Abstract
Environmental epigenetic is an emerging field that studies the cause–effect relationship between environmental factors and heritable trait via an alteration in epigenetic marks. This field has received much attentions since the impact of environmental factors on different epigenetic marks have been shown to be associated with a broad range of phenotypic disorders in natural ecosystems. Chemical pollutants have been shown to affect immediate epigenetic information carriers of several aquatic species but the heritability of the chromatin marks and the consequences for long term adaptation remain open questions. In this work, we investigated the impact of the diuron herbicide on the DNA methylation pattern of spat from exposed Crassotrea gigas genitors. This oyster is one of the most important mollusk species produced worldwide and a key coastal economic resource in France. The whole genome bisulfite sequencing (WGBS, BS-Seq) was applied to obtain a methylome at single nucleotide resolution on DNA extracted from spat issued from diuron exposed genitors comparatively to control spat. We showed that the parental diuron exposure has an impact on the DNA methylation pattern of its progeny. Most of the differentially methylated regions occurred within coding sequences and we showed that this change in methylation level correlates with RNA level only in a very small group of genes. Although the DNA methylation profile is variable between individuals, we showed conserved DNA methylation patterns in response to parental diuron exposure. This relevant result opens perspectives for the setting of new markers based on epimutations as early indicators of marine pollutions.
Keywords: Crassostrea gigas, mollusk, DNA methylation, herbicide, epigenetic
Introduction
The role of environmental stressors in altering the physiological performance of aquatic wildlife is a fundamental topic that has recently attracted strong attention. Chronic environmental exposure to chemical compounds, including metals, pesticides, endocrine disrupting compounds can affect the behavior, and the physiological or developmental performance of aquatic organisms [1, 2]. Environmental epigenetics investigates the cause–effect relationships between specific environmental factors and the subsequent epigenetic modifications [3–7]. Chromatin structure is generally admitted as the epigenetic element of non-genetic inheritance system. Chromatin is a complex of DNA associated to proteins and epigenetic information carriers are the element which influences its structure. Non-coding RNA, methylation of cytosine residues and histone post translational modification are the main epigenetic information bearers known to affect chromatin structure. The epigenotype is mitotically and to some degree meiotically heritable, but unlike in the genotype, changes in the epigenotype are reversible. Therefore, the epigenetic information system (EIS, epigenetics) can provide heritable, novel phenotypes without variation in DNA sequence [8, 9]. The production of this heritable phenotypic variation may be essential for evolution [10] or may result in mal-adaptation of an organism experiencing changing environments as exemplified by several emerging disease disorders [11–13]. Various environmental contaminations have been shown to alter DNA methylation and histone patterns in animal species [14–17]. Currently, most of evidence of the association of altered epigenotypes and phenotypic disorders is provided from laboratory animal models and human epidemiological studies. Despite the extensive use of aquatic organisms in an ecotoxicological context, only a few studies provide information about the epigenetic mechanisms that may influence phenotypic variations in response to environmental factors in aquatic species [2, 18–22].
Amongst hazardous substances, diuron (N0-[3,4-dichlorophenyl]-N,N-dimethyl-urea) is a biologically active and quite persistent pollutant present in soil, water and sediments. This herbicide was used to eradicate weeds in non-agricultural areas (roadsides, railways, parks, etc.) [23]. Diuron is prohibited in France since 2008 as a phytosanitary substance and as a biocide since 2009 (Directive 2008/91/EC). But, since it is not well degraded naturally [24], it is still detected at high concentrations in French coastal and fresh waters [25]. Consequently, it is considered as a potential Persistent Organic Pollutants (POP), and is currently evaluated under the Stockholm Convention (http://chm.pops.int) [26]. Diuron’s ecotoxicological impacts have been studied on several aquatic species [27–29] including invertebrates of economic interest such as the Pacific oyster Crassostrea gigas. This species is reared in coastal areas (estuaries, lagoons and open sea) under the persistent threat of chemical pollution pressure. C. gigas is a successive and irregular protandrous hermaphroditic mollusc. Gonads are diffuse and non-permanent and their development is strongly influenced by seasonal seawater temperature variations upon which oysters release sperm and eggs into the seawater. Within 6 h, fertilized eggs develop into embryo and larvae in a planktonic stage lasting 2–3 weeks before entering metamorphosis leading to settlement to a solid substrate [30]. A few studies have investigated the gonadal and germline establishment in this species during development. However, following an orthologous to the vasa gene (called Oyvlg), Fabioux et al. [31] were able to follow primordial germ cells (PGCs) that would arise from 4 day blastomeres, hence suggesting that the PGCs specification mode in this species is preformation, i.e. maternally inherited [31–33]. Genomic and epigenomic resources for this species have recently become available [34, 35] and DNA methylation has been recently suggested to be involved in phenotypic variation in this organism [36]. The main goal of the present study was to determine how a parental exposure to the herbicide diuron can affect the DNA methylation pattern in C. gigas offspring. Some results were previously obtained demonstrating both direct and indirect effects of the parental herbicide exposure on oyster genome. A genotoxic effect has been detected through the evidence of primary DNA damages in both somatic, germinal and reproductive cells of diuron-exposed genitors [37, 38]. Moreover, the vertical transmission of damaged DNA was confirmed by the detection of chromosomal abnormalities in the offspring (DNA aneuploidy) [38]. By a transcriptional approach, we showed that exposure of oyster genitors to diuron at environmental concentrations affected gene expression including genes encoding enzymes involved in DNA methylation machinery [39]. In the offspring, a RNA-seq analysis demonstrated significant change in the transcriptional profile on F1 spat as well [40] that could explain developmental and growth impairment observed in F1 early life stages [37, 38].
In our study, we showed that a parental diuron exposure strongly affected spat DNA methylation pattern predominantly in intragenic regions and we discussed the possible consequences of this change on molecular phenotypes.
Results
Parental Diuron Exposure Causes Differential DNA Methylation in the Offspring
Whole Genome Bisulfite Sequencing was performed on three biological replicates for two investigated conditions (diuron-exposed (D) and seawater control (SWC). Sequencing of the samples yielded 620 166 410 100 bp Illumina paired-end reads, with an average of 103 361 068 paired-end reads by library (Supplementary Fig. S1). After quality control and alignment, an average of 36 965 778 (17.9%) reads by library was mapped (Supplementary Fig. S1).
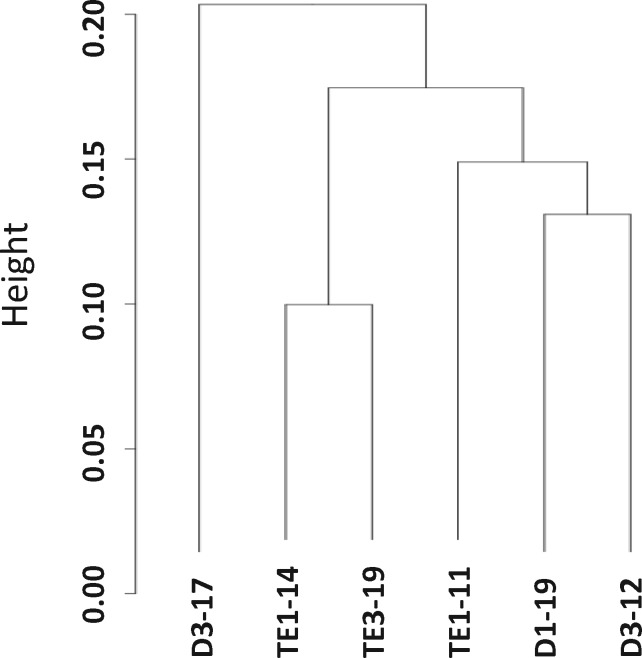
On average, 16.6% of CpGs and 0.5% of CHH were found methylated, and the total methylation was 2.0% (5 mC of total DNA). There was no statistically significant difference in global methylation between D and SWC samples. The clustering analysis showed that the methylation profile obtained in exposure sample did not differ globally compared to the SWC as the different samples did not cluster according to their treatment (Fig. 1). For the three individual samples analyzed per group, parental diuron exposure did not globally modify the DNA methylation. Distance method used to produce the clustering is Pearson. We reasoned that if the epimutation rate would be higher in the diuron treated samples then the control samples should cluster together. Pearson correlation is also considered very sensitive to outliers. Strong differences in epimutation rates within diuron samples should produce such outliers, which is not the case. Consequently, differential methylation was restricted to specific regions. After visual inspection of methylation profiles we decided to use a window size of 500 bp for further analysis. Of the total of 1 124 112 500 bp windows of the genome in average 251 069 fulfilled the criterion of 10× coverage (Supplementary Fig. S1). However, in one sample, due to relatively poor coverage, only 57 819 windows were ≥10-fold covered. Within these 57 819 eligible windows, we found 1967 differentially methylated regions (DMRs) (3.4% of eligible windows): 1060 were hypermethylated and 907 hypomethylated in the D compared to SWC samples (Supplementary Table S2, sheet “MethDiff - Global results”).
