in the game of life when the environmental conditions change [2, 4, 66]. The evolutionary relevance of epigenetically mediated phenotypes would even be far greater if the best suited ones were inherited across generations, selected, and genetically fixed on the long range. In the following, I will first present some examples on the consolidating role of epigenetic mechanisms in speciation and then discuss the possibility of epigenetic mechanisms as triggers of speciation.
Stabilization of New Lineages and Species by Epigenetic Mechanisms
Epigenetic mechanisms are thought to contribute to speciation by stabilizing the genomes of new species [67, 68]. The examples presented below show that epigenetic patterns can markedly change during speciation, particularly in hybrids and polyploids, in which genome rearrangements are most intense. However, these examples do not conclusively answer the question whether the observed epigenetic alterations passively follow genetic changes or whether they play an active role in restructuring of the genome, which I consider possible. The relationship between epigenetics and speciation is better investigated in plants than in animals and is particularly pronounced in polyploids and domesticated species [67–73].
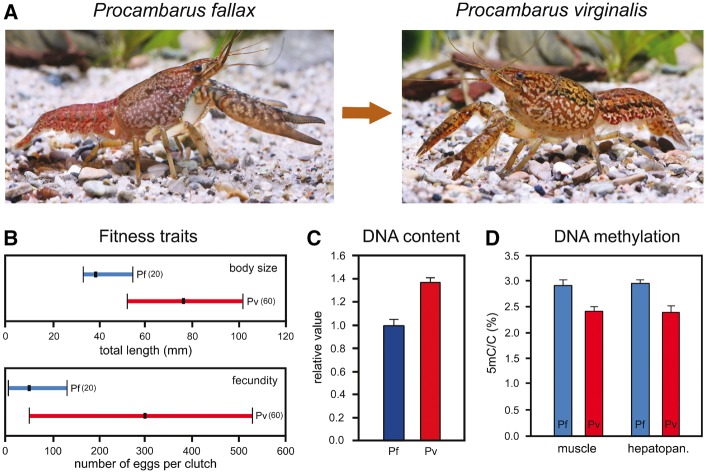
Polyploid speciation is usually saltational and accompanied by reproductive isolation, alteration of life history traits, and modifications of DNA content and DNA methylation level. An example is the marbled crayfish Procambarus virginalis (Fig. 4A), which originated in evolutionarily recent times from slough crayfish Procambarus fallax (Fig. 4A) by autotriploidy [74–76]. Concomitantly, the sexual system shifted from gonochorism to apomictic parthenogenesis. Body size and fecundity were significantly enhanced in the neo-species (Fig. 4B), indicating superior fitness. Mean life span of the adults increased from about 1 year in P. fallax to 2 years in P. virginalis [77, 78].
Figure 4.
Alteration of fitness traits, DNA content, and DNA methylation level in crayfish after saltational speciation. (A) Marbled crayfish Procambarus virginalis originated from slough crayfish P. fallax by autotriploidization and concomitant change of the sexual system from gonochorism to parthenogenesis. (B) Marbled crayfish (Pv) grow considerably bigger and are more fecund than slough crayfish (Pf). Dots and horizontal bars indicate means and ranges, and figures in brackets give numbers of specimens investigated. (C) Marbled crayfish have a 1.4-fold DNA content in blood cells when compared with the parent species. Vertical bars are SDs of three samples measured by flow cytometry. (D) Global DNA methylation of abdominal musculature and hepatopancreas (major organ of metabolism) is about 20% lower in marbled crayfish than in slough crayfish, suggesting marked alterations of DNA methylation during speciation. Vertical bars are SDs of three samples measured by mass spectrometry (redrawn after [76]; photographs by Chris Lukhaup)
Triploid marbled crayfish has a 1.4-fold increased DNA content when compared with its diploid parent species (Fig. 4C), suggesting loss of some DNA after autopolyploidization. Global DNA methylation (5-methylcytosine/total cytosine) is about 20% lower in marbled crayfish (Fig. 4D), which mainly results from hypomethylation of genes and repeats. Comparison of both species at the whole genome level identified larger proportions of genomic information specific to P. fallax as well as differentially expressed and differentially methylated genes, suggesting the involvement of DNA methylation in alteration of gene expression [79].
Reproductive isolation is an important requirement for speciation. In plants, there is multiple evidence for the contribution of DNA methylation to hybrid incompatibility [70]. An animal example is the deer mouse Peromyscus maniculatus species complex, in which the reproductive barrier is provided by epigenetic imprinting of genes involved in placentation [80]. New autopolyploid and allopolyploid (hybrid) genomes are usually unstable and require chromatin rearrangement, which is apparently achieved by the interaction of genetic and epigenetic mechanisms [67, 81, 82]. A common feature in polyploid neo-species is the reduction of the genome size, in extreme cases back to diploidy [83]. DNA loss mostly concerns redundant genes. For example, after doubling of the genome in the stem line of fishes 70–80% of duplicated genes have been lost over time [83].
The global DNA methylation levels in polyploids can either be higher or lower when compared with the parent species. For example, hybrids of kangaroo Macropus eugenii and Wallabia bicolor and autotriploids of crayfish P. virginalis showed genome-wide undermethylation [76, 81], whereas hybrids of red crucian carp Carassius auratus and common carp Cyprinus carpio displayed hypermethylation [84]. In the kangaroo hybrids, the removal of DNA methylation from retrotransposons facilitated their amplification and caused gross changes in genome structure.
In hybrids of the frogs Xenopus laevis and Xenopus muelleri, 364 out of 546 MS-AFLP markers were effective in elucidating the difference in methylation patterns between hybrids and parental species. Hybrids exhibited a significantly higher proportion of methylated fragments relative to both parental species, which may translate into changes of gene expression profiles. Moreover, 76 methylated fragments were diagnostic of hybrids only (Fig. 5). These new epigenetic patterns may indicate the involvement of epigenetic mechanisms in restructuring of the hybrid genome [85]. The parental species and hybrids are all tetraploids but X. muelleri and the hybrids have a DNA content of 113.5% and 112.3% when compared with X. laevis, indicating that DNA methylation levels are not strictly associated with genome size. Hybrid females are fertile and hybrid males are sterile. Differential methylation between sexes (Fig. 5) and misexpression of genes responsible for reproduction in males may account for these differences [85, 86].
Figure 5.
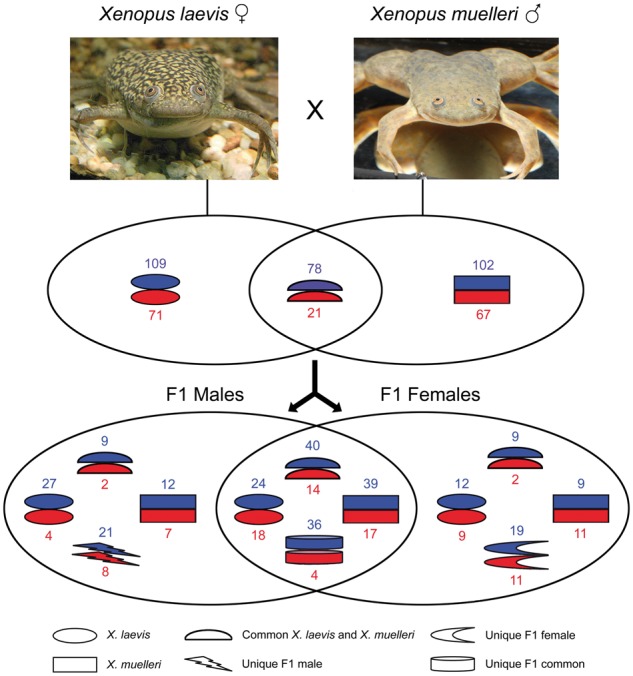
Changes in DNA methylation profiles after hybridization of Xenopus frogs. The graph shows methylated (blue) and unmethylated (red) fragments obtained by MS-AFLP from muscle tissue of parents and their F1 hybrids. Hybrids have higher proportions of methylated fragments than the parental species. Seventy-six methylated fragments are diagnostic of hybrids only suggesting the involvement of DNA methylation in genome reconfiguration after hybridization. Some of the methylated fragments in hybrids are sex-specific and may account for the difference in fertility between females and males (redrawn after [85]; photograph of X. laevis by Benedikt Rauscher, photograph of X. muelleri by Martin Grimm)
Polyploids often have life history traits that are different from those of the parent species as shown for marbled crayfish (Fig. 4B). Growth, number of offspring, and other quantitative traits can either be decreased or increased when compared with the diploid ancestors [76, 87, 88]. In allopolyploids, the increase of life history traits is usually explained as the result of heterozygosity. This explanation is not applicable for autopolyploids, which have the same set of genes as their parent species. In autopolyploids, trait alteration is caused by changes of gene dosage, rearrangement of gene-networks, and modulation of gene expression. All of these features apparently require the contribution of epigenetic mechanisms. As an example, during ancient mammalian gene duplication, DNA methylation apparently played a dominant role in dosage rebalance by inhibiting transcription initiation of duplicate genes [89].
Domesticated animals are particularly suitable for evolutionary epigenetics because their evolution (breeding history) is short and much better known when compared with wild species. Moreover, domesticated species show an exceptionally broad variation in phenotypic traits and their genetics is comparably well investigated. In dogs (Canis familiaris), which descended from the grey wolf (Canis lupus), some of the phenotypic changes have already been linked to specific genes. For example, different alleles involved in the fight-or-flight response have been subject to strong selection and resulted in behavioral differences between dogs and wolves [72]. Janowitz Koch et al. [73] established that domestication of dogs has also been associated with epigenetic alterations. They analyzed methylation differences in >24 000 cytosines distributed across the genomes and revealed species-specific patterns of DMRs at 68 sites. The majority (>66%) of DMRs were associated with repetitive elements, indicative of a genotype-mediated trend. However, DMRs were also often linked to functionally relevant genes (e.g. neurotransmitters), suggesting that selection has not only acted on genes but also on methylation patterns [73].
