Abstract
The brown planthopper (BPH) Nilaparvata lugens is one of the most destructive insect pests in the rice fields of Asia. Like other hemipteran insects, BPH is not susceptible to Cry toxins of Bacillus thuringiensis (Bt) or transgenic rice carrying Bt cry genes. Lack of Cry receptors in the midgut is one of the main reasons that BPH is not susceptible to the Cry toxins. The main Cry-binding proteins (CBPs) of the susceptible insects are cadherin, aminopeptidase N (APN), and alkaline phosphatase (ALP). In this study, we analyzed and validated de novo assembled transcripts from transcriptome sequencing data of BPH to identify and characterize homologs of cadherin, APN, and ALP. We then compared the cadherin-, APN-, and ALP-like proteins of BPH to previously reported CBPs to identify their homologs in BPH. The sequence analysis revealed that at least one cadherin, one APN, and two ALPs of BPH contained homologous functional domains identified from the Cry-binding cadherin, APN, and ALP, respectively. Quantitative real-time polymerase chain reaction used to verify the expression level of each putative Cry receptor homolog in the BPH midgut indicated that the CBPs homologous APN and ALP were expressed at high or medium-high levels while the cadherin was expressed at a low level. These results suggest that homologs of CBPs exist in the midgut of BPH. However, differences in key motifs of CBPs, which are functional in interacting with Cry toxins, may be responsible for insusceptibility of BPH to Cry toxins.
Keywords: de novo assembly, brown planthopper, cadherin, aminopeptidase N, alkaline phosphatase
The brown planthopper (BPH) Nilaparvata lugens is a widely distributed insect pest in the rice fields of Asia. BPH can cause severe damage by feeding on phloem tissues and by transmitting viral diseases (Hibino 1996, Jia et al. 2012). No effective environment-friendly control methods are currently available for BPH. Hence, the use of chemical pesticides is the major means of management of BPH infestations (Lu et al. 2015).
Bacillus thuringiensis (Bt) Cry toxins are well-known biological agents for control of insect pests (Gómez et al. 2007, Vandenborre et al. 2011). Cry toxins have high insecticidal activity against lepidopterans, coleopterans, and mosquitoes (Bravo et al. 2011). However, Cry toxins show no or very weak insecticidal activity on hemipteran insects, such as planthopper, leafhopper, and aphids (Chougule and Bonning 2012, Shao et al. 2013).
Lack of proper receptors in the midgut of target insects has been known to be a key reason for the insusceptibility of insect to Cry toxins. Cry toxins are known to bind to cadherin-like proteins, glycosylphosphatidylinositol (GPI)-anchored aminopeptidase N (APN), GPI-anchored alkaline phosphatase (ALP), and ATP-binding cassette transporters after activation of protoxin in the process of their insecticidal activity (Pardo-López et al. 2013, Bravo et al. 2013). The known Bt receptor proteins play critical roles in insect metabolism and are essential proteins of all insects. Hence, homologous proteins of Cry-binding protein (CBP) are likely to be present in the insects which are not susceptible to Bt toxins.
RNA-deep sequencing technology has become a powerful means for transcript discovery and measurements of transcriptional expression levels. RNA sequencing has also been used in the identification of receptor homologous genes of Bt toxins. For example, Liu et al. (2012) identified five APNs and four ALPs from the midgut of Aphis glycines (Hemiptera: Aphididae) (Liu et al. 2012). Shu et al. (2015) identified an APN from the Holotrichia parallela midgut as a receptor of Cr y8Ea toxin (Shu et al. 2015). Transcriptome sequencing of BPH has also been conducted and resulted in the identification of digestion, detoxification, and immune response-related genes from BPH guts (Peng et al. 2011, Bao et al. 2012). However, homologs of potential Cry toxin receptor-related genes, e.g., cadherins, APNs, and ALPs, in the midgut of BPH have not yet been investigated. We have previously reported that Cry1Ab protoxin could be proteolytically processed by gut proteases of BPH but still be nontoxic to BPH nymphs (Shao et al. 2013). Consequently, lack of proper receptor proteins in the midgut may be one reason for the low toxicity of Cry toxins to BPH. In this research, we investigated transcriptome-sequencing data of BPH to identify putative homologous genes of CBPs, followed by sequence verification by real-time polymerase chain reaction (RT-PCR) and expression-level calculation by RT-qPCR. We also compared characteristics and structure of CBPs with their homologs in BPH. Our results indicated that homolog of CBPs exists in the midgut of BPH showing similar characteristics and structure.