Figure 1:
CpG methylation clustering. DNA methylation pattern clustering of the different samples is based on the Ward’s method and was performed with Bismark. Samples D3-17, D1-19, D3-12 correspond to spats from D samples TE1-14, TE3-19, TE1-11 correspond to spats from SWC
Target-Induced DMRs Occur Predominantly in Genes
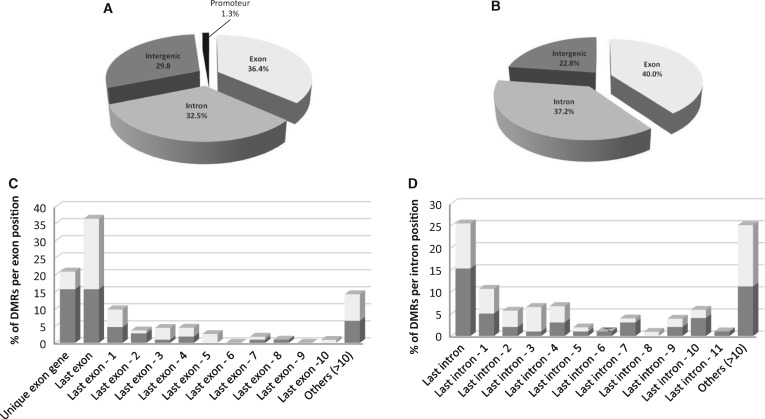
Although the 1967 detected regions were statistically significantly differentially methylated between the D and the SWC individuals, it appeared that in many cases one individual among the three was less impacted. Therefore, we thoroughly examined the 1967 detected regions one by one to retain only those that are affected in the same way in the three biological replicates. This led to the selection of 236 DMRs (12% of DMRs, 0.4% of eligible windows). We therefore identified two types of epimutations: (i) environment-induced epimutations (in 0.4% of the epigenome, epimutation rate 4 × 10−3), and (ii) spontaneous epimutations (in 3% of the epigenome, epimutation rate 3 × 10−2). Following an earlier naming convention, we used the term “targeted epimutation” for type 1 epimutation, and “random epimutations” for type 2 [41]. Among the targeted DMRs, 121 were hypermethyled and 115 hypomethylated in D compared to SWC samples (Supplementary Table S2). The majority of these DMRs (73.2%) were found in genes, with 38.2% and 34.8% localized in exons and introns, respectively (Supplementary Table S2). Intergenic regions contain 30% DMRs among which 0.7% were found in putative promoter regions (only two DMRs). The distribution of hypermethylated and hypomethylated regions follows roughly the same pattern (Fig. 2, Supplementary Table S2). We found 170 DMRs located within genes of which 83 were hypermethylated and 87 hypomethylated (Supplementary Table S2, sheet “MethDiff—targeted epi hyper” and sheet “MethDiff—targeted epi hypo”). We further investigated the accurate location of the DMRs within the genes. 36.4% of the exon specific DMRs occurred in the last exon genes. 25.5% of the intron specific DMRs occurred in the last intron genes (Fig. 2). This overrepresentation of the DMRs was not observed in the first exon of genes if we counted the number of DMRs from the first exon position (data not shown). Therefore, we observed that DMRs preferentially occurred at the end of the coding sequences.
Figure 2:
Genomic feature of the target DMRs. The genomic features of the target DMRs were determined using as a guide the gene annotation obtained from previous transcriptomic data [40]. Hypo/hypermethylation refers to methylation level in D as compared to SWC. A and B represent genomic features for hypermethylated and hypomethylated regions, respectively. C and D represent the number of DMRs per exon or intron, respectively, counting from the last exon or intron position, dark and pale grey represents the hypermethylation and hypomethylation, respectively
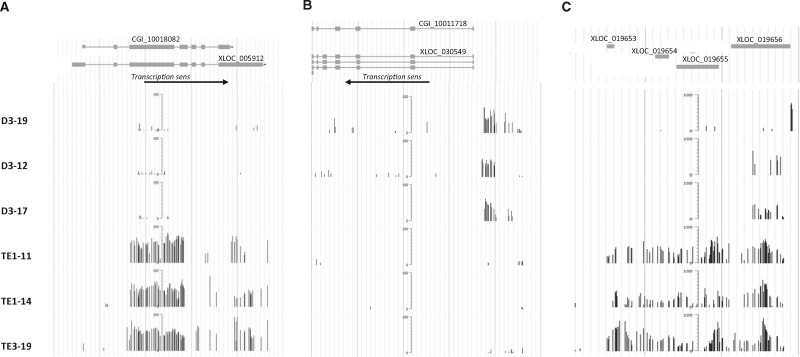
Interestingly, we found several genes strongly affected by the parental diuron exposure (Fig. 3 and Supplementary Fig. S2). For these candidates, several 500 bp DMRs accumulates within the coding sequence. XLOC_005912 (CGI_10018082) encodes an uncharacterized protein with two conserved domains, one found in chromosome partitioning proteins and another one found in mitochondria eating proteins. It contained 63 intragenic CpG sites in a 1.437 kb window which were strongly methylated in the three SWC biological replicates. All these sites were poorly methylated in the three samples from the D individuals. XLOC_030549 (CGI_10011718) encodes a serine protease with domains found in fibrinolytic proteins. 16 strongly methylated CpG sites were found in a 1.132 kb window upstream of the transcriptional start site in the SWC group. These sites were unmethylated in the D group. The scaffold309 from position 131250 to position 137240 contains 67 CpG sites highly methylated in the individuals from D group compared to SWC group. This region contained four transposon-like elements (XLOC_019653, XLOC_019654, XLOC_019655, XLOC _019656) for which transcripts had been detected in previous RNA-Seq experiment [40]. In summary, among the 1967 differentially methylated regions, we found 12% which were targeted epimutations. These epimutations mostly occured within coding sequences and preferentially at the end of genes.
Figure 3:
Example of strongly impacted regions. Extract of the C. gigas jbrowse interface which allows visualization of transcriptomic and DNA methylation data for three regions: (A) Scaffold 1255 from 364500 to 370000 containing XLOC_005912 (CGI_10018082), (B) Scaffold 43664 from 222500 to 235000 containing XLOC_030549 (CGI_10011718), and (C) Scaffold 309 from 130000 to 137500 containing four unique exon genes (XLOC_019653, XLOC_019654, XLOC_019655, XLOC_019656). Upper panel represents the annotation of genes in the current genome assembly V9 (CGI number), and the CDS according to previous transcriptomic data [40]. Bottom panel represents the methylated cytosine position. X-axis represents the position of each methylated cytosine. Y axis (bars) represents the % of methylation observed for each methylated cytosine (scale is 0–100). The six tracks represent the three biological replicates of individual spats from genitors treated with diuron or control (from top to bottom)
About 80% of Environmentally Induced DMRs Are Specifically Targeted
As outlined above we identified 236 targeted DMRs using an epigenome-wide screen in single nucleotide resolution in three individuals. However, roughly 88% or all detected DMRs were random epimutations. It was therefore conceivable that all DMRs were actually random and just by chance, overlapping existing DMRs in our three replicates that were mistakenly assigned to be environmentally induced. We therefore randomly sampled 10 candidate DMRs among those that showed the most pronounced differences in DNA methylation and performed DNA methylation analysis on offspring spat of 10 individuals of each group (D, SC, SWC) by targeted bisulfite sequencing (T-BS-Seq) (Table 1, Supplementary Fig. S2). For 8 out of 10 samples, we observed a correlation between the BS-Seq and target analysis (Supplementary Fig. S2). In one loci (Scaffold42366: 107142.107725, Supplementary Fig. S2) we found good correlation between BS-Seq and T-BS-Seq for the three individuals studied by BS-Seq but not for the others. In one candidate (Scaffold1720:210182.210722), we did not confirm the BS-Seq results. Moreover, we observed several scenarios of diuron/solvent effect (Table 1 and Supplementary Fig. S2): a diuron specific effect was observed in a unique region. Otherwise, we observed a solvent specific effect in another region, a synergistic/same/antagonist effect was observed in two, three and one regions, respectively. And finally, two regions displayed really complex profile where the diuron/solvent mixture effect relied on each CpG site individually. Finally, we barely found the same DNA methylation profile between individuals whatever the environmental conditions (Supplementary Fig. S2). Despite this individual variation, we found two consistent loci for which DNA methylation pattern was really conserved between individuals. The diuron/solvent suppressed the methylation of all the CpG sites in all the tested individuals in the loci XLOC_005912/CGI_10018082 present on scaffold1255 (365000–370000), whereas this mixture induced an overall increase in DNA methylation on several CpG sites of a region spanning 3 kb of the scaffold33832 (12000–15000). These consistent results within the 10 tested individuals led us to calculate a positive predictive value (PPV). The PPV tests the performance of a diagnostic based on the proportion of positive and negative results. It aims at checking if the two candidate regions can be used as potential environmental contamination marker based on DNA methylation profile. A PPV of 85.7% was obtained when taking into account the combined results of these two loci (Supplementary Fig. S3).
Table 1:
DNA methylation analysis on target loci
| Targeted loci | Diuron/solvent effect |
|---|---|
| Scaffold1154:309231.309691 | Diuron specific effect |
| Scaffold433: 896527.897085 | Solvent specific effect |
| Scaffold33832: 13205.13768 | Solvent effect, further increased with diuron |
| Scaffold1255: 75.750 | |
| Scaffold41174: 81270.81829 | Same effect for solvent and diuron |
| Scaffold42366: 108172.108673 | |
| Scaffold43170: 116309.116863 | |
| Scaffol37178: 4921.5527 | Antagonist effect between solvent and diuron |
| Scaffold1720: 210182.210722 | Diuron or solvent effect rely on CpG sites |
| Scaffold42366: 107142.107725 |
Ten loci were selected as indicated in column 1 and targeted by PCR after DNA bisulfite conversion. Ten individuals were analyzed per condition (SWC, SC, D). Detailed results are given in Supplementary Fig. S2.