Triggering of Evolutionary Change by Epigenetic Mechanisms—A Provocative Idea
Genetics and epigenetics are closely linked, simply because epigenetic marks are on the nucleotides and histones that built up the DNA and chromatin. Therefore, when analyzing genetic and epigenetic diversity in populations it is difficult to identify what was first, genetic variation or epigenetic variation. Probably, most scientists tend to interpret epigenetic changes as followers of genetic changes [90, 91] rejecting the possibility of a leading role of epigenetic mechanisms in evolution (genetics-first hypothesis). However, some authors including myself consider the latter alternative possible [4, 7, 18, 92] (epigenetics-first hypothesis), which would considerably enhance the relevance of development and environment for evolution.
Support for the genetics-first hypothesis comes from the methylome analysis of a geographically dispersed Arabidopsis thaliana population of near-isogenic lines that diverged for at least a century from a common ancestor [91]. Methylome variation largely reflected genetic distance and was in many aspects similar to that of lines raised in uniform conditions. The authors concluded from their results that even when plants are grown in varying and diverse natural sites genome-wide epigenetic variation accumulates in a clock-like manner, suggesting that epigenetic divergence parallels the pattern of genome-wide DNA sequence divergence.
Support for the epigenetics-first hypothesis comes from the investigation of the well-known radiation of Darwin’s finches [92]. Darwin’s finches evolved over a period of 2–3 million years from a single invader and yielded 14 extant species that fill distinct ecological niches. In the five species investigated, epimutations (DMRs) were more common than genetic mutations (copy number variants, CNVs). Moreover, the number of DMRs increased monotonically with phylogenetic distance, whereas the number of CNVs did not (Fig. 6). The number, chromosomal locations, regional clustering, and lack of overlap of epimutations and genetic mutations suggest that epigenetic changes are distinct and may cause evolutionary change independently from genetic change.
Figure 6.
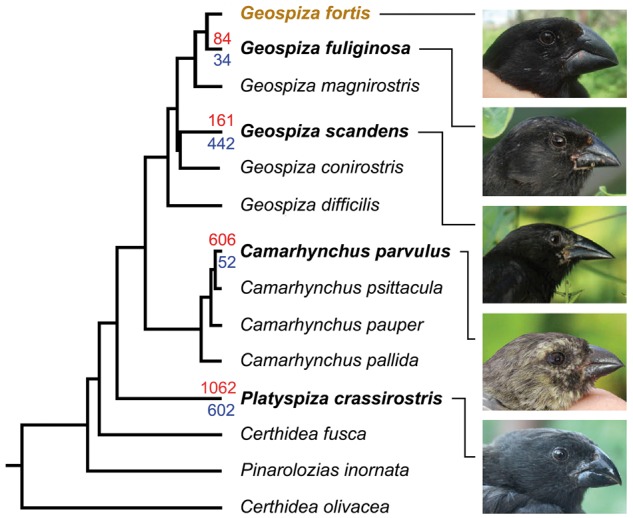
Correlation of genetic and epigenetic changes with phylogenetic distance in five species of Darwin’s finches. Red numbers on branches are epimutations (DMRs) and blue numbers are genetic mutations (CNVs) for four species compared with a reference species (Geospiza fortis). The phylogram is based on allele length variation at 16 polymorphic microsatellite loci. Photographs show variation in bill size and shape. The graph shows that the number of DMRs increases consistently with phylogenetic distance, whereas the number of CNVs does not (redrawn after [92]; photographs by Jennifer A. H. Koop and Sarah A. Knutie)
The idea of epigenetic variation as a driving force of evolution requires transgenerational inheritance of adaptive epigenetic patterns, selection of the epigenetically determined phenotypes, and genetic fixation of these phenotypes on the long term. The concept of transgenerational epigenetic inheritance (TEI) is still controversial [93] but there are several theoretical models and convincing empirical examples published for plants and animals [7, 94–98]. Major arguments against the involvement of epigenetics in animal evolution come from Weismann's germ plasm theory [94, 99] and the observation that DNA methylation is erased in the primordial germ cells and the zygote of mammals [100, 101], preventing epigenetic profiles from being inherited to the next generation. However, both objections were inferred from mammals only and do not apply to all animal groups. In nematodes, insects, and vertebrates, there is indeed an early separation of the germline from the soma [102], but in sponges, cnidarians, bryozoans, flatworms, annelids, ascidians, and echinoderms, somatic cells can transform into germ cells in later life stages, and thus, somatic epimutations acquired during early life stages may reach the germs cells to be trangenerationally inherited. Moreover, many animals can reproduce asexually by budding, fragmentation, and parthenogenesis [103, 104], sometimes alternating with sexual reproduction. Genome-wide DNA demethylation and remethylation has so far only been demonstrated for the germ cells and zygote of sexually reproducing mammals but not for the germ cells and zygote of sexually reproducing invertebrates and the reproductive units of asexually reproducing animals. And even in mammals, there are increasing examples of incomplete DNA methylation in the zygote [105] and epigenetic inheritance via microRNAs and DMRs in sperm [35, 106–108].
Natural selection acts on phenotypes, and therefore, epigenetic variation is thought to provide a substrate for selection similarly to genetic variation [109]. Ideally, only those epigenetic profiles should be preserved and selected that accomplish adaptation to new environmental conditions or enhance fitness in the prevailing environment. Herman et al. [110] emphasized that the stability of epigenetic states beyond a single generation depends on the degree of environmental variation and the costs of epigenetic resetting. Becker et al. [111] investigated spontaneous epimutations in 10 lineages of A. thaliana over 30 generations under constant greenhouse conditions and found that under these conditions not many epimutations were inherited over the long term. Some sites seemed to go through recurrent cycles of forward and reverse epimutations. This seemingly low stability of epimutations in constant environments was often used to argue against an evolutionary role of epigenetic inheritance.
Recent theory established that epigenetic flexibility can buy time for a genetic response to evolve. Epigenetic variants can be exposed to selection so that they follow dynamics similar to those of ordinary genetic variants [112–115]. However, under some circumstances epigenetic inheritance can impede adaptation. Kronholm and Collins [114] modeled cases where epigenetic mutations speed adaptation resulting in populations with higher fitness and cases where they slow adaptation resulting in populations with lower fitness. The effect of epigenetic mutations on adaptation seems to depend on their stability and fitness effects relative to genetic mutations. Geoghegan and Spencer [116] investigated the conditions under which an epigenetically modifiable allele can invade a population. They found that epialleles were less likely to invade when mismatch between environment and phenotype was too costly. Moreover, for a wide range of parameters, a higher rate of germline epigenetic resetting decreased the likelihood of epiallele invasion. Furrow and colleagues [53, 117] predicted from their model that epigenetic variation may be less responsive to natural selection than genetic variation. On the other hand, simple deterministic selection models showed that newly arising epimutations are stable enough to respond effectively to long-term selection, yielding epimutation-selection equilibria that are close to those expected for DNA sequence mutation rates [43]. Appropriate experiments should be designed to empirically test these conflicting model predictions.
Epigenetically determined phenotypes can be genetically accommodated or assimilated on the long term by a series of quantitative genetic changes [9, 10, 118–120]. This mechanism requires pre-existing genetic variation in the population and is thus not applicable to genetically uniform populations. There, epigenetically caused phenotypic diversity may be genetically fixed either by awaiting suitable random mutations or, probably with much higher speed, by facilitated mutations at epigenetically modified sites. Potential mechanisms are the well-established high mutability of methylated CpGs [121], the transposable element-epigenetic component engine [122], or the promotion of genetic mutations in CNVs via epimutations [92]. Methylated CpGs have a much higher mutation rate when compared with cytosine in other contexts. Such sites can even have mutational effects on the surrounding DNA [121]. Transposons are often silenced by DNA methylation, which is responsive to environmental cues. If activated by new environmental signals they can multiply and shift to new sites in the genome generating genetic diversity [68, 123]. Transgenerationally inherited sperm epimutations in rat were shown to promote genome instability such that CNV mutations were acquired in later generations [92].
The potential role of TEI for evolution is discussed in depth in Jablonka and Raz [94]. The authors stressed that epigenetic control mechanisms and heritable epigenetic variations are relevant for evolution because they affect both the processes of adaptation and divergence. Adaptation can occur through the selection of heritable epialleles without any genetic change, which is of particular importance when populations are small and lack genetic variability. Bonduriansky et al. [124] reviewed models and empirical examples of adaptation to changing environments that included environmentally induced as well as spontaneously arising phenotypic variants. They predicted that non-genetic inheritance can increase the rate of both genetic and phenotypic change and, in some cases, alter the direction of change. Empirical evidence confirms that a diversity of epigenetically determined phenotypes, spanning a continuum from adaptive to pathological, can be transmitted non-genetically [124].
Branciamore et al. [125, 126] revealed with their model that epigenetic modifications of gene duplicates and transposons with rates consistent with experimental observation can have significant micro- and macro-evolutionary effects. Epigenetic modifications can drive neo- and subfunctionalization of gene duplicates thus dramatically improving the efficacy of evolution. Epigenetic mechanisms can also rapidly fix transposons and cis-located gene alleles. Interestingly, the model further suggests that epigenetic silencing, even if strictly transient (being reset at each generation), can still have significant macro-evolutionary effects.