Materials and Methods
Insects
The BPH strain used for this work was originally collected from rice fields of Fuzhou, Fujian, and has been reared in the laboratory conditions by the Institute of Plant Virology, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China for more than 5 yr. The BPH colony was maintained on rice (Oryza sativa L.) seedlings and kept in a growth chamber at 28°C with a photoperiod of 14:10 (light:dark) h.
Tissue Collection, RNA Isolation, and Illumina Sequencing
Over 200 third- to fourth-instar BPH nymphs were anesthetized on ice for 10 min and dissected under an SMZ-B4 microscope (Chongqing Optec Instrument, China) to isolate the digestive tract. The anterior diverticulum, hindgut, malpighian tubules, and esophagus were then removed before rinsing the midgut tissue with ice-cold diethyl pyrocarbonate (DEPC)-treated phosphate-buffered saline (PBS) solution. The obtained midguts were stored in DEPC-treated PBS solution and kept in a −80°C freezer for further use. Midgut collection was repeated for three times. The midgut tissues were homogenized in a Dounce tissue grinder (Wheaton), and total RNA was extracted using an HP Total RNA Kit (Omega, USA) according to the manufacturer’s protocol. The quantity of extracted RNA was measured with a NanoDrop (Bio-Rad, USA), and the quality of RNA was verified by an Agilent 2100 (Agilent, Germany) and electrophoresis gel analysis. Total RNA of the BPH body was prepared from 50 of the third- to fourth-instar nymphs by the previously described procedures with three replications. Small aliquots of RNA extracted from samples were stored at −80°C for further analysis by RT-qPCR assays. The rest of the RNA samples of either midgut or body were pooled together, and 30 μg of each pooled sample was used for RNA library preparation.
Purification of mRNA was performed using NEBNext Poly (A) mRNA Magnetic Isolation Module (NEB, E7490). Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) following the manufacturer’s recommendations. Fragment sizes of the prepared cDNA library were selected by agarose gel electrophoresis and then were quantitated through qPCR by the use of Library Quantification Kit-Illumina GA Universal (Kapa, KK4824). The qualified cDNA libraries were clustered through the Illumina cbot system and sequenced on an Illumina HiSeq 2500 platform to generate 125 nt paired-end reads (Biomarker Technologies Co., Ltd., Beijing, China). The original image data were processed with Illumina GA Pipeline v1.3 to clean reads, followed by the removal of adapter sequences, empty reads, and low-quality reads. A short reads assembling program from Trinity (version 2.1.1) was used for sequence assembly (Grabherr et al. 2011). To evaluate the completeness of the assembly result, assembled contigs were analyzed by the BUSCO (Benchmarking Universal Single-Copy Orthologs) program, based on the arthropod gene sets.
Sequencing Data Analysis
The OrfPredictor program (Min et al. 2005) was used to obtain the open reading frame (ORF) of each unigene. Three ORF predictions were made for nucleotide sequence of each unigene, starting from the first, the second, and the third nucleotide with termination at a stop codon, respectively. The ORF with the maximum size was selected as the final output for each sequence. Cleaned reads were mapped to unigenes by the Bowtie software (Langmead 2010), and then the number of reads mapping to each unigene as RPKM (Reads Per Kilobase per Million mapped reads) was calculated through the RSEM software (Li and Dewey 2011). Annotation of each unigene was conducted by searching all unigenes in the National Center of Biotechnology Information (NCBI) nr (nonredundant) database through the BLAST software (Altschul et al. 1997) with an E-value cutoff of 10−5. Functional annotation by gene ontology (GO) was conducted by searching unigenes against the GO database (Ashburner et al. 2000) through the Blast2GO software (http://www.blast2go.org/). Sequences of putative cadherins, APNs, and ALPs have been deposited in the GenBank (the accession number of each gene is shown in Supp Table 1).