No Indication of Genetic Selection Following Parental Diuron Exposure
Diuron exposure could have led to selection of genetically encoded phenotypes and the observed DNA methylation (epiallele frequency) differences could be an expression of such underlying genetic (allele frequency [AF]) difference. We used the genetic variants obtained with MethylExtract to search for signs of selection on a genome wide level. We used a total of 11 425 759 SNPs (control) and 9 517 133 SNPs (diuron) to calculate Tajima’s D in 200 kb sliding windows. Genome wide scan’s of Tajima’s D in sliding windows can identify regions whose D greatly deviates from the bulk of the empirical distribution and such windows are reported as significant. Frequency distribution of Tajima’s D indicates that most 200 kb windows have a value of around 0 and that there are no major differences in D distribution between SWC and D conditions. For a high-resolution analysis, we also cloned and sequenced a 2.198 kb amplified cDNA fragment from the locus XLOC_005912/CGI_10018082 in which DNA methylation is strongly affected by the diuron parental exposure. 248 SNPs and 14 Indels were detected in this 2.198 kb fragment of XLOC_005912/CGI_10018082 cDNA. The clustering analysis performed groups according to individuals rather than condition (Supplementary Fig. S4). While we could not formally exclude that minor changes in allele frequency occurred during parental diuron exposure, it is sure that there was no strong and systematic AF distortion.
Relationship between DNA Methylation and RNA Abundance
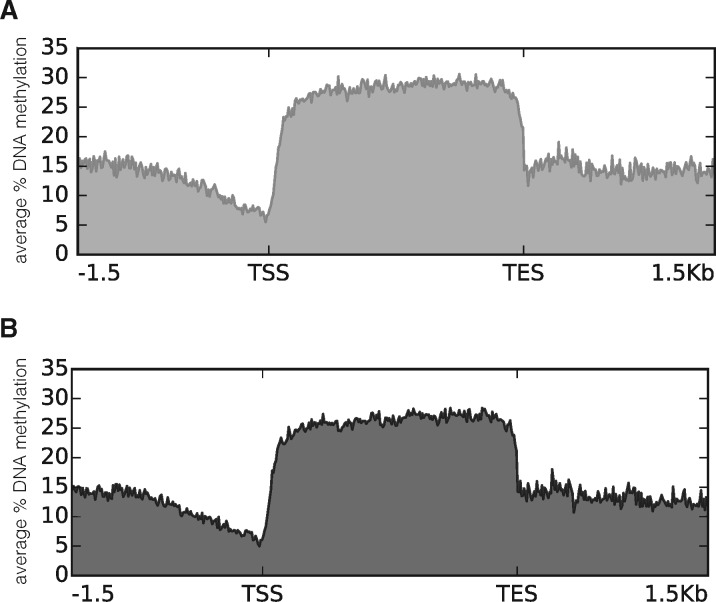
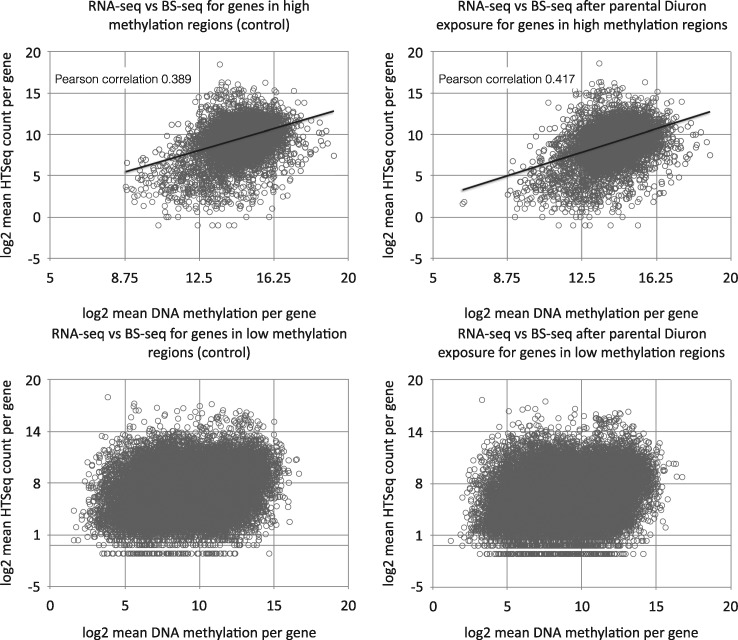
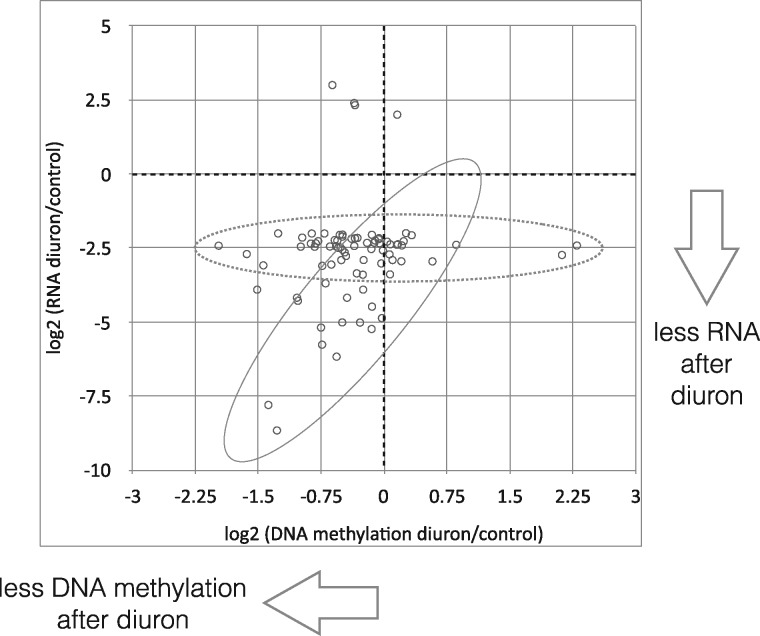
After having firmly established that parental diuron exposure led to either random or targeted changes in DNA methylation profiles we wondered if such changes could affect the level of RNA abundance. To establish the regions of methylation enrichment around genes we produced average methylation profiles for all 42 463 genes (Fig. 4). As expected, gene bodies were highly enriched for methylation and no difference could be detected between SWC and D offspring. Visual inspection of genome-wide methylation profiles confirmed the mosaic-style methylation with large blocks of high methylation separated by blocks with no or weak methylation. We established the following criteria for both methylation types: regions with at least 5 CpGs that are all at least 70% methylated, and having a maximum distance of 2 kb between them (read coverage ≥ 10) were considered as “high methylation region” (HMR). All other regions were considered as “low methylation region” (LMR) (Supplementary Fig. S1). HMRs contain 5 655 genes (13%) whereas LMRs contain 36 808 genes (87%). We then used our earlier RNA-Seq data [40] to evaluate the relationship between gene body DNA methylation and RNA abundance. For genes located in HMRs, we observed a weak correlation (Pearson coefficient 0.4) between degree of methylation and RNA abundance (Fig. 5). No correlation was found for genes located in the LMRs. For genes in the high methylation compartment, it was therefore in principle conceivable that changes in methylation could translate into changes in RNA level (or vice versa). Among the top 25 483 differentially expressed genes (abs(log2) fold change ≥2) only 80 were located in HMRs. Within these genes we observed two populations: (i) those that showed only moderate changes in RNA abundance and this despite strong changes in DNA methylation, and (ii) a smaller fraction where increase in DNA methylation correlated with increase in RNA amounts (Fig. 6). Amongst the above mentioned conserved 170 DMGs, high methylation compartment contains 84 (49%) (54 hypomethylated after diuron, and 30 hypermethylated). However, their abs(log2) expression differences were <2. In summary, it appeared that DNA methylation changes correlated directly with RNA level only in a very small group of genes, and only for genes located in the high methylation compartment of the (epi)genome. For this analysis, we used pooled BS-Seq data and compared them with pooled RNA-Seq data. A potential caveat of this approach is that potential individual differences in RNA abundance and DNA methylation were leveled out by absence of such differences in the replicates. Therefore, we performed qPCR experiments targeting arbitrarily selected candidate genes in DMRs using cDNA from the same individuals used for the BS-Seq experiment. We chose different contexts of DMR position: promoter, within genes, 3′ distant regulatory region (Table 2). Among the 12 tested genes, only two (XLOC_005912/CGI_10018082 and XLOC_032365/CGI_10010585) showed a significant difference in their expression levels between control and parental diuron exposure. XLOC_005912 was 1.80 times more expressed in the D than in the SWC sample. This gene was hypomethylated in the D sample all along its coding sequence (Table 2 and Fig. 3). Reversely, XLOC_032365 was 1.66 times less expressed in the D than in the SWC sample although it was hypomethylated in the D sample in an intronic region (Table 2). The other candidates did not show significant differences in their expression profiles although DNA methylation pattern was sometimes strongly affected by the parental diuron exposure (Fig. 3).