Clonal Invaders as Promising Models for Environmental and Evolutionary Epigenetics: Theoretical Considerations and Research Agenda
The different outcomes and interpretations of theoretical models, laboratory experiments, and field studies complicate our understanding of the role of epigenetics in ecology and evolution, requiring further studies with simple model systems. Clonal invaders seem to be particularly useful because they combine the advantages of clonal organisms and invaders and can adapt to different environments despite genetic uniformity. There are several interesting clonal animal invaders known, among them crustaceans, insects, snails, and fishes [5, 64, 127–131]. In this section, I will first highlight the advantages of clonal species and successful invaders for research on environmental and evolutionary epigenetics. Thereafter, I will sketch a simple scenario on the invasion of a new environment by a monoclonal invader to hypothesize about the role of epigenetic phenotype diversification in adaptation and speciation. Finally, I will describe two highly successful monoclonal invaders, which I regard particularly suitable to empirically test some of the controversial issues of environmental and evolutionary epigenetics.
The Advantages of Clonal Lineages
The epigenetic proportion of phenotypic variation can more easily be determined in clonal lineages than in sexually reproducing species because of the paucity or absence of genetic variation. Therefore, asexually reproducing plants and animals have repeatedly been used to investigate the role of epigenetics in ecology and evolution [2, 64, 127–133]. Natural asexual populations are mostly polyclonal and only rarely monoclonal. Polyclonality is the result of repeated hybridization events, backcrossing with related sexual species, and long-lasting accumulation of genetic mutations [64, 134]. Monoclonal populations are of single origin and genetically identical with the exception of random mutations. In evolutionarily young monoclonal populations genetic diversity is virtually absent.
The number of potentially disturbing genetic mutations is particularly small among asexually produced batch-mates. Therefore, they are preferred subjects for laboratory studies on epigenetic variation that is independent of genetic variation. SPV and EPV can be distinguished by raising batch-mates either in the same or different environments. Laboratory experiments with clonal lineages are also suitable to identify the environmental cues that induce alterations of epigenetic profiles and the conditions under which phenotype modulating epigenetic marks are transgenerationally inherited. Moreover, they are useful to test whether epigenetic diversity can be transformed into genetic diversity in the long term. The investigation of wild clonal populations, particularly monoclonal ones, gives an idea on the extent and relevance of epigenetic phenotype variation under real life conditions.
Latzel et al. [135] presented evidence from greenhouse experiments with genetically identical but epigenetically diverse populations of Arabidopsis thaliana that the generation of epigenetically different phenotypes from the same genome can markedly modify fitness and functional diversity. They recorded up to 40% more biomass in epigenetically diverse populations when compared with epigenetically uniform populations. Interestingly, the positive effects of epigenetic diversity were strongest under stress conditions, for instance, when populations were grown together with competitors or infected with pathogens. Such studies are not yet available for animals. Dodd and Douhovnikoff [136] speculated that the expansion of phenotypic diversity by epigenetic mechanisms may provide a buffer against the hazards of global warming.
The Advantages of Invasions and Adaptive Radiations
Above, I have already elaborated on the advantages of animal invasions for ecological epigenetics. Respective references on plants invasions are found in [137, 138]. In this section, I want to highlight the advantages of invasions and adaptive radiations for evolutionary epigenetics.
Phylogenetically young invasions are expected to provide information on the role of epigenetic mechanisms in adaptation of the first generations to the new environment whereas older radiations may provide information on the role of epigenetics in evolutionary adaptation and speciation. Examples of relatively young animal invasions are the marbled crayfish in Europe and Madagascar (∼10 years) [131], the snail Potamopyrgus antipodarum in America (∼30 years) [129], the house sparrows in Kenya (∼50 years) [60], the American water flea Daphnia pulex in Africa (∼90 years) [128], the house sparrows in Florida (∼150 years) [139], and P. antipodarum in Europe (∼175 years) [127]. The ages of these invasions are proven by historical documents. Evolutionarily older invasions and radiations are easiest found on islands and in lakes of known geological age. Examples are the cichlid fish Amphilophus citrinellus in crater lakes in Nicaragua (∼20 000 years) [140], the cyprinid fish Chrosomus. eos–neogaeus in lakes and streams of Canada (∼54 000 years) [64], the Darwin’s finches on Galapagos (∼2 million years) [92], and the cichlid fish in East Africa’s large rift lakes (500 000–12 million years, depending on lake) [141].
Preliminary genetic and epigenetic analyses are available for only three of these examples, the Kenyan house sparrows, the Canadian C. eos–neogaeus and the Darwin’s finches [5, 60, 92]. The results seem to suggest that epigenetic diversity may compensate for reduced genetic diversity in the first period after invasion and promote speciation later on. All examples show that alleles and epigenetic patterns can vary independently from each other to some degree. Support for the independent variation also comes from the radiation of human populations [48], suggesting that epigenetic variation may contribute to adaptation and evolution. However, all of these studies lack information on the dynamics of genetic and epigenetic patterns over time, which makes it difficult to decide what was first, genetic or epigenetic variation. This issue could be best investigated by long-term laboratory experiments or field studies with animals that deposit dormant stages, which remain viable for decades and centuries, as detailed below.
Scenario on the Invasion of a New Geographical Area by a Monoclonal Invader and Subsequent Radiation
In order to illustrate the possible role of epigenetic phenotype variation for adaptation and evolution I use a simple thought experiment, namely the invasion of a new geographical region by a single gravid apomictic parthenogenetic female. The embryos carried by this female consist of genetically identical but epigenetically different phenotypes that have been generated by SPV under the conditions of the old environment (Fig. 7A). If the epigenetic differences concern ecologically relevant traits like food processing efficiency or tolerance to extreme temperatures, then they increase the chance of survival of the first generation and facilitate establishment of a founder population.
Figure 7.
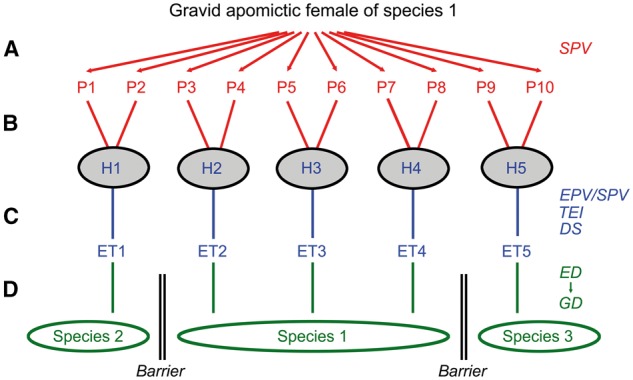
Thought experiment on the role of epigenetic phenotype variation in environmental adaptation and evolution. (A) Starting point is the invasion of a new geographical region by a gravid parthenogenetic female and the subsequent release of genetically uniform but epigenetically and phenotypically diverse progeny (P1–P10). The diversity of the offspring was produced by SPV under the conditions of the old environment. (B) The epigenetically different offspring then occupy different habitats (H1–H5) and are now exposed to different environmental cues. (C) In the following generations, different ecotypes (ET1–ET5) evolve by the interplay of EPV and SPV, TEI of evolutionarily relevant epigenetic signatures, and differential selection (DS) on the epigenetically mediated phenotypes. (D) The conversion of epigenetic diversity (ED) into genetic diversity (GD) and the establishment of ecological barriers may finally lead to the origin of new species
Differences in the preference of food, substrate or temperature may then lead to the occupation of different habitats and ecological niches (Fig. 7B). Exposure to different environmental cues may speed up phenotype diversification via EPV. In the following generations, the interplay of EPV and SPV, transgenerational inheritance of epigenetic patterns with phenotypic effects, and differential selection on the epigenetically determined phenotypes may pave the way to the evolution of different ecotypes (Fig. 7C). Finally, epigenetic diversity may be converted into genetic diversity by facilitated mutations in those DMRs that underpin epigenetic phenotype variation (Fig. 7D). This idea on the conversion of epigenetic diversity into genetic diversity differs from genetic accommodation and assimilation in as far as it does not require preexisting genetic variation. The long-term accumulation of genetic differences and the establishment of ecological and reproductive barriers may finally lead to new species.
Speciation is often believed to need thousands of generations but there are also examples of speciation within less than 20 generations, mainly resulting from ecological speciation [142]. It has to be noted here that speciation in parthenogens and other asexual reproducers is a problematic issue because they do not fit into the classical concepts of species, as discussed in detail by Mayr [143]. Asexual descendants of sexually reproducing parent species are often regarded as biotypes or lineages of the parent species rather than as separate species, despite having different fitness traits and different evolutionary trajectories. In order to overcome this unsatisfactory situation, Barraclough and colleagues [144, 145] developed the Evolutionary Genetic Species Concept. This theory focuses on the criterion that the individuals of the parent species and neo-species form discrete clusters of phenotypes. The new cluster should be of single origin, and both clusters must be separated from each other by a gap of evolutionarily relevant phenotypic traits and ecological, geographical, or reproductive isolation so that natural selection can ensure a divergent evolution over time.
Scenarios on the invasion of new environments by clonal invaders similar to that in Figure 7 have been published earlier by researchers dealing with phenotypic plasticity [55, 146, 147]. In these scenarios phenotypic plasticity was seen as an important facilitator of adaptation to new environments, and genetic assimilation of the environmentally induced phenotypes was used to explain evolutionary diversification. I have modified this scenario by introducing SPV, disentangling the role of SPV and EPV, and explaining genetic fixation of epigenetically mediated phenotypes by facilitated mutation at and around DMRs.
Real life support for the scenario of Figure 7 may come from ‘ancient asexual scandals’ [104, 148–151]. These are the bdelloid rotifers, darwinulid ostracods, some groups of oribatid mites and stick insects, and strains of the brine shrimp Artemia salina. Ancient asexuals are considered to have reproduced and radiated without males for almost 100 million years. For instance, the darwinulid ostracods and bdelloid rotifers revealed dozens and hundreds of species although there is convincing evidence that they never had sex [104, 151]. The scenario sketched in Figure 7 could be an explanation for the evolution of these groups.