Quantitative Real-Time PCR (RT-qPCR) Analysis and Reverse PCR (RT-PCR)
RT-qPCR was performed using the total RNA isolated from either the midgut or body of BPH, and assays were repeated with three biological replications. Total RNA of each replication from either the midgut or body, extracted as described previously, was used for first-strand cDNA synthesis using a random primer and AMV reverse transcriptase (TaKaRa, Japan). Efficiencies of each primer pair were verified to be around 96–110% before RT-qPCR assay. RT-qPCRs were carried out on an ABI 7500 real-time PCR detection system using 100 ng of the cDNA template or the nontemplate control (NTC), 0.2 μM of primers (Supp Table 2), and SYBR Premix Ex Taq (TaKaRa, Japan). PCR was performed with an initial denaturation step at 95°C for 30 s, followed by 40 cycles at 95°C for 5 s, 55°C for 30 s, and a dissociation step added at the end. A melting-curve analysis was performed upon completion of the real-time PCR runs for each sample to examine the specificity of the PCR reactions. The melting curves from all sample running in this study showed a single peak. After expression evaluation by the RefFinder online software (http://fulxie.0fees.us) (Xie et al. 2012), the BPH 18s rRNA gene (GenBank Accession No. JN662398) was selected as an internal control by using the primers previously reported by Bao et al. (2012). RT-qPCR was repeated three times, and the results were standardized to the expression level of the BPH 18s rRNA gene. An NTC sample was run to detect any contamination and to determine the degree of dimer formation. Statistical analysis of the data was performed using one-way analysis of variance running by SPSS software (version 22.0.0), along with online statistical tools (http://www.xuru.org/st/ DS.asp).
To assess the de novo-assembled sequence of selected bphCadherins, bphAPNs, and bphALPs, RT-PCR was conducted using Premix Taq (Ex Taq Version 2.0 plus dye) (TaKaRa, Japan) with the midgut cDNA as the template. Primer pairs used for PCR reactions were designed based on the assembled unigene sequences and are shown in Supp Table 2. The PCR fragments were isolated from the agarose gel and shotgun sequenced to verify the de novo-assembled sequence of selected bphAPNs, bphALPs, and bphCadherins. Verified sequences were submitted to the GenBank. The accession number of each gene is shown in Supp Table 3.
Identification of Homologs of Cry Receptors and Phylogenetic Analysis
To obtain the homologs of Cry receptor proteins expressed in the midgut of BPH, unigenes annotated with the keyword ‘cadherin’, ‘alkaline phosphates’ or ‘aminopeptidase N’ were screened through custom Perl script from all NCBI nr-annotated unigenes. The full-length genes were determined if the predicted ORF was inside the unigene sequence, together with the start codon, stop codon, 5ʹ-UTR and 3ʹ-UTR. Cry-binding cadherins, APNs, and ALPs in Cry toxin-susceptible insects and aphids used for sequence identity, sequence alignment, and phylogenetic analysis were randomly selected from the GenBank. Protein alignment was performed with the MEGA 6.06 software (Tamura et al. 2013) using the built-in MUSCLE program. The poorly aligned positions and divergent regions of the alignment were eliminated by the Gblocks program (Castresana 2000, Talavera and Castresana 2007) to obtain conserved blocks. A sequence identity matrix was calculated by BioEdit 7.0.9. The multiple sequence alignments and phylogenetic trees (maximum-likelihood trees) were generated using the MEGA 6.06 with a bootstrap value of 1000.