Figure 4:
Average DNA methylation for transcripts. The average DNA methylation pattern around genes in offspring from diuron exposed parents (A) and from sea water control parents (B) was generated using Deeptool based on 67 754 transcripts produced earlier by RNA-Seq [40]. X axis represents the gene context; gene body was scaled to 2 kb. Y axis represents the average percentage of DNA methylation
Figure 5:
Correlation between DNA methylation and gene expression. Correlation was performed using the HTSeq counts from previous RNA-Seq experiment [40] and average DNA methylation level per gene from the three biological replicates of each condition
Figure 6:
Correlation between the change in methylation level and change in expression level. The fold change in expression observed between the diuron and sea water control conditions (output from DESeq analysis [40]) was compared with the fold change in DNA methylation between the D and SWC conditions
Table 2:
Effect of DNA methylation on gene transcription
| CGI number | XLOC number | Annotation | DMR positions | Context | Fold change |
|---|---|---|---|---|---|
| CGI_10018082 | XLOC_005912 | Centromeric protein | Scaffold 1255 | Intronic and exonic | 1.80 |
| 366001–367000 | Hypomethylation | ||||
| CGI_10026162 | XLOC_004001 | Thap domain-containing protein 9-like | Scaffold 1154 | Exonic and intronic | No significant difference |
| 309001–311000 | Hypermethylation | ||||
| XLOC_020586 | Myb-related transcription factor, partner of profilin | Scaffold 33832 | Exonic and 3′ distant reg. region | No significant difference | |
| 12001–12500 | Hypermethylation | ||||
| CGI_10009103 | XLOC_029410 | Hypothetical protein | Scaffold 43170 | Exonic | No significant difference |
| 116501–117000 | Hypomethylation | ||||
| XLOC_030549 | Hepatocyte growth factor-like protein | Scaffold 43664 | Promoter | No significant difference | |
| 232001–232500 | Hypermethylation | ||||
| CGI_10011372 | XLOC_017268 | Reverse transcriptase | Scaffold 206 | Exonic | No significant difference |
| 81001–81500 | Hypomethylation | ||||
| CGI_10010582 | XLOC_032353 | Synaptic vesicle glycoprotein 2c-like | Scaffold 473 | Exonic | No significant difference |
| 121501–122000 | Hypomethylation | ||||
| CGI_10010585 | XLOC_032365 | 28s ribosomal protein mitochondrial-like | Scaffold 473 | Intronic | 0.60 |
| 130001–130500 | Hypomethylation | ||||
| CGI_10017761 | XLOC_037874 | Beta-arrestin1 | Scaffold 713 | Exonic (3′UTR) | No significant difference |
| 415001–415500 | Hypomethylation | ||||
| XLOC_019656 | Retrovirus polyprotein | Scaffold 309 | Exonic | No significant difference | |
| 136001–136500 | Hypomethylation | ||||
| CGI_10026723 | XLOC_019464 | Paired box pox meso protein | Scaffold 301 | Exonic | No significant difference |
| 516001–516500 | Hypermethylation | ||||
| CGI_10023654 | XLOC_034086 | Solute carrier family 13 member 2 | Scaffold 54 | Intronic | No significant difference |
| 79501–80000 | Hypermethylated |
Genes have been chosen according to different context of DMR position. DMRs context are indicated in the “context” column. The accurate position on C. gigas genome assembly V9 is indicated in column “DMRs position.” Hypo/hypermethylation and difference in expression refers to D compared to SWC. Fold changes are indicated only in the case of significant P value (P < 0.01).
Since it has been suggested that intragenic DNA methylation plays a role in splice variant regulation [42], we investigated the occurrence of different splice variants between the D and SWC group. We amplified by conventional PCR long fragments that covered almost the full CDS for three candidate genes (XLOC_005912/CGI_10018082, XLOC_032353/CGI_10010582 and XLOC_032353/CGI_10010585) in 15 individuals per condition including the individuals which had been used for the BS-Seq experiment. The size of amplified products was checked on agarose gels and we observed a unique fragment of the expected size in both conditions for each targeted locus (data not shown), suggesting that differential methylation did not systematically lead to splice variants. Taken together, our data indicated that although the diuron exposure strongly affected the DNA methylation profile of spat from exposed genitors, differential methylation had generally no direct impact on RNA abundance and splicing.
Discussion
A growing body of evidence shows that environmental stressors can be responsible for changes in epigenetic marks, which may result in heritable phenotypic outcomes and changes in life history trait of living species [43–45]. The increasing data regarding the influence of the environment on chromatin structure has led to the emergence of the “environmental epigenetics” discipline [7, 19, 46, 47]. Recently, this field has received many attentions since DNA methylation and histone modifications have been shown to be associated with a broad range of disease outcomes, including cancer, metabolic disorders, allergies, asthma, and reproductive disabilities [16, 48–50]. Although the majority of the efforts are oriented to the context of human health, growing evidences suggest that epigenetic based events play also a major role in natural ecosystems [18, 51]. Substantial efforts have been devoted to the study of multigenerational impacts of the climate change (ocean acidification and temperature increase) [52, 53] and chemical pollution on several marine species [2], including mollusks [54, 55]. Diverse examples illustrate how parental exposure could benefit to the offspring which exhibit traits with increased fitness in the environmental conditions experienced by their parents [54, 55]. Other studies have shown how chemical pollutants such as endocrine disrupting compounds can have a negative impact on different life history traits such as fertilization success, egg viability, reproductive behaviors, hatch success and development [2, 56]. Although, there are evidences for an impact of the environment, including chemical pollutants, on DNA methylation content of several aquatic species [57–59], no studies have investigated the multigenerational or intergenerational effect of pollutants on epigenetic marks of the offspring of aquatic relevant species. In this work, we investigated for the first time the impact of a parental exposure to the herbicide diuron on epigenetic marks of C. gigas. The effect of this herbicide on different C. gigas life history traits has been demonstrated several times on exposed individuals but also on the progeny of exposed parents [37, 38, 60–62]. In the present study, we showed by BS-seq analysis on a restricted number of samples the existence of 1967 differentially methylated regions (DMRs) after a parental diuron exposure. These DMRs could be produced by the parental stress, but some of these differences could be the product of an interindividual variability of DNA methylation in the spats used. Among the 1967 DMRs, 88% are spontaneous epimutations and therefore the results of interindividual variability whereas 12% of these methylation differences are target-induced epimutations and therefore those regions are the results of the diuron-solvent mixture effect on the DNA methylation pattern. We then established the CpG methylation profile of 10 regions in a more subsequent number of samples by conventional PCR target analysis after DNA conversion with a bisulfite treatment. This analysis reveals the complexity of the diuron/solvent mixture effect on C. gigas methylome as we barely observed the same consequences depending on the considered loci, CpG within each loci and individuals. A diuron specific effect was observed in an unique case and we rather showed that the differential CpG methylation pattern were the result of different scenarios, including solvent effect only and a synergistic or antagonist effect of the diuron/solvent combination. Although we observed a strong individual variation in response to the exposure, two regions were consistently affected in the 10 tested individuals by the solvent/diuron combination. Using the combination of DNA methylation in both these markers, we calculated a positive predictive value which suggests that these two regions could indicate pollution in marine environment by a confidence level of 85.7% and could be used as potential biomarkers for marine risk assessment. The use of DNA methylation patterns as an early indicator of general disease disorder is already applied successfully in cancer diagnostics [63–65]. Further application as biomarkers in the evaluation of the carcinogenic potential of environmental factors has also been suggested [66]. The same kind of application in an ecologically relevant context to detect contamination in the environment on sentinel species has been already discussed but never applied [19, 51, 67]. In this context, these two regions represent relevant candidates which could be tested for the biomonitoring of pollution in the marine environment although further appropriate validations are required. Especially, it will be important to study whether the impact of parental diuron exposure on the DNA methylation profile of its progeny mirrors a general stress effect or corresponds to specific effect of this class of chemical pollutants.
Most of the studies conducted so far in the field of environmental epigenetic have been centered on DNA methylation. This covalent modification is conserved in most major eukaryotic groups [68–70], although it has been lost in certain model species [69, 71, 72]. The levels, patterns and function of DNA methylation vary among species. In invertebrates, it occurs in many taxa [69, 73–79] whose genomes are characterized by interspaced regions of methylated and unmethylated DNA. Intragenic (exon and intron) methylation is a general feature of invertebrate animals while methylation of repetitive elements and intergenic regions occurs only at moderate level [80]. The role of intragenic DNA methylation in invertebrate species remains an open question. It is more abundant in the 5′ region of open reading frame and associated with histone marks indicative of open chromatin structure in insect species [74, 81]. Our work confirmed previous data on the DNA methylation pattern in C. gigas [35, 77]. We showed that 2% of total cytosine was methylated predominantly in the CpG context, DNA methylation was organized as a mosaic feature and was higher in the coding region. Furthermore, our work highlighted that this pattern was sensitive to environmental condition and could be involved in intergenerational effects. The majority of DMRs that we identified was essentially found in intragenic regions. We investigated the consequences of these DMRs which occurred within genes on their corresponding transcripts. Some studies have shown that intragenic methylation is likely to occur in genes associated with stable transcription [74, 80] while other studies have suggested that DNA methylation may regulate gene expression by altering the landscape of splice variants [73, 82]. In our case, we observed that DNA methylation changes correlated with RNA level only in a very small group of genes, located in the high methylated regions. The target expression analysis confirmed the absence of obvious impact on gene expression and splice variant generation, although, some of these candidates displayed a striking difference in their CpG methylation profile and in one case a strong difference was observed in a promoter region. This lack of obvious phenotype raised the question of the function of DNA methylation for molecular phenotypes, if not playing a role in controlling gene expression as clearly demonstrated in flowering plants and mammals [68]. Another striking result of our study was the severe impact of the diuron parental exposure on several unique exon genes that encode transposon relic. These mobile genetic elements seemed to be active since we could detect their transcripts. However, there were no differences in their level of expression between the SWC and the D groups which further questions the role of CpG methylation for transposon silencing in C. gigas. Previous work has shown that DNA methylation only targeted some classes of repetitive elements in C. gigas [77] and therefore mobile elements were not a preferential target of DNA methylation as observed in plants and mammals for which their role as a genome defense has been largely demonstrated [68]. Consequently, our study confirmed previous observations that DNA methylation in invertebrates do not follow the general paradigm concerning its role for gene silencing [83–85]. We suggest that the admitted role of DNA methylation for heterochromatization of DNA must be revisited and its function in controlling gene expression in invertebrates, if there is any, may be restricted to really conserved pathway such as those encountered for developmental and key signaling pathways [86–89]. Our analysis further highlighted that a bias exists in the position of the DMRs which preferentially occurred in the 3′ end region of the genes. Given that little information exists on the possible role of invertebrate intragenic DNA methylation, the meaning of such a bias certainly deserves further attention. An important issue to address is whether the observed effect is linked to the genotoxic effects of diuron observed in parallel during this experiment or whether it is a general feature that is observed whatever the stress experienced. As a matter of fact, some studies pointed out a possible interaction between genotoxicity and DNA methylation mechanisms [90].