Research Agenda with Monoclonal Invaders
Long-term laboratory experiments with monoclonal lineages combined with the analysis of respective wild populations should reveal whether the epigenetics-first scenario is principally possible or not. This approach is also expected to solve the chicken and eggs dilemma described above by comparing the chronology of genetic, epigenetic, and phenotypic changes in a population. It should also provide information on the extent of stochastic and environmentally induced epigenotype diversification and the heritability and selectability of epigenetic patterns. Finally, it should reveal whether epimutations can induce genetic mutations, which permanently fix phenotypes that were initially epigenetically determined. In the following, I will introduce two highly successful monoclonal invaders that I consider particularly suitable to test the first steps of the scenario in Figure 7 and several of the above questions. In order to highlight the special advantages of these model systems I need to describe their biology and invasion history in some detail.
Obligately Parthenogenetic Crayfish Clone Invasive in Europe and Madagascar
The parthenogenetic marbled crayfish Procambarus virginalis has evolved from sexually reproducing slough crayfish P. fallax by autotriploidy as described earlier. It is regarded as an independent species for reasons detailed in [76]. In the year 2013, we have launched a project at the German Cancer Research Center (DKFZ, Heidelberg) to establish the complete genome sequence and a genome-wide reference methylome [152], which are basic requirements for using this clonal species for research in ecological and evolutionary epigenetics. Whole genome and whole methylome comparison between marbled crayfish and slough crayfish is expected to reveal how much genetic and epigenetic change is necessary to create a new species and which genes and epigenetic markers underlie the transition from gonochorism to parthenogenesis. Comparison of the genomes and methylomes of marbled crayfish adapted to different environments is expected to establish whether DNA methylation is involved in ecological adaptation and the evolution of different ecotypes from the same genotype.
Meanwhile, both crayfish have been sequenced on an Illumina HiSeq platform for de novo assembly of the genomes and whole-genome bisulfite sequencing has been applied to establish genome-wide methylation maps at single-base resolution [76, 153, 154]. The genome of the marbled crayfish has a size of 3.5 GB and includes more than 21 000 genes [153]. A draft genome is accessible at the Marbled Crayfish Genome Server (http://marmorkrebs.dkfz.de/). The transcriptome revealed 22 338 transcripts of which 12 855 could be automatically annotated. Proteome analysis yielded a total of 43 783 peptides [154]. The global DNA methylation level is 2.4% (Fig. 4D) [76]. Genome-wide methylome analysis revealed mosaic methylation, gene body methylation, and hypomethylation of repeats. Methylation is particularly prominent in housekeeping genes, suggesting a role in fine-tuning of gene expression and perhaps environmental adaptation [154].
Marbled crayfish have an adult size of 4–11 cm, a generation time of 6–7 months and a maximum age of about 4.5 years [77, 155–159]. The relatively large size allows whole genome and methylome analyses of individuals and even individual organs. Marbled crayfish produce up to seven clutches per lifetime, each clutch comprising some 50–400 genetically identical siblings (Fig. 8A) [24, 78, 160]. The high number of batch-mates enables intense experimentation with isogenic specimens. Comparison of genomes and methylomes among brooded batch-mates that have not yet experienced different environments (Fig. 8A) are particularly suitable to obtain information on the frequency of random mutations and stochastic epimutations per generation. Since marbled crayfish can be raised in a variety of housing systems at water temperatures between 3 °C and 30 °C and fed with a variety of food [161, 162], they are also well suitable for the investigation of environmentally induced phenotype variation. Long term experiments in the laboratory under different stable and unstable conditions are expected to reveal whether epigenetic profiles can be inherited and genetically fixed and under which conditions this might happen.
Figure 8.
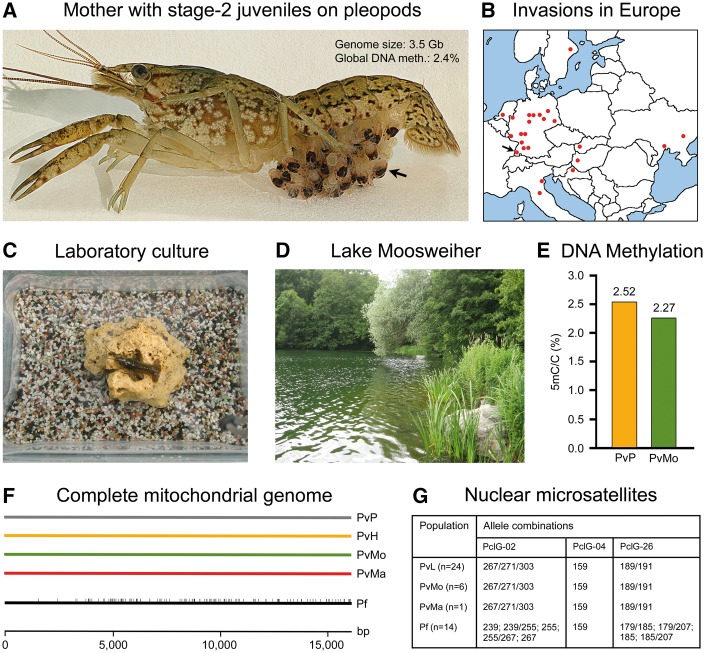
Marbled crayfish Procambarus virginalis (Pv) as promising model of environmental and evolutionary epigenetics. (A) Marbled crayfish produce numerous genetically identical offspring by apomictic parthenogenesis. The lecithotrophic offspring are brooded on the maternal pleopods (arrow) until the 3rd juvenile stage. Genome size and global DNA methylation level are in the order of magnitude of humans (photograph from [174]). (B) In Europe, marbled crayfish has invaded more than 20 water bodies including Lake Moosweiher in southern Germany (arrow) (redrawn after [131], updated). (C, D) Marbled crayfish thrive well in both very simple laboratory settings and natural habitats (photographs from [152]). (E) First global DNA methylation measurement revealed a lower level in wild (Lake Moosweiher, PvMo) than in cultured specimen (Petshop, PvP) (data from [76]). (F) Specimens from the Petshop and Heidelberg (PvH) lineages and from Lake Moosweiher and Madagascar (PvMa), which evolved separately for at least 10 generations, show identity of the complete mitochondrial genomes, demonstrating single origin and monoclonality. They differ from their parent species P. fallax by numerous SNPs (vertical bars). bp, base pairs (redrawn after [76]). (G) Monoclonality of cultured (PvL) and wild marbled crayfish is confirmed by nuclear microsatellites PclG-02 and PclG-26. Sexually reproducing P. fallax shows considerable variation in these microsatellites (redrawn after [76])
In order to investigate whether epigenetic mechanisms facilitate ecological adaptation, laboratory-raised reference lineages should be compared with wild populations of known age that have evolved under clearly different conditions. In the years 2003 and 2004, I have founded two indoor lineages from single individuals, which are well suitable as reference lineages [76]. These lineages (Heidelberg and Petshop) have consistently been raised in highly standardized settings (Fig. 8C) and were fed throughout life and over generations with a single pellet food (Tetra WaferMix). A 3-year-old individual of the Petshop lineage was used for sequencing of the reference genome. Wild invasive populations are known from Europe (Fig. 8B) and Madagascar. They have established themselves since 2003 as the result of introductions [131, 163–167].
Apparently, all invasive populations originate from the same source as our laboratory populations, namely an individual or small isogenic group that first appeared in the German aquarium trade in about 1995 [155, 157]. Comparison of the complete mitochondrial genomes, nuclear microsatellites, and larger segments of genomic DNA between members of the two reference lineages and two wild populations from Lake Moosweiher (Germany) and Madagascar revealed complete genetic identity (Fig. 8F and G), demonstrating that all populations are of single origin and monoclonal [76, 153]. The wild populations have evolved independently from our laboratory lineages for some 10–20 generations and have adapted to different habitats such as creeks, rivers, oligotrophic to eutrophic lakes (Fig. 8D), thermal and ice-covered lakes, acidic and polluted ponds, and swamps and rice fields. Adaptation to these highly diverse ecosystems cannot be explained by genetic variation and may have been achieved by epigenetic variation instead. The first comparison of global DNA methylation between Heidelberg and Moosweiher specimens revealed a higher value in the former (Fig. 8E). However, meaningful information on the involvement of DNA methylation in environmental adaptation is only expected from the ongoing comparison of genome-wide methylomes.
Obligately Parthenogenetic American Water Flea Clone Invasive in Africa
The second monoclonal invader that could contribute significantly to ecological and evolutionary epigenetics is an obligately parthenogenetic American Daphnia pulex clone (Fig. 9A) that invaded Africa about 90 years ago [128]. Genotyping of dormant eggs sampled from sediment layers of known age allowed the reconstruction of the dynamics of this invasion in Lake Naivasha, Kenya. In about 1930, the new genotype appeared first in the lake and increased then steadily in relative frequency until 1955 when it accounted for about 60% of the total water flea population (Fig. 9B). After a 30-year period of relative stability the invasive clone expanded further until 1998 when it had completely displaced the native D. pulex genotypes. Genetic analysis of dormant eggs from different decades (Fig. 9C) and laboratory rearing of resurged specimens identified the invading clone as an obligately parthenogenetic D. pulex × D. pulicaria hybrid, which is common in North America [128, 168]. The displaced native water flea was genetically more closely related to European representatives of the D. pulex species complex (Fig. 9C). The absence of variation in the mitochondrial NADH dehydrogenase subunit 5 gene and 10 hypervariable nuclear markers of several other African populations of the invasive clone strongly suggest single origin and monoclonality. Most likely, this clone was accidentally introduced into Lake Naivasha in the years 1927–29 during stocking of largemouth bass from the USA [128].
Figure 9.