The amino acid sequence of a selected BPH protein was input to SMART (http://smart.embl-heidelberg.de) for motif scanning. Prediction of a signal peptide at the N-terminus of each protein was conducted with SignalP 4.1 (Petersen et al. 2011). GPI-anchor signal at the C-terminus was predicted using PredGPI, FragAnchor, and big-PI Predictor. N-glycosylation and O-glycosylation sites were predicted by the NetNglyc 1.0 Server (Blom et al. 2004) and NetOglyc 4.0 Server (Julenius et al. 2005), respectively. Transmembrane helices of each protein were predicted by the TMHMM 2.0 Server (http://www.cbs.dtu.dk/services/TMHMM/).
Three-Dimensional Modeling of Selected APN and ALP
Three-dimensional structure modeling of APN and ALP was based on templates 4wz9.1.A and 1zef.1.A, respectively. Models were built by online software SWISS-MODEL (Arnold et al. 2006, Guex et al. 2009, Kiefer et al. 2009, Biasini et al. 2014) and displayed by the PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC.
Results
Sequencing, de novo Assembly, and Functional Annotation
Illumina sequencing of the midgut and whole-body transcriptome generated over 39 million and 33 million raw reads, respectively (Supp Table 4). The raw reads of both midgut and whole body were assembled by Trinity assembler with the default parameter setting, resulting in an assembly of 91,536 transcripts (≥200 nt) with a mean length of 1,177 bp. From the transcripts, 53,502 unigenes were predicted with a mean size of 856 bp (Supp Table 5). To assess the completeness of the assembled data, the assembled transcripts were analyzed using the arthropod gene sets in the BUSCO database as reference by the BUSCO program (Simão et al. 2015). The results showed that out of 2,675 BUSCO searched genes, 84.11% (2,250 BUSCOs) were complete, 5.08% (136 BUSCOs) were complete and duplicated, 4.49% (120 BUSCOs) were fragmented, and 11.40% (305 BUSCOs) were not found, suggesting that the quality of the assembly was completely acceptable. The ORF sequences were predicted from the resultant unigenes by using the OrfPredictor program (Min et al. 2005). The numbers and the average length of the predicted ORF sequences have been summarized in Supp Table 5. The transcriptome sequencing data set has been deposited in NCBI as BioProject ID PRJNA383084.
Annotation of the assembled transcripts was conducted by using the BLAST program against the NCBI nr database with a cutoff E-value of 10−5. Only 18,810 of 53,502 (35%) unigenes hit the reference genes from the NCBI nr database (Supp Table 6). Statistics from the BLAST results (E-value, sequence similarity and hit species distributions) are shown in Supp Fig. 1.
Identification of Cadherin, APN, and ALP Genes
To identify putative homologous genes of Cry-binding cadherin, APN, and ALP, we searched for the transcripts that hit cadherins, APNs, and ALPs from the BLAST annotation and identified eight putative cadherins, five putative APNs, and six putative ALPs, respectively. Among those identified genes, one APN and two ALPs had been previously reported but with slight sequence variations (Supp Table 1). We then conducted RT-PCR to verify sequences of the assembled genes that hit CBPs. The cDNA fragments amplified by RT-PCR matched the expected lengths of the assembled sequences that were tested (Supp Figs. 2–4). The assembled sequences were then confirmed by Sanger sequencing of the PCR fragments with only a few nucleotide variations compared to the assembled sequences (results not shown).
Comparison of the CBPs With Their Putative Homologs in BPH
To identify Cry receptor homologs in the BPH midgut, we first analyzed the sequence similarity between the known Cry receptors derived from Bt-susceptible insects and their potential counterparts in BPH. We then analyzed putative motif regions and compared sequence variations of the motifs between Cry receptors and their potential homologs of BPH.