In conclusion, we have investigated the impact of a parental diuron exposure on the DNA methylation profile of C. gigas. We showed that a parental chemical stress effect can have an impact on the DNA methylation pattern of the progeny. This is the first evidence for the implication of epigenetic marks for a parental effect in aquatic species. Our work showed that epigenetic changes might represent an important pathway by which environmental factors influence phenotypic outcomes, within individuals and across generations in invertebrate aquatic species of economic and ecological importance.
Methodology
Genitor Origin, Parental Diuron Exposure, and Production of the Next Generation
The biological material used for the present study was obtained at the experimental hatchery of Ifremer La Tremblade in the frame of the GIMEPEC project funded by the French National Agency for Research (ANR-CESA-01601) [37]. Briefly, wild adult oysters from Marennes-Oléron Bay (France) were transferred from the field to the hatchery. Seawater was pumped directly from the Seudre river Estuary, filtered through a sand filter (40 μm) and passed through UV rays before draining the tanks in a continuous oxygenated flow system. Oysters were fed daily with a mixture of four marine microalgae and the water temperature was maintained at 8°C (±1°C) throughout the 1 month acclimatization period. In order to initiate the oyster gonad maturation process, the temperature was then raised by two degrees per day for 1 week, to reach 19.8°C (±0.3°C) at the start of the experiment. A total of 720 individuals were divided into three experimental groups (three replicate tanks per group): a diuron-exposed group (D), a solvent-exposed group as diuron was prepared in 0.005% acetonitrile (SC) and a seawater control group (SWC). At the start and the mid-course of gametogenesis, the oysters were exposed during two 7-day periods to pulses of 0.2 and 0.3 µg/l of diuron, respectively, and of 0.005% (50 µl/l) acetonitrile for the SC group. The effective diuron concentrations reached into the assay tanks were measured thanks to the use of Polar Organic Chemical Integrative Samplers (POCIS) [37]. This exposure scenario is expected to be environmentally realistic with an exposure time which mimics rain events at diuron concentrations close to those detected in coastal waters [25, 91]. In France, Diuron has been detected at concentrations up to 0.25 µg l−1 [92]. At time of spawning, male and female gametes from each genitor lots (“SWC”, “SC” and “D”) were recovered and used to produce the next generation as previously described [37].
Genomic Isolation
Individual whole tissue samples of spat oysters previously frozen in liquid nitrogen were homogenized in RL1 lysis buffer supplied by the NucleoSpin 96 RNA Tissue Core kit from Macherey-Nagel. Genomic DNA (gDNA) was isolated individually as previously described [93]. Briefly, 100 µl of oyster tissue were incubated overnight at 55 °C in 500 µl of DNA extraction buffer (100 mM NaCl, 10 mM Tris–HCl pH 8, 25 mM EDTA pH 8, 0.5% SDS, 0.1 mg/ml protease K). Following a step of phenol/chloroform extraction, gDNA samples were precipitated by addition of cold ethanol and treated with 50 µg/ml RNase A (Invitrogen) for 30 min at 37 °C to eliminate contaminating RNA.
Genome Wide Methylome Analysis
gDNA (3.02–5.44 µg) was sent to GATC (Germany) for bisulfite conversion, BS-seq paired-end library construction and sequencing. Bisulfite conversion rate was 99.5%. Three libraries per oyster group (SWC and D) were constructed as biological triplicates. One library corresponds to whole DNA sample from a single individual. Individual D3-17, D1-19, and D3-12 were chosen for the D group and individuals TE1-14, TE3-19, and TE1-11 were chosen for the SWC group. These individuals are from the same study for which we previously generated a RNA-Seq experiment except that in this RNA-Seq study, reverse transcription has been realized on pool of RNA from 10 individuals including the one analyzed here [40]. The workflow applied for BS-Seq analysis is shown in Supplementary Fig. S1. Fastq reads of six libraries (three per condition) were filtered separately based on quality phred-score. Reads with <95% of nucleotides with a quality phred-score equal or higher than 26 were discarded with the Filter by quality tool. Interlace of forward and reverse libraries per sample were performed to discard unpaired reads. De-interlace step was performed per sample to separate forward and reverse libraries previous to mapping. The reference genome, CgigasV9 [34], was modified for the subsequent mapping step by discarding contigs ≤ 5 Kb. Bismark [94] was used to map the filtered reads to the modified genome with zero mismatch allowed in the 100 bp reads. Bismark sorted alignment output (BAM file) were converted to SAM file on a local Galaxy instance. Coverage was analyzed using “bedtools makewindows” [95] and “samtools bedcov” [96]. The methylkit software (R packages) [97] was used for comparative statistical analysis between the samples using the SAM files as input. Criteria used to determine differentially methylated regions (DMRs) of 500 bp were as follow: a minimum coverage of 10× in all replicates, a minimum of 25% of methylation difference between conditions with a corresponding adjusted P value (FDR) of 0.01. Methylkit defines methylation difference as absolute value of methylation percentage change between test and control. Since there is no data in oyster about the degree of methylation change that is necessary to obtain a biological impact we tentatively used the 25% default value. MethylExtract 1.8.1 [98] was used to generate Methylation Wiggle and BED files that were displayed in IGV [99] for visual inspection of the data genomic regions (promoter, exon, intron or intergenic region) affected by parental exposure to diuron. MethylExtract served also to generate genetic variant calls in VCF format. VCF files were analyzed with VCFTools v0.1.12b [100]. Average methylation profiles over transcribed regions were generated with DeepTools [101] on the public Galaxy instance (https://usegalaxy.org/). We used our earlier produced RNA-Seq based annotation of the genome [40]. Correlation analysis between DNA methylation and gene expression was performed using the Pearson and Apos correlation tool under the public galaxy instance (https://usegalaxy.org/). Average HTSeq count per gene [40] was used for the gene expression level. Average DNA methylation profile per gene for the three biological replicates was generated. The fold change in expression observed between the diuron and seawater control conditions was the result of the DESeq analysis [40]. The fold change in DNA methylation between the diuron and seawater control conditions was the result of the Bismark analysis. For the bioinformatic pipeline, scripts are available upon request.
Visual inspection of the methylation profile of each replicates in IGV software was performed in order to identify DMRs for which all biological replicates were affected in the same way. We determined the genomic features of these target DMRs as being localized either within coding region (exon or intron), promoter or intergenic region using the gene annotation obtained with previous RNA-seq data [40]. A 500 bp DMRs which was encompassing two genomic features were counted as being located in the two contexts. Promoter regions were considered to be the 1 kb region upstream of the first exon of an annotated gene. For accurate exonic position within the genes, we defined 13 possible positions: position in unique exon, in last exon, in last exon −1 to last exon −10 and others which concerns genes with more than 10 exons. The same definition was used to localize the accurate intronic position within the genes.