Invasive American clone of water flea Daphnia pulex in Africa as promising model to study epigenotype, genotype, and phenotype diversification across space and time. (A) Obligately parthenogenetic female carrying ephippium with dormant eggs (arrow) in brood chamber (from [128], photograph by Joachim Mergeay). (B) The American clone invaded Lake Naivasha in Kenya in about 1927. It successively replaced native D. pulex as revealed by microsatellite analysis of dormant eggs from dated sediments since the year 1920. Stippled line shows gradual decrease of genetic diversity over time (bars indicate 95% confidence). (C) The analysis of 12S rRNA genes of dormant eggs from sediments of Lake Naivasha (LN) dated to the years 1925, 1955 and 2003 demonstrates close genetic relationship of the pre-invasive D. pulex to European strains and identity of the invasive clone with a Canadian strain. (D) The invasive clone has meanwhile spread across the cooler regions of Eastern and Southern Africa and replaced the native populations. Arrow indicates Lake Naivasha (graphs modified after [128])
Meanwhile, the invasive clone has spread from Lake Naivasha throughout Eastern and Southern Africa and displaced native D. pulex populations (Fig. 9D). The invader was found in different types of freshwater habitat including small temporary ponds, eutrophic sewage ponds, shallow turbid lakes, reservoirs, and clear lakes rich in aquatic macrophytes [128]. The absence of genetic variation did not hamper its continent-wide establishment or the effective displacement of the well adapted and genetically diverse native sibling species, which reproduced by cyclic parthenogenesis (alternation of sexual and asexual reproduction). Cyclic parthenogenetic Daphnia populations have been shown to genetically adapt to changing environmental conditions within a few years [169]. The invasive D. pulex clone may have used epigenetic mechanisms instead.
The main advantages of the invasive water flea clone for research on environmental and evolutionary epigenetics are short generation time, good knowledge of its genetics and epigenetics, and the production of dormant eggs. In natural habitats, D. pulex reproduce several times per year suggesting an African invasion history of a few hundred generations. Under optimal laboratory conditions, these water fleas have generation times of only 10 days allowing monitoring of epigenetic and genetic signatures over many generations in reasonable time. The genome of the species is fully sequenced and annotated [170] and analysis of the methylome was recently performed [171], albeit not in the invasive clone. The DNA methylation data suggest that epigenetic modifications in D. pulex are involved in regulation of gene expression and may thus contribute to the generation of phenotypic variation and environmental adaptation [171, 172]. An outstanding feature of D. pulex is the regular production of dormant eggs, which accumulate in the sediment and remain viable for up to 700 years [173]. DNA from these dormant eggs enables the investigation of epigenetic and genetic diversification of the invader across time and space. Resurged individuals from dormant eggs additionally allow the identification of corresponding morphological and physiological trait alterations [173]. A disadvantage of the water flea model system is the small body size, which currently allows whole genome and methylome analyses only for pooled samples and not for individuals or single organs as in marbled crayfish.
Conclusions
Phenotypes are determined by the genome and the epigenome. Since phenotypes provide the raw material for adaptation and natural selection, epigenetic phenotype variation may play an important yet overlooked role in ecology and evolution. There is evidence from experiments with genetically identical organisms that epigenetic phenotype variation can be generated either by developmental stochasticity or environmental induction. SPV contributes to bet-hedging and may be regarded as a Darwinian mechanism whereas EPV serves for directed phenotype optimization and may be regarded as Lamarckian. Usually, both mechanisms act conjointly but their relative contribution to epigenetic phenotype variation depends on species, environment, and trait and is apparently subject to evolutionary change.
The production of epigenetically diverse phenotypes from the same genome is assumed to contribute significantly to environmental adaptation because it broadens the range of phenotypes and thus enhances the chance to stay in the game of life when the environment changes. The advantages of epigenetic phenotype variation are particularly obvious in asexually reproducing populations, bottlenecked populations, and genetically uniform invasive groups, providing a mechanistic explanation for the ‘invasion paradox’ and the ‘general-purpose-genotype’. However, epigenetic phenotype variation is probably also important in sexually reproducing species and may help populations to cope with environmental changes in the short term. In times of global warming, it may become particularly relevant.
There is no doubt that epigenetic patterns change during speciation but it is not yet clear whether they passively follow genetic changes and help to consolidate the new genome or whether they can also trigger speciation, or in other words, whether they can be leaders rather than followers in evolution. Epigenetic mechanisms can contribute to reproductive isolation, chromatin reconfiguration, and alteration of gene expression in the neo-species. The possibility of a triggering role of epigenetic mechanisms in speciation would require transgenerational inheritance of phenotype-determining epigenetic patterns and final genetic fixation of the novelties. There are hypotheses and models on these issues but empirical evidence is scarce and contradictory.
Verhoeven et al. [18] have recently discussed what we know and what we need to know about the role of epigenetics in ecological adaptation and evolution. The authors emphasized that assessing the importance of epigenetic effects to ecology and evolution requires the surveillance of epigenetic variation in natural populations and a mechanistic understanding of the relation of epigenotypes to the underlying genotype, the linked phenotypes, and environmental signals. The authors further stressed that high-resolution genome-wide screening of genetic and epigenetic variation combined with expression analysis will be necessary to pinpoint the contributions of epigenetic variation to ecology and evolution. Combined long-term laboratory studies on genomics, epigenomics, transcriptomics, and phenomics of clonal lineages exposed to different environmental conditions and analyses of natural monoclonal invasions seem particularly suitable to address controversial key issues of ecological and evolutionary epigenetics.
Acknowledgements
I thank Chris Lukhaup and Martin Grimm for providing pictures as indicated in the figure legends and three anonymous referees for valuable comments that helped to improve the article.
Conflict of interest statement. None declared.
References
- 1. Mayr E, Provine WB (ed.). The Evolutionary Synthesis: Perspectives on the Unification of Biology. Cambridge, MA: Harvard University Press, 1998. [Google Scholar]
- 2. Verhoeven KJF, Preite V.. Epigenetic variation in asexually reproducing organisms. Evolution 2014;68:644–55. [DOI] [PubMed] [Google Scholar]
- 3. Skinner MK. Environmental epigenetics and a unified theory of the molecular aspects of evolution: a neo-Lamarckian concept that facilitates neo-Darwinian evolution. Genome Biol Evol 2015;7:1296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vogt G. Stochastic developmental variation, an epigenetic source of phenotypic diversity with far-reaching biological consequences. J Biosci 2015;40:159–204. [DOI] [PubMed] [Google Scholar]
- 5. Leung C, Breton S, Angers B.. Facing environmental predictability with different sources of epigenetic variation. Ecol Evol 2016;6:5234–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Feinberg AP, Irizarry RA.. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA 2010;107(Suppl1):1757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jablonka E, Lamb MJ. Epigenetic Inheritance and Evolution: The Lamarckian Dimension. Oxford, UK: Oxford University Press, 1995. [Google Scholar]
- 8. Jablonka E, Lamb MJ. Evolution in Four Dimensions: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life, revised edn. Cambridge, MA: MIT Press, 2014. [Google Scholar]
- 9. Nanjundiah V. Phenotypic plasticity and evolution by genetic assimilation In: Müller GB, Newman SA (eds.), Origination of Organismal Form: Beyond the Gene in Development and Evolutionary Biology. Cambridge, MA: MIT Press, 2003, 245–64. [Google Scholar]
- 10. West-Eberhard MJ. Developmental Plasticity and Evolution. New York, NY: Oxford University Press, 2003. [Google Scholar]
- 11. Pigliucci M, Müller GB (ed.). Evolution: The Extended Synthesis. Cambridge, MA: MIT Press, 2010. [Google Scholar]
- 12. Laland KN, Uller T, Feldman MW, et al. The extended evolutionary synthesis: its structure, assumptions and predictions. Proc R Soc B 2015;282:20151019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaenisch R, Bird A.. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 2003;33(Suppl):245–54. [DOI] [PubMed] [Google Scholar]
- 14. Feinberg AP. Epigenetic stochasticity, nuclear structure and cancer: the implications for medicine. J Intern Med 2014;276:5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hallgrímsson B, Hall BK (ed.). Epigenetics: Linking Genotype and Phenotype in Development and Evolution. Berkeley, CA: University of California Press, 2011. [Google Scholar]