Sequence Analysis of the Putative BPH Cadherin, APN, and ALP Proteins
Comparisons of protein sequence identities between Cry receptor-related cadherins, APNs, and ALPs derived from lepidopteran, coleopteran, and dipteran insects and those of BPH are summarized in Supp Tables 7–9, respectively. Overall, sequence identities of the previously reported Cry-binding cadherins were around 50% and even over 70% among insects of the same order. The only exception was bmCadherin2, which had a 36.5 and 31.1% identity with the known Cry-binding cadherins in B. mori (bmCadherin1) and T. ni (tnCadherin), respectively (Supp Table 7). However, the sequence identities between the putative BPH cadherins and most of the selected Cry-binding cadherins were less than 30% (bphCadherin 5–8) to ~40% (bphCadherin 1–4). Only bphCadherin1 showed over a 50% sequence identity (53.8%) with a cadherin originating from B. mori (bmCadherin2) (Supp Table 7). The amino acid identities of APN proteins showed similar diversities (~35 to 40%) among the Cry-sensitive insects and between Cry target insects or nontarget insects (BPH or aphids) (Supp Table 8). Compared to cadherin and APN, sequence identity of ALP isolated from BPH and Bt-susceptible insects was relatively higher (>50%) (Supp Table 9).
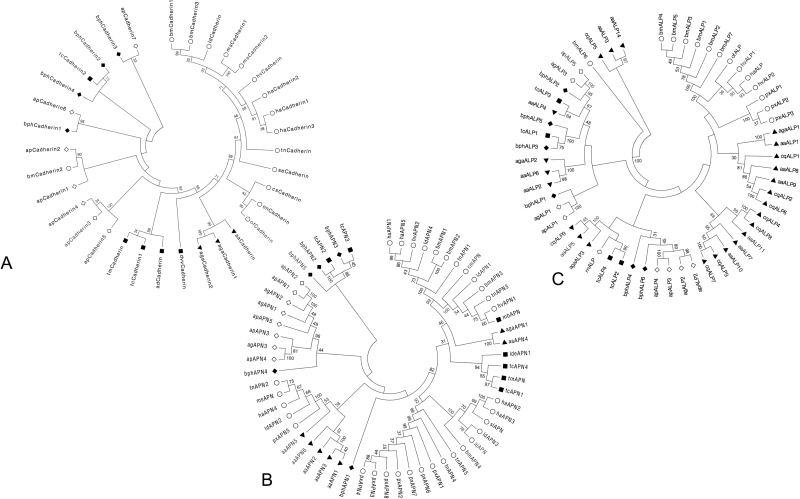
To conduct phylogenetic analysis, the BPH proteins (bphCadherins, bphAPNs, and bphALPs) which showed higher sequence identity with those of the Bt-susceptible insects were selected for construction of phylogenetic trees. It is not surprising that the protein sequences derived from the same species or the same insect orders were mainly clustered into the same lineages with some exceptions (Fig. 1). For example, most of the cadherins from moths, mosquitoes, and beetles were grouped based on orders of the insects, while bphCadherins and apCadherins of hemipteran insects showed phylogenetic distance from the Cry-binding cadherins. However, a cadherin derived from Tribolium castaneum (Coleoptera: Tenebrionidae) (tcCadherin2) and bmCadherin2 was grouped together with bphCadherins and apCadherins. Sequence identities of bmCadherin2 to apCadherins were up to 80.3% (Fig. 1A). But bmCadherin2 and tcCadherin2 were not found to be associated with the insecticidal activity of Cry toxins. Phylogenetic analysis showed that the five deduced bphAPNs were either clustered with two coleopteran APNs or grouped into the same lineage of the APNs derived from aphids. Interestingly, bphAPN1 showed distance from other APNs of hemipteran insects but was closer to the root of APNs from the vast majority of the lepidopteran and coleopteran insects (Fig. 1B). Similar to APNs, the BPH ALPs were clustered with either ALPs of aphids or ALPs of coleopterans (Fig. 1C).
Fig. 1.