DNA Methylation of Target Gene Analysis
Bisulfite treatment was done as described before [102] (http://ihpe.univ-perp.fr/methods/methods/bisulphite_engl_mc.html) on gDNA extracted from SWC, SC and D groups. Ten individuals were analyzed for each experimental condition. Briefly, 300 ng of DNA was denaturated with 3 M NaOH, treated with a solution of sodium-bisulfite and hydroquinone at pH 5 in the dark for 4 h at 55 °C; desalted (Amicon Ultra column, UFC510024 Millipore), desulfonated by adding 350 µl of 0.1 M NaOH to the DNA in the Microcon column, and finally dissolved in 10 mM Tris/Cl pH 8. The genomic sequence of 10 DMRs was targeted by nested PCR on 10 individuals per group (SWC, SC, D). Primer pairs (Supplementary Table S1) were designed using MethPrimer [103]. PCR amplification was performed in 50 µl using 1.25 U of the M0495L-Hot start Taq DNA polymerase (Biolabs), 1× Hot start Taq buffer, 0.2 µM of primers, 200 µM of dNTP. PCR cycles were:94 °C for 2 min, five cycles of 94 °C for 1 min, 50 °C for 2 min, and 72 °C for 3 min; followed by 25 cycles of 94 °C for 30 s, 50 °C for 2 min, and 72 °C for 1 min 30 s and finally 72 °C for 10 min on a thermocycler (Electrocon corporation HBPX2220). 2.5 µl of the bisulfite treated DNA was used as template for the first round of PCR using external primers, then 2.5 µl of the PCR product was used as template for the nested PCR using internal primers. Primers were used at 0.2 µM and dNTP at 200 µM. Products were sequenced using the genoscreen sequencing facilities (http://www.genoscreen.fr/fr/). Chromatograms were edited using MEGA 6 [104] and DNA Chromatogram Explorer (http://www.dnabaser.com/download/chromatogram-explorer/). For each methylated cytosine, the percentage of methylation was determined according to five levels (0, 25, 50, 75, and 100%) depending on the height of the chromatogram peaks for the cytosine (non-converted methylated cytosine) compared to the height of the thymine (converted non-methylated cytosine) overlapping at the same position on the forward strand (and reversely on the reverse strand). For positive predictive value (PPV) calculation, the two regions: scaffold1255:365901.366597 and scaffold33832:13205.13768 were considered “diuron exposed profile” when individuals displayed percentage of methylation of 0 and above 25, respectively. The PPV value was calculated using a MedCalc based interface (https://www.medcalc.org/calc/diagnostic_test.php) and true positive took into account a positive occurrence in the two markers. The disease group was the 10 D samples; the non-disease group was the 10 SWC samples. True positives (a) are individuals for which DNA methylation profile is typical of “diuron exposed profile” in the D group. False positives (c) are individuals for whom DNA methylation profile is typical of “diuron exposed profile” but those individuals are from the control group. PPV equals a/(a + c) (Supplementary Fig. S3).
Expression Analysis of Target Genes
RNA extraction and cDNA synthesis were performed as previously described [40]. Briefly, individual whole tissue samples of oyster spat previously frozen in liquid nitrogen were homogenized in RL1 lysis buffer supplied by the NucleoSpin 96 RNA Tissue Core kit (Macherey-Nagel, France). Total RNA was extracted according to the manufacturer protocol. RNA samples were treated with DNase to prevent DNA contamination. cDNAs were synthesized from 300 ng of total individual RNA samples using VILO Superscript enzyme according to manufacturer’s instruction (Invitrogen). Primer specificity was tested performing a blastN against C. gigas genome version 9 (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Primer sequences and expected PCR product sizes are listed in Supplementary Table S1.
End point Polymerase Chain Reaction (PCR) was performed to check for the presence of alternative transcripts in the three candidates XLOC_005912 (CGI_10018082), XLOC_032353 (CGI_100 10582) and XLOC_032365 (CGI_10010585) and amplified, respectively, with the following primer pairs XLOC_005912.3for/XLOC_ 005912.5rev, XLOC_032353.2for/XLOC_032353.1rev and XLOC_0 32365.1for and XLOC_032365.3rev. A total of 20 individuals were tested per condition (SWC, SC and D). PCR amplification was performed in 25 µl using 0.625 U of GoTaq DNA (Promega kit: GoTaq G2 Hot Start Polymerase), 1× GoTaq buffer, 0.2 µM of each primer, 0.2 mM dNTPs, 2 mM MgCl2. PCR cycles were: 95 °C for 3 min, 35 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min and finally 72 °C for 10 min. PCR were accomplished on MastercycleEp Gradient Eppendorf thermocycler. 3 µl of the cDNA matrix were used. PCR product sizes were checked by electrophoresis (TAE 1%). For high resolution analysis on cDNA sequence, a 2.198 kb fragment from CGI_10018082 was amplified with XLOC _005912.3for and XLOC_005912.5rev. 1 µl of this PCR product was cloned into pCR4 (TOPO TA Cloning kit, Invitrogen) and the inserts (17 inserts per condition) derived from amplified cDNA of D (D3-17, D3-12, D3-19) and SWC (TE1-11, TE1-14, TE1-19) individuals were sequenced with primers (M13F, M13R and XLOC_005912.1rev) using the genoscreen sequencing facilities (http://www.genoscreen.fr/fr/). Chromatograms were edited using Sequencher Software, and a total of 34 sequences (combining several clones of each individual from each condition) were thoroughly examined for final analysis. Phylogeny analysis was performed using the on line phylogeny analysis tools [105].
Real time PCR (qPCR) were performed in duplicates in a total volume of 10 µl with 1.5 µl of H2O, 0.5 µl of each primers, 5 µl of MelTaq and 2.5 µl of cDNA (Roche kit: LightCycler® 480 SYBR Green I Master). qPCR were accomplished on the LightCycler480 II (Roche Diagnostics). For each reaction, the crossing point (Cp) was determined using the second derivative maximum method using Light Cycler Software version 3.3 (Roche Diagnostics). For each reaction, the level of transcription was normalized using the mean geometric transcription rate of three reference sequences: Elongation factor 1: EKC33063.1 Ubiquitin 60S ribosomal protein L40: EKC19428.1 40S ribosomal protein S6: AFJ91756.1 as previously described [106] (Supplementary Table S1). Difference between SWC and D individuals was tested statistically using Student’s test on R software.
Supplementary Material
Acknowledgements
This work was supported by the French National Agency for Research (GIMEPEC project, ANRCESA-01601), by the CNRS program EC2CO (GENEPIH), by the Région Languedoc Roussillon (TRANSGIGAS, Chercheur d’avenir 2015), the “Institut Français de Recherche pour l’Exploitation de la Mer” (“French Research Institute for Sea Exploration”-Ifremer), the “Centre National de la Recherche Scientifique” (National Centre for Scientific Research-CNRS) and the University of Perpignan via Domitia. R. Rondon was supported by a PhD grant from Ifremer.
Conflict of interest statement. None declared.
Data Availability
BS-Seq and RNA-Seq reads are available at the NCBI-SRA under the BioProject accession number PRJNA291364.
Author’s Contributions
F. Akcha, C. Cosseau, E. Bachere, C. Grunau designed research; R. Rondon, M. Fallet, C. Chaparro and N. Charlemagne performed research; R. Sussarellu, R. Rondon, C. Grunau and C. Cosseau analyzed the data; C. Grunau, C. Montagnani, R. Rondon C. Cosseau and G. Mitta wrote the paper.
Authorship
E.B. and F.A. designed the study. R.R., M.F., C.C., R.S., and N.C. performed the experiments and acquired the data. R.R., C.G., C.C., and C.C. analyzed the data. R.R., C.M., G.M., C.G., and C.C. wrote the manuscript. All authors edited and approved the manuscript.
Supplementary data
Supplementary data is available at EnvEpig online.
References
- 1. Gonzalez-Romero R, Rivera-Casas C, Fernandez-Tajes J, Ausio J, Mendez J, Eirin-Lopez JM.. Chromatin specialization in bivalve molluscs: a leap forward for the evaluation of Okadaic Acid genotoxicity in the marine environment. Comp Biochem Physiol C Toxicol Pharmacol 2011; 155:175–81. [DOI] [PubMed] [Google Scholar]
- 2. Schwindt AR. Parental effects of endocrine disrupting compounds in aquatic wildlife: is there evidence of transgenerational inheritance? Gen Comp Endocrinol 2015; 219:152–64. [DOI] [PubMed] [Google Scholar]
- 3. Burdge GC, Lillycrop KA.. Nutrition, epigenetics, and developmental plasticity: implications for understanding human disease. Annu Rev Nutr 2010; 30:315–39. [DOI] [PubMed] [Google Scholar]
- 4. Champagne FA. Epigenetic mechanisms and the transgenerational effects of maternal care. Front Neuroendocrinol 2008; 29:386–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Angers B, Castonguay E, Massicotte R.. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol Ecol 2010; 19:1283–95. [DOI] [PubMed] [Google Scholar]
- 6. Hala D, Huggett DB, Burggren WW.. Environmental stressors and the epigenome. Drug Discov Today Technol 2014; 12:e3–8. [DOI] [PubMed] [Google Scholar]
- 7. Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, Haile RW, Laird PW.. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 2012; 131:1565–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Day T, Bonduriansky R.. A unified approach to the evolutionary consequences of genetic and nongenetic inheritance. Am Nat 2011; 178:E18–36. [DOI] [PubMed] [Google Scholar]
- 9. Geoghegan JL, Spencer HG.. Population-epigenetic models of selection. Theor Popul Biol 2012; 81:232–42. [DOI] [PubMed] [Google Scholar]