- 16. Bossdorf O, Richards CL, Pigliucci M.. Epigenetics for ecologists. Ecol Lett 2008;11:106–15. [DOI] [PubMed] [Google Scholar]
- 17. Zhang Y-Y, Fischer M, Colot V, et al. Epigenetic variation creates potential for evolution of plant phenotypic plasticity. New Phytol 2013;197:314–22. [DOI] [PubMed] [Google Scholar]
- 18. Verhoeven KJF, Vonholdt BM, Sork VL.. Epigenetics in ecology and evolution: what we know and what we need to know. Mol Ecol 2016;25:1631–8. [DOI] [PubMed] [Google Scholar]
- 19. Vogt G. Evolution of epigenetic mechanisms in animals In: Tollefsbol T. (ed.), Handbook of Epigenetics: The New Molecular and Medical Genetics, 2nd edn New York, NY: Academic Press, 2017, in press. [Google Scholar]
- 20. Falconer DS, Mackay TFC. Introduction to Quantitative Genetics, 4th edn Harlow, UK: Longman, 1996. [Google Scholar]
- 21. Finch CE, Kirkwood TBL. Chance, Development, and Aging. New York, NY: Oxford University Press, 2000. [Google Scholar]
- 22. Scheiner SM. Genetics and evolution of phenotypic plasticity. Annu Rev Ecol Syst 1993;24:35–68. [Google Scholar]
- 23. Scheiner SM. The genetics of phenotypic plasticity. XIII. Interactions with developmental instability. Ecol Evol 2014;4:1347–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vogt G, Huber M, Thiemann M, et al. Production of different phenotypes from the same genotype in the same environment by developmental variation. J Exp Biol 2008;211:510–23. [DOI] [PubMed] [Google Scholar]
- 25. Schlichting CD, Wund MA.. Phenotypic plasticity and epigenetic marking: an assessment of evidence for genetic accommodation. Evolution 2014;68:656–72. [DOI] [PubMed] [Google Scholar]
- 26. Sultan SE. Organism and Environment: Ecological Development, Niche Construction, and Adaptation. Oxford, UK: Oxford University Press, 2015. [Google Scholar]
- 27. Snell-Rood EC. An overview of the evolutionary causes and consequences of behavioural plasticity. Anim Behav 2013;85:1004–11. [Google Scholar]
- 28. Schlichting CD, Pigliucci M. Phenotypic Evolution: A Reaction Norm Perspective. Sunderland, MA: Sinauer Associates, 1998. [Google Scholar]
- 29. Cridge AG, Leask MP, Duncan EJ, et al. What do studies of insect polyphenisms tell us about nutritionally-triggered epigenomic changes and their consequences? Nutrients 2015;7:1787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mallon EB, Amarasinghe HE, Ott SR. Acute and chronic gregarisation are associated with distinct DNA methylation fingerprints in desert locusts. Sci Rep 2016;6:35608.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lyko F, Foret S, Kucharski R, et al. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010;8:e1000506.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herb BR, Wolschin F, Hansen KD, et al. Reversible switching between epigenetic states in honeybee behavioral subcastes. Nat Neurosci 2012;15:1371–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol Cell Endocrinol 2014;398:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guillette LJ Jr, Parrott BB, Nilsson E, et al. Epigenetic programming alterations in alligators from environmentally contaminated lakes. Gen Comp Endocrinol 2016;238:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pavelka J, Poláková S, Pavelková V An epigenetic change in a moth is generated by temperature and transmitted to many subsequent generations mediated by RNA. bioRxiv2017; doi:10.1101/099259. [DOI] [PMC free article] [PubMed]
- 36. Gärtner K. Commentary: Random variability of quantitative characteristics, an intangible epigenomic product, supporting adaptation. Int J Epidemiol 2012;41:342–6. [DOI] [PubMed] [Google Scholar]
- 37. Aranda-Anzaldo A, Dent MAR.. Developmental noise, ageing and cancer. Mech Ageing Dev 2003;124:711–20. [DOI] [PubMed] [Google Scholar]
- 38. Martin GM. Nature, nurture, and chance: their roles in interspecific and intraspecific modulations of aging. Annu Rev Gerontol Geriat 2014;34:267–84. [Google Scholar]
- 39. Burggren WW, Mueller CA.. Developmental critical windows and sensitive periods as three-dimensional constructs in time and space. Physiol Biochem Zool 2015;88:91–102. [DOI] [PubMed] [Google Scholar]
- 40. Baldanzi S, Watson R, McQuaid CD, et al. Epigenetic variation among natural populations of the South African sandhopper Talorchestia capensis. Evol Ecol 2017;31:77–91. [Google Scholar]
- 41. Smith TA, Martin MD, Nguyen M, et al. Epigenetic divergence as a potential first step in darter speciation. Mol Ecol 2016;25:1883–94. [DOI] [PubMed] [Google Scholar]
- 42. Liu S, Sun K, Jiang T, et al. Natural epigenetic variation in bats and its role in evolution. J Exp Biol 2015;218:100–6. [DOI] [PubMed] [Google Scholar]
- 43. Van der Graaf A, Wardenaar R, Neumann DA, et al. Rate, spectrum, and evolutionary dynamics of spontaneous epimutations. Proc Natl Acad Sci USA 2015;112:6676–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ossowski S, Schneeberger K, Lucas-Liedó JI, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 2010;327:92–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Atwell S, Huang YS, Vilhjálmsson BJ, et al. Genome-wide association study of 107 phenotypes in a common set of Arabidopsis thaliana inbred lines. Nature 2010;465:627–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stranger BE, Stahl EA, Raj T.. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics 2011;187:367–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cortijo S, Wardenaar R, Colomé-Tatché M, et al. Mapping the epigenetic basis of complex traits. Science 2014;343:1145–8. [DOI] [PubMed] [Google Scholar]
- 48. Heyn H, Moran S, Hernando-Herraez I, et al. DNA methylation contributes to natural human variation. Genome Res 2013;23:1363–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weigel D, Colot V.. Epialleles in plant evolution. Genome Biol 2012;13:249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Quadrana L, Almeida J, Asís R, et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat Commun 2014;5:3027. [DOI] [PubMed] [Google Scholar]
- 51. Pignatta D, Erdmann RM, Scheer E, et al. Natural epigenetic polymorphisms lead to intraspecific variation in Arabidopsis gene imprinting. eLife 2014;3:e03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rakyan VK, Blewitt ME, Druker R, et al. Metastable epialleles in mammals. Trends Genet 2002;18:348–51. [DOI] [PubMed] [Google Scholar]
- 53. Furrow RE. Epigenetic inheritance, epimutation, and the response to selection. PLoS One 2014;9:e101559.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Prentis PJ, Wilson JRU, Dormontt EE, et al. Adaptive evolution in invasive species. Trends Plant Sci 2008;13:288–94. [DOI] [PubMed] [Google Scholar]
- 55. Dybdahl MF, Kane SL.. Adaptation vs. phenotypic plasticity in the success of a clonal invader. Ecology 2005;86:1592–601. [Google Scholar]
- 56. Sax DF, Brown JH.. The paradox of invasion. Global Ecol Biogeogr 2000;9:363–71. [Google Scholar]
- 57. Allendorf FW, Lundquist LL.. Introduction: Population biology, evolution, and control of invasive species. Conserv Biol 2003;17:24–30. [Google Scholar]
- 58. Frankham R. Resolving the genetic paradox in invasive species. Heredity 2005;94:385.. [DOI] [PubMed] [Google Scholar]
- 59. Fridley JD, Stachowicz JJ, Naeem S, et al. The invasion paradox: reconciling pattern and process in species invasions. Ecology 2007;88:3–17. [DOI] [PubMed] [Google Scholar]
- 60. Liebl AL, Schrey AW, Richards CL, et al. Patterns of DNA methylation throughout a range expansion of an introduced songbird. Integr Comp Biol 2013;53:351–8. [DOI] [PubMed] [Google Scholar]
- 61. Parker IM, Rodriguez J, Loik ME.. An evolutionary approach to understanding the biology of invasions: local adaptation and general-purpose genotypes in the weed Verbascum thapsus. Conserv Biol 2003;17:59–72. [Google Scholar]
- 62. Lundmark M. Otiorhynchus sulcatus, an autopolyploid general-purpose genotype species? Hereditas 2010;147:278–82. [DOI] [PubMed] [Google Scholar]
- 63. Caron V, Ede FJ, Sunnucks P.. Unravelling the paradox of loss of genetic variation during invasion: superclones may explain the success of a clonal invader. PLoS One 2014;9:e97744.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Angers B, Schlosser IJ.. The origin of Phoxinus eos-neogaeus unisexual hybrids. Mol Ecol 2007;16:4562–71. [DOI] [PubMed] [Google Scholar]
- 65. Massicotte R, Angers B.. General-purpose genotype or how epigenetics extend the flexibility of a genotype. Genet Res Int 2012;2012:317175.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Castonguay E, Angers B.. The key role of epigenetics in the persistence of asexual lineages. Genet Res Int 2012;2012:534289.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Madlung A, Wendel JF.. Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet Genome Res 2013;140:270–85. [DOI] [PubMed] [Google Scholar]
- 68. De Boer JG, Yazawa R, Davidson WS, et al. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics 2007;8:422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sehrish T, Symonds VV, Soltis DE, et al. Gene silencing via DNA methylation in naturally occurring Tragopogon miscellus (Asteraceae) allopolyploids. BMC Genomics 2014; 15:701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lafon-Placette C, Köhler C.. Epigenetic mechanisms of postzygotic reproductive isolation in plants. Curr Opin Plant Biol 2015;23:39–44. [DOI] [PubMed] [Google Scholar]
- 71. Parisod C, Holderegger R, Brochmann C.. Evolutionary consequences of autopolyploidy. New Phytol 2010;186:5–17. [DOI] [PubMed] [Google Scholar]