Molecular phylogenetic analysis of selected cadherins (A), APNs (B), and ALPs (C) from BPH transcriptome by maximum-likelihood method. Amino acid sequence alignment for each analysis was conducted by the MEGA 6.06 built-in MUSCLE program followed by the screening of conserved blocks by the Gblocks program. The evolutionary history was inferred by using the maximum-likelihood method based on the Le_Gascuel_2008 model (Le and Gascuel 2008). The bootstrap consensus tree inferred from 1,000 replicates (Felsenstein 1985) is taken to represent the evolutionary history of the taxa analyzed (Tamura et al. 2013). Initial trees for the heuristic search were obtained by applying the neighbor-joining method to a matrix of pairwise distances estimated using a JTT model. A discrete gamma distribution was used to model evolutionary rate differences among sites [four categories (+G, parameter = 3.2778)]. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA 6.06. Information about amino acid sequences included in each tree is shown in Additional Table 9.
Domain Characterization of the Selected Cadherin, APN, and ALP in the BPH Midgut
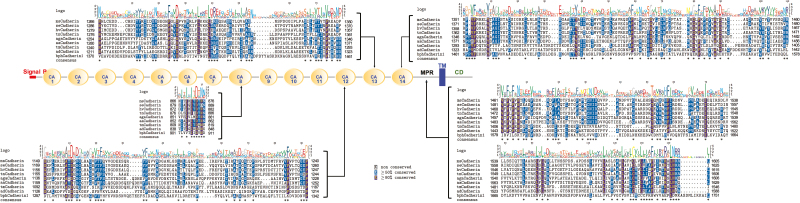
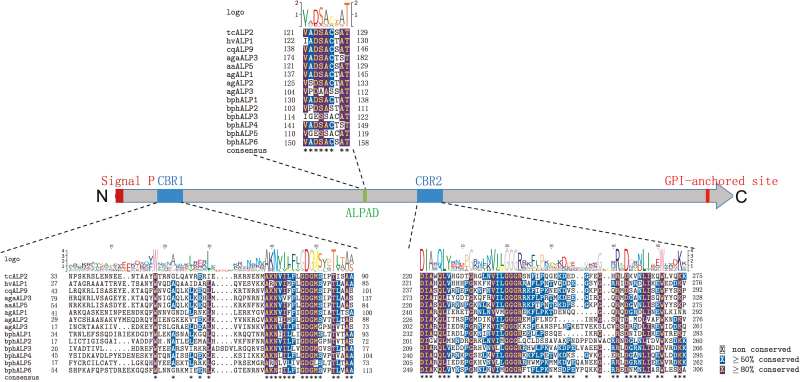
Signal peptide and transmembrane helices are essential sequence components of the transmembrane proteins. In the four selected bphCadherins, signal peptides were predicted from bphCadherin1, 2, and 3 by an online program SignalP (http://www.cbs.dtu.dk/). The Cry-binding cadherins contain various cadherin repeats (CAs), a membrane-proximal region (MPR), a transmembrane domain (TM), and a cytoplasmic domain (CD) (Zhang et al. 2012). Prediction of transmembrane helices through TMHMM suggested that all the selected bphCadherins contain at least one transmembrane helix. In addition, more than 10 potential N-glycosylation sites and various numbers of O-glycosylation sites in each bphCadherin were predicted (Table 1). We also conducted motif scanning of the bphCadherins. The results suggested that only bphCadherin1 contained 14 CAs, an MPR, a TM, and a CD, which is similar to the motifs found in Cry-binding cadherins (Fig. 2). Illustrations of the signal peptides, 14 CAs, and TM of bpCadherin1 as examples of the analysis are shown in Supp Fig. 5.
Table 1.