- 10. Skinner MK, Gurerrero-Bosagna C, Haque MM, Nilsson EE, Koop JA, Knutie SA, Clayton DH.. Epigenetics and the evolution of Darwin’s Finches. Genome Biol Evol 2014; 6:1972–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH.. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A 2008; 105:17046–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Steenaard RV, Ligthart S, Stolk L, Peters MJ, van Meurs JB, Uitterlinden AG, Hofman A, Franco OH, Dehghan A.. Tobacco smoking is associated with methylation of genes related to coronary artery disease. Clin Epigenetics 2015; 7:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xin F, Susiarjo M, Bartolomei MS.. Multigenerational and transgenerational effects of endocrine disrupting chemicals: A role for altered epigenetic regulation? Semin Cell Dev Biol 2015; 43:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skinner MK. Endocrine disruptors in 2015: epigenetic transgenerational inheritance. Nat Rev Endocrinol 2015; 12:68–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collotta M, Bertazzi PA, Bollati V.. Epigenetics and pesticides. Toxicology 2013; 307:35–41. [DOI] [PubMed] [Google Scholar]
- 16. Vaiserman A. Epidemiologic evidence for association between adverse environmental exposures in early life and epigenetic variation: a potential link to disease susceptibility? Clin Epigenetics 2015; 7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alonso-Magdalena P, Rivera FJ, Guerrero-Bosagna C.. Bisphenol-A and metabolic diseases: epigenetic, developmental and transgenerational basis. Environ. Epigenetics 2016; 2(3): dvw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Head JA. Patterns of DNA methylation in animals: an ecotoxicological perspective. Integr Comp Biol 2014; 54:77–86. [DOI] [PubMed] [Google Scholar]
- 19. Suarez-Ulloa V, Gonzalez-Romero R, Eirin-Lopez JM.. Environmental epigenetics: a promising venue for developing next-generation pollution biomonitoring tools in marine invertebrates. Mar Pollut Bull 2015; 98:5–13. [DOI] [PubMed] [Google Scholar]
- 20. Pierron F, Baillon L, Sow M, Gotreau S, Gonzalez P.. Effect of low-dose cadmium exposure on DNA methylation in the endangered European eel. Environ Sci Technol 2014; 48:797–803. [DOI] [PubMed] [Google Scholar]
- 21. Pierron F, Bureau du, Colombier S, Moffett A, Caron A, Peluhet L, Daffe G, Lambert P, Elie P, Labadie P, Budzinski H, et al. Abnormal ovarian DNA methylation programming during gonad maturation in wild contaminated fish. Environ Sci Technol 2014; 48:11688–95. [DOI] [PubMed] [Google Scholar]
- 22. Tran TK, MacFarlane GR, Kong RY, O’Connor WA, Yu RM.. Potential mechanisms underlying estrogen-induced expression of the molluscan estrogen receptor (ER) gene. Aquat Toxicol 2016; 179:82–94. [DOI] [PubMed] [Google Scholar]
- 23. Giacomazzi S, Cochet N.. Environmental impact of diuron transformation: a review. Chemosphere 2004; 56:1021–32. [DOI] [PubMed] [Google Scholar]
- 24. Tixier C, Sancelme M, Bonnemoy F, Cuer A, Veschambre H.. Degradation products of a phenylurea herbicide, diuron: synthesis, ecotoxicity, and biotransformation. Environ Toxicol Chem 2001; 20:1381–9. [DOI] [PubMed] [Google Scholar]
- 25. Caquet T, Roucaute M, Mazzella N, Delmas F, Madigou C, Farcy E, Burgeot T, Allenou JP, Gabellec R.. Risk assessment of herbicides and booster biocides along estuarine continuums in the Bay of Vilaine area (Brittany, France). Environ Sci Pollut Res Int 2013; 20:651–66. [DOI] [PubMed] [Google Scholar]
- 26. Scheringer M, Strempel S, Hukari S, Ng CA, Blepp M, Hungerbuhler K.. How many persistent organic pollutants should we expect? Atmos Pollut Res 2012; 3:383–91. [Google Scholar]
- 27. Korkaric M, Xiao M, Behra R, Eggen RI.. Acclimation of Chlamydomonas reinhardtii to ultraviolet radiation and its impact on chemical toxicity. Aquat Toxicol 2015; 167:209–19. [DOI] [PubMed] [Google Scholar]
- 28. Alyürük H, Çavas L.. Toxicities of diuron and irgarol on the hatchability and early stage development of Artemia salina. Turkish J Biol 2013; 37:151–7. [Google Scholar]
- 29. Nélieu S, Bonnemoy F, Bonnet JL, Lefeuvre L, Baudiffier D, Heydorff M, Quéméneur A, Azam D, Ducrot PH, Lagadic L, et al. Ecotoxicological effects of diuron and chlorotoluron nitrate-induced photodegradation products: monospecific and aquatic mesocosm-integrated studies. Environ Toxicol Chem 2010; 29:2644–52. [DOI] [PubMed] [Google Scholar]
- 30. Guo XM, Hedgecock D, Hershberger WK, Cooper K, Allen SK.. Genetic determinants of protandric sex in the Pacific oyster, Crassostrea gigas Thunberg. Evolution 1998; 52:394–402. [DOI] [PubMed] [Google Scholar]
- 31. Fabioux C, Pouvreau S, Le Roux F, Huvet A.. The oyster vasa-like gene: a specific marker of the germline in Crassostrea gigas. Biochem Biophys Res Commun 2004; 315:897–904. [DOI] [PubMed] [Google Scholar]
- 32. Naimi A, Martinez AS, Specq ML, Diss B, Mathieu M, Sourdaine P.. Molecular cloning and gene expression of Cg-Foxl2 during the development and the adult gametogenetic cycle in the oyster Crassostrea gigas. Comp Biochem Physiol B Biochem Mol Biol 2009; 154:134–42. [DOI] [PubMed] [Google Scholar]
- 33. Obata M, Komaru A.. The mechanisms of primordial germ cell determination during embryogenesis in molluscan species. Isj-Invert Surviv 2012; J9:223–9. [Google Scholar]
- 34. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012; 490:49–54. [DOI] [PubMed] [Google Scholar]
- 35. Olson CE, Roberts SB.. Genome-wide profiling of DNA methylation and gene expression in Crassostrea gigas male gametes. Front Physiol 2014; 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roberts SB, Gavery MR.. Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front Physiol 2012; 2:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barranger A, Akcha F, Rouxel J, Brizard R, Maurouard E, Pallud M, Menard D, Tapie N, Budzinski H, Burgeot T, et al. Study of genetic damage in the Japanese oyster induced by an environmentally-relevant exposure to diuron: evidence of vertical transmission of DNA damage. Aquat Toxicol 2014; 146:93–104. [DOI] [PubMed] [Google Scholar]
- 38. Barranger A, Benabdelmouna A, Degremont L, Burgeot T, Akcha F.. Parental exposure to environmental concentrations of diuron leads to aneuploidy in embryos of the Pacific oyster, as evidenced by fluorescent in situ hybridization. Aquat Toxicol 2015; 159:36–43. [DOI] [PubMed] [Google Scholar]
- 39. Akcha F, Barranger A, Bachére E, Berthelin CH, Piquemal D, Alonso P, Sallan RR, Dimastrogiovanni G, Porte C, Menard D, et al. Effects of an environmentally relevant concentration of diuron on oyster genitors during gametogenesis: responses of early molecular and cellular markers and physiological impacts. Environ Sci Pollut Res Int 2016; 23:8008–20. [DOI] [PubMed] [Google Scholar]
- 40. Rondon R, Akcha F, Alonso P, Menard D, Rouxel J, Montagnani C, Mitta G, Cosseau C, Grunau C.. Transcriptional changes in Crassostrea gigas oyster spat following a parental exposure to the herbicide diuron. Aquat Toxicol 2016; 175:47–55. [DOI] [PubMed] [Google Scholar]
- 41. Roquis D, Rognon A, Chaparro C, Boissier J, Arancibia N, Cosseau C, Parrinello H, Grunau C.. Frequency and mitotic heritability of epimutations in Schistosoma mansoni. Mol Ecol 2016; 25:1741–58. [DOI] [PubMed] [Google Scholar]
- 42. Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, Maleszka R.. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A 2012; 109:4968–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Youngson NA, Whitelaw E.. Transgenerational epigenetic effects. Annu Rev Genomics Hum Genet 2008; 9:233–57. [DOI] [PubMed] [Google Scholar]
- 44. Kucharski R, Maleszka J, Foret S, Maleszka R.. Nutritional control of reproductive status in honeybees via DNA methylation. Science 2008; 319:1827–30. [DOI] [PubMed] [Google Scholar]
- 45. Verhoeven KJ, Jansen JJ, van Dijk PJ, Biere A.. Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol 2010; 185:1108–18. [DOI] [PubMed] [Google Scholar]
- 46. Talbert PB, Henikoff S.. Environmental responses mediated by histone variants. Trends Cell Biol 2014; 24:642–50. [DOI] [PubMed] [Google Scholar]
- 47. Baccarelli A, Bollati V.. Epigenetics and environmental chemicals. Curr Opin Pediatr 2009; 21:243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Skinner MK, Manikkam M, Guerrero-Bosagna C.. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol Metab 2010; 21:214–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DeVries A, Vercelli D.. Epigenetics in allergic diseases. Curr Opin Pediatr 2015; 27(6):719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huo X, Chen D, He Y, Zhu W, Zhou W, Zhang J.. Bisphenol-A and female infertility: a possible role of gene-environment interactions. Int J Environ Res Public Health 2015; 12:11101–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vandegehuchte MB, Janssen CR.. Epigenetics in an ecotoxicological context. Mutat Res Genet Toxicol Environ Mutagen 2014; 764-765:36–45. [DOI] [PubMed] [Google Scholar]
- 52. Foo SA, Byrne M.. Acclimatization and adaptive capacity of marine species in a changing ocean. Adv Mar Biol 2016; 74:69–116. [DOI] [PubMed] [Google Scholar]
- 53. Calosi P, De Wit P, Thor P, Dupont S.. Will life find a way? evolution of marine species under global change. Evol Appl 2016; 9:1035–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parker LM, O’Connor WA, Raftos DA, Portner HO, Ross PM.. Persistence of positive carryover effects in the oyster, Saccostrea glomerata, following transgenerational exposure to ocean acidification. PLoS One 2015; 10:e0132276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ross PM, Parker L, Byrne M.. Transgenerational responses of molluscs and echinoderms to changing ocean conditions. ICES J Mar Sci 2016; 73:537–49. [Google Scholar]
- 56. Nice HE, Morritt D, Crane M, Thorndyke M.. Long-term and transgenerational effects of nonylphenol exposure at a key stage in the development of Crassostrea gigas. Possible endocrine disruption? Mar Ecol Prog Ser 2003; 256:293–300. [Google Scholar]