- 72. Cagan A, Blass T.. Identification of genomic variants putatively targeted by selection during dog domestication. BMC Evol Biol 2016;16:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janowitz Koch I, Clark MM, Thompson MJ, et al. The concerted impact of domestication and transposon insertions on methylation patterns between dogs and grey wolves. Mol Ecol 2016;25:1838–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Martin P, Dorn NJ, Kawai T, et al. The enigmatic Marmorkrebs (marbled crayfish) is the parthenogenetic form of Procambarus fallax (Hagen, 1870). Contrib Zool 2010;79:107–18. [Google Scholar]
- 75. Martin P, Thonagel S, Scholtz G.. The parthenogenetic Marmorkrebs (Malacostraca: Decapoda: Cambaridae) is a triploid organism. J Zool Syst Evol Res 2016;54:13–21. [Google Scholar]
- 76. Vogt G, Falckenhayn C, Schrimpf A, et al. The marbled crayfish as a paradigm for saltational speciation by autopolyploidy and parthenogenesis in animals. Biol Open 2015;4:1583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Van der Heiden CA, Dorn NJ.. Benefits of adjacent habitat patches to the distribution of a crayfish population in a hydro-dynamic wetland landscape. Aquat Ecol 2017; in press. [Google Scholar]
- 78. Vogt G. Suitability of the clonal marbled crayfish for biogerontological research: a review and perspective, with remarks on some further crustaceans. Biogerontology 2010;11:643–69. [DOI] [PubMed] [Google Scholar]
- 79. Gatzmann FB, Falckenhayn C, Gutekunst J, et al. Saltational speciation in the marbled crayfish P. virginalis. Abstracts BioGenomics 2017. Washington, DC: Smithsonian National Museum of Natural History, 2017, 42. [Google Scholar]
- 80. Vrana PB. Genomic imprinting as a mechanism of reproductive isolation in mammals. J Mammal 2007;88:5–23. [Google Scholar]
- 81. O’Neill RJW, O’Neill MJ, Graves JAM.. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 1998;393:68–72. [DOI] [PubMed] [Google Scholar]
- 82. Chen X, Ge X, Wang J, et al. Genome-wide DNA methylation profiling by modified reduced representation bisulfite sequencing in Brassica rapa suggests that epigenetic modifications play a key role in polyploid genome evolution. Front Plant Sci 2015;6:836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Inoue J, Sato Y, Sinclair R, et al. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci USA 2015;112:14918–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Xiao J, Song C, Liu S, et al. DNA methylation analysis of allotetraploid hybrids of red crucian carp (Carassius auratus red var.) and common carp (Cyprinus carpio L.). PLoS One 2013;8:e56409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koroma AP, Jones R, Michalak P.. Snapshot of DNA methylation changes associated with hybridization in Xenopus. Physiol Genomics 2011;43:1276–80. [DOI] [PubMed] [Google Scholar]
- 86. Malone JH, Chrzanowski TH, Michalak P.. Sterility and gene expression in hybrid males of Xenopus laevis and X. muelleri. PLoS One 2007;2:e781.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Xiang J, Li F, Zhang C, et al. Evaluation of induced triploid shrimp Penaeus (Fenneropenaeus) chinensis cultured under laboratory conditions. Aquaculture 2006;259:108–15. [Google Scholar]
- 88. Krois NR, Cherukuri A, Puttagunta N, et al. Higher rate of tissue regeneration in polyploid asexual versus diploid sexual freshwater snails. Biol Lett 2013;9:20130422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chang AY-F, Liao B-Y.. DNA methylation rebalances gene dosage after mammalian gene duplications. Mol Biol Evol 2012;29:133–44. [DOI] [PubMed] [Google Scholar]
- 90. Seymour DK, Koenig D, Hagmann J, et al. Evolution of DNA methylation patterns in the Brassicaceae is driven by differences in genome organization. PLoS Genet 2014;10:e1004785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hagmann J, Becker C, Müller J, et al. Century-scale methylome stability in a recently diverged Arabidopsis thaliana lineage. PLoS Genet 2015;11:e1004920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Skinner MK, Gurerrero-Bosagna C, Muksitul Haque M, et al. Epigenetics and the evolution of Darwin’s finches. Genome Biol Evol 2014;6:1972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Crews D, Gore AC. Controversies and debate of generational epigenetic inheritance In: Tollefsbol T. (ed.), Transgenerational Epigenetics: Evidence and Debate. Waltham, MA: Academic Press, 2014, 369–90. [Google Scholar]
- 94. Jablonka E, Raz G.. Transgenerational epigenetic inheritance: prevalence, mechanisms, and implications for the study of heredity and evolution. Quart Rev Biol 2009;84:131–76. [DOI] [PubMed] [Google Scholar]
- 95. Robertson M, Richards C.. Non-genetic inheritance in evolutionary theory—the importance of plant studies. Non-Genet Inherit 2015;2:3–11. [Google Scholar]
- 96. Mesoudi A, Blanchet S, Charmantier A, et al. Is non-genetic inheritance just a proximate mechanism? A corroboration of the extended evolutionary synthesis. Biol Theory 2013;7:189–95. [Google Scholar]
- 97. Burggren W. Epigenetic inheritance and its role in evolutionary biology: re-evaluation and new perspectives. Biology 2016;5:24.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Triantaphyllopoulos KA, Ikonomopoulos I, Bannister AJ.. Epigenetics and inheritance of phenotype variation in livestock. Epigenet Chrom 2016;9:31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Haig D. Weismann rules! Ok? Epigenetics and the Lamarckian temptation. Biol Phil 2007;22:415–28. [Google Scholar]
- 100. Seisenberger S, Peat JR, Hore TA, et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Phil Trans R Soc B 2012;368:20110330.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Miyoshi N, Stel JM, Shioda K, et al. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc Natl Acad Sci USA 2016;113:9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Extavour CG, Akam M.. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 2003;130:5869–84. [DOI] [PubMed] [Google Scholar]
- 103. De Meeûs T, Prugnolle F, Agnew P.. Asexual reproduction: genetics and evolutionary aspects. Cell Mol Life Sci 2007;64:1355–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Schön I, Rossetti G, Martens K. Darwinulid ostracods: ancient asexual scandals or scandalous gossip In: Schön I, Martens K, van Dijk P (eds.), Lost Sex: The Evolutionary Biology of Parthenogenesis. Dordrecht, Netherlands: Springer, 2009, 217–40. [Google Scholar]
- 105. Van Otterdijk SD, Michels KB.. Transgenerational epigenetic inheritance in mammals: how good is the evidence? FASEB J 2016;30:2457–65. [DOI] [PubMed] [Google Scholar]
- 106. Rassoulzadegan M, Cuzin F.. Epigenetic heredity: RNA-mediated modes of phenotypic variation. Ann NY Acad Sci 2015;1341:172–5. [DOI] [PubMed] [Google Scholar]
- 107. Skinner MK, Guerrero-Bosagna C, Haque MM.. Environmentally induced epigenetic transgenerational inheritance of sperm epimutations promote genetic mutations. Epigenetics 2015;10:762–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Chen Q, Yan W, Duan E.. Epigenetic inheritance of acquired traits through sperm RNAs and sperm RNA modifications. Nat Rev Genet 2016;17:733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Hirsch S, Baumberger R, Grossniklaus U.. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harb Symp Quant Biol 2012;77:97–104. [DOI] [PubMed] [Google Scholar]
- 110. Herman JJ, Spencer HG, Donohue K, et al. How stable ‘should’ epigenetic modifications be? Insights from adaptive plasticity and bet hedging. Evolution 2014;68:632–43. [DOI] [PubMed] [Google Scholar]
- 111. Becker C, Hagmann J, Müller J, et al. Spontaneous epigenetic variation in the Arabidopsis thaliana methylome. Nature 2011;480:245–9. [DOI] [PubMed] [Google Scholar]
- 112. Leimar O, McNamara JM.. The evolution of transgenerational integration of information in heterogeneous environments. Am Nat 2015;185:E55–69. [DOI] [PubMed] [Google Scholar]
- 113. McNamara JM, Dall SRX, Hammerstein P, et al. Detection vs. selection: integration of genetic, epigenetic and environmental cues in fluctuating environments. Ecol Lett 2016;19:1267–76. [DOI] [PubMed] [Google Scholar]
- 114. Kronholm I, Collins S.. Epigenetic mutations can both help and hinder adaptive evolution. Mol Ecol 2016;25:1856–68. [DOI] [PubMed] [Google Scholar]
- 115. Wang J, Fan C.. A neutrality test for detecting selection on DNA methylation using single methylation polymorphism frequency spectrum. Genome Biol Evol 2014;7:154–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Geoghegan JL, Spencer HG.. Exploring epiallele stability in a population-epigenetic model. Theor Popul Biol 2013;83: 136–44. [DOI] [PubMed] [Google Scholar]
- 117. Furrow RE, Christiansen FB, Feldman MW. Epigenetic variation, phenotypic heritability, and evolution In: Naumova AK, Greenwood CMT (eds.), Epigenetics and Complex Traits. New York, NY: Springer, 2013, 233–46. [Google Scholar]
- 118. Crispo E. The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 2007;61:2469–79. [DOI] [PubMed] [Google Scholar]
- 119. Pigliucci M, Murren CJ, Schlichting CD.. Phenotypic plasticity and evolution by genetic assimilation. J Exp Biol 2006;209:2362–7. [DOI] [PubMed] [Google Scholar]
- 120. Ehrenreich IM, Pfennig DW.. Genetic assimilation: a review of its potential proximate causes and evolutionary consequences. Ann Bot 2015;117:769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Walser J-C, Furano AV.. The mutational spectrum of non-CpG DNA varies with CpG content. Genome Res 2010;20:875–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Rey O, Danchin E, Mirouze M, et al. Adaptation to global change: a transposable element-epigenetics perspective. Trends Ecol Evol 2016;31:514–26. [DOI] [PubMed] [Google Scholar]
- 123. Rebollo R, Horard B, Hubert B, et al. Jumping genes and epigenetics: towards new species. Gene 2010;454:1–7. [DOI] [PubMed] [Google Scholar]