Characters of potential Cry receptor in BPH
| Predicted proteins | Signal P | GPI-anchored sites | Transmembrane helix† | Number of N-glycosylation site | Number of O-glycosylation site | ||
|---|---|---|---|---|---|---|---|
| PredGPI | fragAnchor | big-PI Predictor | |||||
| bphCadherin1 | ○ | N/A | N/A | N/A | o1687-1709i | 15 | 2 |
| bphCadherin2 | ○ | N/A | N/A | N/A | i7-26o1369-1391i | 14 | 2 |
| bphCadherin3 | ○ | N/A | N/A | N/A | o1376-1398i | 10 | 9 |
| bphCadherin4 | × | N/A | N/A | N/A | o1579-1601i | 17 | 3 |
| bphAPN1 | ○ | × | × | × | o | 10 | 3 |
| bphAPN2 | ○ | × | × | × | o | 6 | 2 |
| bphAPN3 | × | × | × | × | i33-55o | 7 | 5 |
| bphAPN4 | ○ | ○ | ○ | × | o | 18 | 26 |
| bphAPN5 | × | × | × | × | o | 4 | 1 |
| bphALP1 | ○ | × | × | × | o | 3 | 11 |
| bphALP2 | × | ○ | ○ | ○ | i12-34o508-530i | 2 | 8 |
| bphALP3 | ○ | × | × | × | o | 3 | 12 |
| bphALP4 | ○ | ○ | ○ | ○ | o | 3 | 9 |
| bphALP5 | × | × | × | × | i7-29o | 0 | 8 |
| bphALP6 | ○ | ○ | × | × | o | 2 | 11 |
N/A indicates character prediction was not conducted.
† i and o in the row of transmembrane helix indicated the domain inside and outside the cell membrane, respectively. Numbers indicate the starting amino acids of domains.
Fig. 2.
Overview of sequence components in bphCadherin1 and multiple alignments of specific motifs in bphCadherin1 against Cry receptor cadherins. The signal peptide (Signal P), transmembrane domain (TM), membrane proximal region (MPR), cytoplasmic domain, and numbered cadherin repeats (CAs) are illustrated. The sequence of each CA containing previously reported Cry-binding regions is shown as insets. Amino acids equal to or greater than 50% conserved are shaded in blue while those 80% conserved are shaded in red.
Similar to bphCadherins, a signal peptide in the N-terminus of bphAPN1, 2, and 4 was predicted (Supp Fig. 6). However, a putative transmembrane helix was only found in bphAPN3 (Table 1) but not from bphAPN1, 2, and 4. The GPI-anchored sites are common motifs found in Cry-associated APNs. Analysis of GPI-anchored signals of bphAPNs by using three online prediction programs (PredGPI, FragAnchor, and big-PI Predictor) suggested that only near the C-terminus of bphAPN4 is there a high probability of finding a GPI-anchored site (Supp Fig. 6; Table 1). In addition, both N- and O-glycosylation sites were predicted in five deduced bphAPNs with the bphAPN4 containing the most abundant N- and O-glycosylation sites (18 N-linked glycosylation sites and 26 O-linked glycosylation sites) (Table 1). All predicted N- or O-glycosylation sites are illustrated in Supp Fig. 6.
Analysis of the bphALPs detected a signal peptide at the N-terminus from all bphALPs except for bphALP2 and 5 (Table 1). The transmembrane helices were only predicted from bphALP2 (two sites) and bphALP5 (one site) by TMHMM (Table 1). In addition, bphALP2 and bphALP4 were predicted as GPI-anchored proteins, while bphALP6 was a putative GPI-anchored protein (Table 1, Supp Fig. 7). Prediction of N- or O- glycosylation sites showed that all bphALPs contain at least two N-glycosylation sites except for bphALP5, while all six bphALPs contain 8–12 putative O-glycosylation sites (Table 1). The predicted N- or O-glycosylation sites are highlighted in Supp Fig. 7.