- 57. Metzger DC, Schulte PM.. Epigenomics in marine fishes. Mar Genomics 2016; 30: 43–54. [DOI] [PubMed] [Google Scholar]
- 58. Putnam HM, Davidson JM, Gates RD.. Ocean acidification influences host DNA methylation and phenotypic plasticity in environmentally susceptible corals. Evol Appl 2016; 9:1165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Marsh AG, Pasqualone AA.. DNA methylation and temperature stress in an Antarctic polychaete, Spiophanes tcherniai. Front Physiol 2014; 5:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Akcha F, Spagnol C, Rouxel J.. Genotoxicity of diuron and glyphosate in oyster spermatozoa and embryos. Aquat Toxicol 2012; 106–107:104–13. [DOI] [PubMed] [Google Scholar]
- 61. Mai H, Morin B, Pardon P, Gonzalez P, Budzinski H, Cachot J.. Environmental concentrations of irgarol, diuron and S-metolachlor induce deleterious effects on gametes and embryos of the Pacific oyster, Crassostrea gigas. Mar Environ Res 2013; 89:1–8. [DOI] [PubMed] [Google Scholar]
- 62. Bouilly K, Bonnard M, Gagnaire B, Renault T, Lapegue S.. Impact of diuron on aneuploidy and hemocyte parameters in Pacific oyster, Crassostrea gigas. Arch Environ Contam Toxicol 2007; 52:58–63. [DOI] [PubMed] [Google Scholar]
- 63. Dirks RA, Stunnenberg HG, Marks H.. Genome-wide epigenomic profiling for biomarker discovery. Clin Epigenetics 2016; 8:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Y, Breitling LP, Balavarca Y, Holleczek B, Schottker B, Brenner H.. Comparison and combination of blood DNA methylation at smoking-associated genes and at lung cancer-related genes in prediction of lung cancer mortality. Int J Cancer 2016; 139:2482–92. [DOI] [PubMed] [Google Scholar]
- 65. Partin AW, Van Neste L, Klein EA, Gee JR, Troyer DA, Rieger-Christ K, Jones JS, Magi-Galluzzi C, Mangold LA, Trock BJ, et al. Clinical validation of an epigenetic assay to predict negative histopathological results in repeat prostate biopsies. J Urol 2014; 192:1081–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. LeBaron MJ, Rasoulpour RJ, Klapacz J, Ellis-Hutchings RG, Hollnagel HM, Gollapudi BB.. Epigenetics and chemical safety assessment. Mutat Res 2010; 705:83–95. [DOI] [PubMed] [Google Scholar]
- 67. Mirbahai L, Chipman JK.. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposures. Mutat Res Genet Toxicol Environ Mutagen 2014; 764–765:10–7. [DOI] [PubMed] [Google Scholar]
- 68. Schubeler D. Function and information content of DNA methylation. Nature 2015; 517:321–6. [DOI] [PubMed] [Google Scholar]
- 69. Zemach A, McDaniel IE, Silva P, Zilberman D.. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 2010; 328:916–9. [DOI] [PubMed] [Google Scholar]
- 70. Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A 2010; 107:8689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Capuano F, Mulleder M, Kok R, Blom HJ, Ralser M.. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem 2014; 86:3697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Simpson VJ, Johnson TE, Hammen RF.. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res 1986; 14:6711–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol 2012; 22:1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hunt BG, Glastad KM, Yi SV, Goodisman MA.. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol Evol 2013; 5:591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Beeler SM, Wong GT, Zheng JM, Bush EC, Remnant EJ, Oldroyd BP, Drewell RA.. Whole-genome DNA methylation profile of the jewel wasp (Nasonia vitripennis). G3 (Bethesda) 2014; 4:383–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gao F, Wang R, Liu M.. Trichinella spiralis, potential model nematode for epigenetics and its implication in metazoan parasitism. Front Physiol 2014; 4:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wang X, Li Q, Lian J, Li L, Jin L, Cai H, Xu F, Qi H, Zhang L, Wu F, et al. Genome-wide and single-base resolution DNA methylomes of the Pacific oyster Crassostrea gigas provide insight into the evolution of invertebrate CpG methylation. BMC Genomics 2014; 15:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dabe EC, Sanford RS, Kohn AB, Bobkova Y, Moroz LL.. DNA methylation in basal metazoans: insights from ctenophores. Integr Comp Biol 2015; 55(6):1096–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fneich S, Dheilly N, Adema C, Rognon A, Reichelt M, Bulla J, Grunau C, Cosseau C.. 5-methyl-cytosine and 5-hydroxy-methyl-cytosine in the genome of Biomphalaria glabrata, a snail intermediate host of Schistosoma mansoni. Parasit Vectors 2013; 6:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sarda S, Zeng J, Hunt BG, Yi SV.. The evolution of invertebrate gene body methylation. Mol Biol Evol 2012; 29:1907–16. [DOI] [PubMed] [Google Scholar]
- 81. Glastad KM, Hunt BG, Goodisman MA.. DNA methylation and chromatin organization in insects: insights from the ant Camponotus floridanus. Genome Biol Evol 2015; 7:931–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R.. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol 2010; 8:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhu A, Chen M, Zhang X, Storey KB.. Gene structure, expression, and DNA methylation characteristics of sea cucumber cyclin B gene during aestivation. Gene 2016; 594:82–8. [DOI] [PubMed] [Google Scholar]
- 84. Patalano S, Vlasova A, Wyatt C, Ewels P, Camara F, Ferreira PG, Asher CL, Jurkowski TP, Segonds-Pichon A, Bachman M, et al. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc Natl Acad Sci U S A 2015; 112:13970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Galbraith DA, Yang X, Nino EL, Yi S, Grozinger C.. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog 2015; 11:e1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Riviere G, Wu GC, Fellous A, Goux D, Sourdaine P, Favrel P.. DNA methylation is crucial for the early development in the Oyster C. gigas. Mar Biotechnol (NY) 2013; 15:739–53. [DOI] [PubMed] [Google Scholar]
- 87. Li Y, Huang X, Guan Y, Shi Y, Zhang H, He M.. DNA methylation is associated with expression level changes of galectin gene in mantle wound healing process of pearl oyster, Pinctada fucata. Fish Shellfish Immunol 2015; 45:912–8. [DOI] [PubMed] [Google Scholar]
- 88. Saint-Carlier E, Riviere G.. Regulation of Hox orthologues in the oyster Crassostrea gigas evidences a functional role for promoter DNA methylation in an invertebrate. FEBS Lett 2015; 589(13):1459–66. [DOI] [PubMed] [Google Scholar]
- 89. Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER.. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell 2012; 149:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Denissenko MF, Chen JX, Tang MS, Pfeifer GP.. Cytosine methylation determines hot spots of DNA damage in the human P53 gene. Proc Natl Acad Sci U S A 1997; 94:3893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Munaron D, Tapie N, Budzinski H, Andral B, Gonzalez JL.. Pharmaceuticals, alkylphenols and pesticides in Mediterranean coastal waters: results from a pilot survey using passive samplers. Estuar Coast Shelf Sci 2012; 114:82–92. [Google Scholar]
- 92. Buisson S, Bouchart V, Guerlet E, Malas JP, Costil K.. Level of contamination and impact of pesticides in cupped oyster, Crassostrea gigas, reared in a shellfish production area in Normandy (France). J Environ Sci Health B 2008; 43:655–64. [DOI] [PubMed] [Google Scholar]
- 93. Rosa RD, Alonso P, Santini A, Vergnes A, Bachere E.. High polymorphism in big defensin gene expression reveals presence-absence gene variability (PAV) in the oyster Crassostrea gigas. Dev Comp Immunol 2015; 49:231–8. [DOI] [PubMed] [Google Scholar]
- 94. Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011; 27:1571–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Quinlan AR, Hall IM.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010; 26:841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. The sequence alignment/map format and SAMtools. Bioinformatics 2009; 25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Akalin A, Kormaksson M, Li S, Garrett-Bakelman FE, Figueroa ME, Melnick A, Mason CE.. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol 2012; 13:R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Barturen G, Rueda A, Oliver JL, Hackenberg M.. MethylExtract: High-Quality methylation maps and SNV calling from whole genome bisulfite sequencing data. F1000Res 2013; 2:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Thorvaldsdottir H, Robinson JT, Mesirov JP.. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 2012; 14:178–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. The variant call format and VCFtools. Bioinformatics 2011; 27:2156–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016; 44:W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Grunau C, Clark SJ, Rosenthal A.. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 2001; 29:E65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li LC, Dahiya R.. MethPrimer: designing primers for methylation PCRs. Bioinformatics 2002; 18:1427–31. [DOI] [PubMed] [Google Scholar]
- 104. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013; 30:2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008; 36:W465–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Rosa RD, de Lorgeril J, Tailliez P, Bruno R, Piquemal D, Bachere E.. A hemocyte gene expression signature correlated with predictive capacity of oysters to survive Vibrio infections. BMC Genomics 2012; 13:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BS-Seq and RNA-Seq reads are available at the NCBI-SRA under the BioProject accession number PRJNA291364.