- 124. Bonduriansky R, Crean AJ, Day T.. The implications of nongenetic inheritance for evolution in changing environments. Evol Appl 2012;5:192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Branciamore S, Rodin AS, Riggs AD, et al. Enhanced evolution by stochastically variable modification of epigenetic marks in the early embryo. Proc Natl Acad Sci USA 2014;111:6353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Branciamore S, Rodin AS, Gogoshin G, et al. Epigenetics and evolution: transposons and the Stochastic Epigenetic Modification Model. AIMS Genet 2015;2:148–62. [Google Scholar]
- 127. Städler T, Frye M, Neiman M, et al. Mitochondrial haplotypes and the New Zealand origin of clonal European Potamopyrgus, an invasive aquatic snail. Mol Ecol 2005;14:2465–73. [DOI] [PubMed] [Google Scholar]
- 128. Mergeay J, Verschuren D, De Meester L.. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proc R Soc B 2006;273:2839–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Dybdahl MF, Drown DM.. The absence of genotypic diversity in a successful parthenogenetic invader. Biol Invasions 2011;13:1663–72. [Google Scholar]
- 130. Forbes AA, Rice LA, Stewart NB, et al. Niche differentiation and colonization of a novel environment by an asexual parasitic wasp. J Evol Biol 2013;26:1330–40. [DOI] [PubMed] [Google Scholar]
- 131. Chucholl C. Marbled crayfish gaining ground in Europe: the role of the pet trade as invasion pathway In: Kawai T, Faulkes Z, Scholtz G (eds.), Freshwater Crayfish: A Global Overview. Boca Raton, FL: CRC Press, 2016, 83–114. [Google Scholar]
- 132. Latzel V, Klimešová J.. Transgenerational plasticity in clonal plants. Evol Ecol 2010;24:1537–43. [Google Scholar]
- 133. Douhovnikoff V, Dodd RS.. Epigenetics: a potential mechanism for clonal plant success. Plant Ecol 2015;216:227–33. [Google Scholar]
- 134. Hebert PDN, Finston TL.. Macrogeographic patterns of breeding system diversity in the Daphnia pulex group from the United States and Mexico. Heredity 2001;87:153–61. [DOI] [PubMed] [Google Scholar]
- 135. Latzel V, Allan E, Silveira AB, et al. Epigenetic diversity increases the productivity and stability of plant populations. Nat Commun 2013;4:2875.. [DOI] [PubMed] [Google Scholar]
- 136. Dodd RS, Douhovnikoff V.. Adjusting to global change through clonal growth and epigenetic variation. Front Ecol Evol 2016;4:86. [Google Scholar]
- 137. Richards CL, Schrey AW, Pigliucci M.. Invasion of diverse habitats by few Japanese knotweed genotypes is correlated with epigenetic differentiation. Ecol Lett 2012;15:1016–25. [DOI] [PubMed] [Google Scholar]
- 138. Gillies S, Clements DR, Grenz J.. Knotweed (Fallopia spp.) invasion of North America utilizes hybridization, epigenetics, seed dispersal (unexpectedly), and an arsenal of physiological tactics. Invasive Plant Sci Manag 2016;9:71–80. [Google Scholar]
- 139. Schrey AW, Coon CAC, Grispo MT, et al. Epigenetic variation may compensate for decreased genetic variation with introductions: a case study using house sparrows (Passer domesticus) on two continents. Genet Res Int 2012;2012:979751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Franchini P, Xiong P, Fruciano C, et al. The role of microRNAs in the repeated parallel diversification of lineages of Midas cichlid fish from Nicaragua. Genome Biol Evol 2016;8:1543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Elmer KR, Meyer A.. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol Evol 2011;26:298–306. [DOI] [PubMed] [Google Scholar]
- 142. Hendry AP, Nosil P, Rieseberg LH.. The speed of ecological speciation. Funct Ecol 2007;21:455–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Mayr E. What is a species, and what is not? Philos Sci 1996;63:262–77. [Google Scholar]
- 144. Barraclough TG, Birky CW Jr, Burt A.. Diversification in sexual and asexual organisms. Evolution 2003;57:2166–72. [DOI] [PubMed] [Google Scholar]
- 145. Birky CW Jr, Barraclough TG. Asexual speciation In: Schön I, Martens K, van Dijk P (eds.), Lost Sex: The Evolutionary Biology of Parthenogenesis. Dordrecht, Netherlands: Springer, 2009, 201–16. [Google Scholar]
- 146. Pfennig DW, Wund MA, Snell-Rood EC, et al. Phenotypic plasticity’s impacts on diversification and speciation. Trend Ecol Evol 2010;25:459–67. [DOI] [PubMed] [Google Scholar]
- 147. Lande R. Adaptation to an extraordinary environment by evolution of phenotypic plasticity and genetic assimilation. J Evol Biol 2009;22:1435–46. [DOI] [PubMed] [Google Scholar]
- 148. Judson OP, Normark BB.. Ancient asexual scandals. Trends Ecol Evol 1996;11:41–6. [DOI] [PubMed] [Google Scholar]
- 149. Neiman M, Schwander T.. Using parthenogenetic lineages to identify advantages of sex. Evol Biol 2011;38:115–23. [Google Scholar]
- 150. Rossetti G, Pinto RL, Martens K.. Description of a new genus and two new species of Darwinulidae (Crustacea, Ostracoda), from Christmas Island (Indian Ocean) with some considerations on the morphological evolution of ancient asexuals. Belg J Zool 2011;141:55–74. [Google Scholar]
- 151. Butlin R. The costs and benefits of sex: new insights from old asexual lineages. Nat Rev Genet 2002;3:311–7. [DOI] [PubMed] [Google Scholar]
- 152. Vogt G, Raddatz G, Falckenhayn C, et al. Ökologische Anpassung invasiver Arten durch epigenetische Mechanismen: ein Teilaspekt des Marmorkrebs-Projekts am Deutschen Krebsforschungszentrum Heidelberg In: Tagungsband Internationale Flusskrebstagung 2013. Aachen (Germany: ): Biologische Station StädteRegion Aachen, 2013, 109–13. [Google Scholar]
- 153. Gutekunst J, Falckenhayn C, Gatzmann F, et al. Assembly and annotation of the marbled crayfish genome In: Abstracts BioGenomics 2017. Washington, DC: Smithsonian National Museum of Natural History, 2017, 44. [Google Scholar]
- 154. Falckenhayn C. The methylome of the marbled crayfish Procambarus virginalis Ph.D. Thesis. Combined Faculties of Natural Sciences and Mathematics. Heidelberg, Germany: University of Heidelberg, 2017.
- 155. Scholtz G, Braband A, Tolley L, et al. Parthenogenesis in an outsider crayfish. Nature 2003;421:806.. [DOI] [PubMed] [Google Scholar]
- 156. Vogt G, Tolley L, Scholtz G.. Life stages and reproductive components of the Marmorkrebs (marbled crayfish), the first parthenogenetic decapod crustacean. J Morphol 2004;261: 286–311. [DOI] [PubMed] [Google Scholar]
- 157. Vogt G. The marbled crayfish: a new model organism for research on development, epigenetics and evolutionary biology. J Zool 2008;276:1–13. [Google Scholar]
- 158. Seitz R, Vilpoux K, Hopp U, et al. Ontogeny of the Marmorkrebs (marbled crayfish): a parthenogenetic crayfish with unknown origin and phylogenetic position. J Exp Zool A 2005;303:393–405. [DOI] [PubMed] [Google Scholar]
- 159. Vogt G. Investigation of hatching and early post-embryonic life of freshwater crayfish by in vitro culture, behavioral analysis, and light and electron microscopy. J Morphol 2008;269:790–811. [DOI] [PubMed] [Google Scholar]
- 160. Martin P, Kohlmann K, Scholtz G.. The parthenogenetic Marmorkrebs (marbled crayfish) produces genetically uniform offspring. Naturwiss 2007;94:843–6. [DOI] [PubMed] [Google Scholar]
- 161. Jimenez SA, Faulkes Z.. Establishment and care of a colony of parthenogenetic marbled crayfish, Marmorkrebs. Invert Rearing 2010;1:10–8. [Google Scholar]
- 162. Kaldre K, Meženin A, Paaver T, et al. A preliminary study on the tolerance of marble crayfish Procambarus fallax f. virginalis to low temperature in nordic climate In: Kawai T, Faulkes Z, Scholtz G (eds.), Freshwater Crayfish: A Global Overview. Boca Raton, FL: CRC Press, 2016, 54–62. [Google Scholar]
- 163. Lipták B, Mrugała A, Pekárik L, et al. Expansion of the marbled crayfish in Slovakia: beginning of an invasion in the Danube catchment? J Limnol 2016;75:305–12. [Google Scholar]
- 164. Lökkös A, Müller T, Kovács K, et al. The alien, parthenogenetic marbled crayfish (Decapoda: Cambaridae) is entering Kis-Balaton (Hungary), one of Europe’s most important wetland biotopes. Knowl Manag Aquat Ecosyst 2016; 417:16. [Google Scholar]
- 165. Kotovska G, Khrystenko D, Patoka J, et al. East European crayfish stocks at risk: arrival of non-indigenous crayfish species. Knowl Manag Aquat Ecosyst 2016;417:37. [Google Scholar]
- 166. Jones JPG, Rasamy JR, Harvey A, et al. The perfect invader: a parthenogenic crayfish poses a new threat to Madagascar’s freshwater biodiversity. Biol Invasions 2009;11:1475–82. [Google Scholar]
- 167. Kawai T, Scholtz G, Morioka S, et al. Parthenogenetic alien crayfish (Decapoda: Cambaridae) spreading in Madagascar. J Crust Biol 2009;29:562–7. [Google Scholar]
- 168. Hebert PDN, Beaton MJ, Schwartz SS, et al. Polyphyletic origins of asexuality in Daphnia pulex. I. Breeding-system variation and levels of clonal diversity. Evolution 1989;43:1004–15. [DOI] [PubMed] [Google Scholar]
- 169. Hairston NG Jr, Lampert W, Cáceres CE, et al. Rapid evolution revealed by dormant eggs. Nature 1999;401:446. [Google Scholar]
- 170. Colbourne JK, Pfrender ME, Gilbert D, et al. The ecoresponsive genome of Daphnia pulex. Science 2011;331:555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Asselman J, De Coninck DIM, Pfrender ME, et al. Gene body methylation patterns in Daphnia are associated with gene family size. Genome Biol Evol 2016;8:1185–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Strepetkaitė D, Alzbutas G, Astromskas E, et al. Analysis of DNA methylation and hydroxymethylation in the genome of crustacean Daphnia pulex. Genes 2016;7:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Frisch D, Morton PK, Chowdhury PR, et al. A millennial-scale chronicle of evolutionary responses to cultural eutrophication in Daphnia. Ecol Lett 2014;17:360–8. [DOI] [PubMed] [Google Scholar]
- 174. Vogt G, Tolley L.. Brood care in freshwater crayfish and relationship with the offspring’s sensory deficiencies. J Morphol 2004;262:566–82. [DOI] [PubMed] [Google Scholar]