Comparison of the Protein Functional Domains
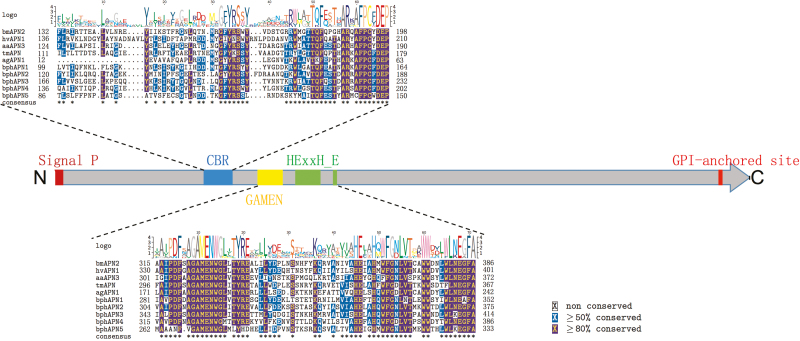
Cry-binding regions (CBRs) of Cry-binding cadherins, APNs, and ALPs have previously been identified (Nakanishi et al. 2002, Budatha et al. 2007, Fernandez et al. 2009, Gómez et al. 2012, Kaur et al. 2014). The above sequence analysis suggests that bphCadherin1 may contain homologous domains of the Cry-binding cadherins derived from susceptible insects. The sequence of bphCadherin1 was then aligned to previously reported Cry-binding cadherins derived from Manduca sexta (Lepidoptera: Sphingidae), Spodoptera exigua (Lepidoptera: Noctuidae), Heliothis virescens (Lepidoptera: Noctuidae), T. ni, Anopheles gambiae, Aedes aegypt (Diptera: Culicidae), Alphitobius diaperinus (Coleoptera: Tenebrionidae), and Tenebrio molitor (Coleoptera: Tenebrionidae) to identify putative homologous domains. The results of sequence alignment suggested that the CA12-14 and MPR of bphCadherin1 were homologous to some CBR domains of the Cry-binding cadherins (Fig. 2). Majority of the highly conserved (≧80% conserved) amino acids among Cry-binding cadherins could be identified from CA12-14 in bphCadherin1, while no obviously conserved site was found by comparing MPR of bphCadherin1 to its counterparts of Cry-binding cadherins (Fig. 2). There are varied numbers of CBRs in the Cry-binding cadherins of different insects. Two small CBRs (865NITIHITDTNN875 and 1331IPLPASILTVTV1342) were found in the Cry-binding cadherin of M. sexta (Zhang et al. 2013). Homologous sequences of these two CBRs were mapped to the CA8 and CA13 regions of bphCadherin1, respectively. However, low identity between both CBRs of Cry-binding cadherins and the corresponding regions in bphCadherin1, although three conserved amino acids (906I, 908D, and 910N) were identified (Fig. 2). There are three amino acid residues, 1425L, 1429F, and 1430Q, in Cry-binding cadherin of H. virescens (Xie et al. 2005) and a 1362R to 1582L region in the cadherin of T. ni (Zhang et al. 2013, Badran et al. 2016), which showed particular importance for toxin binding to Cry1A toxins. These regions were aligned to CA14 and MPR in bphCadherin1. But very low identity was found in these sites of bphCadherin1 compared to that of the Cry-binding cadherins (Fig. 2).
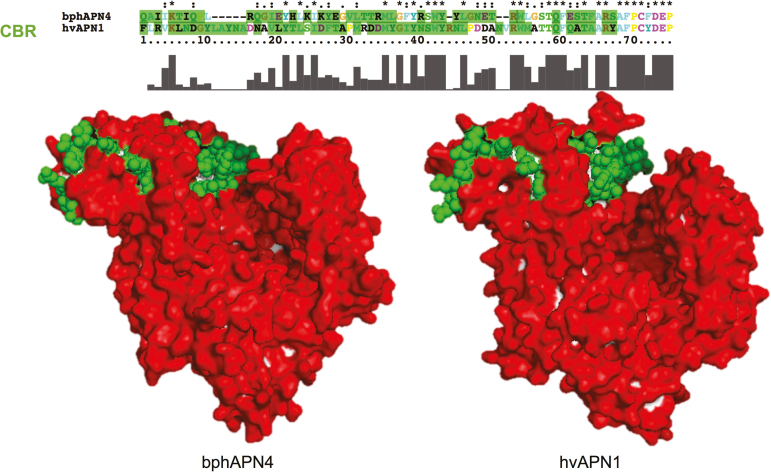
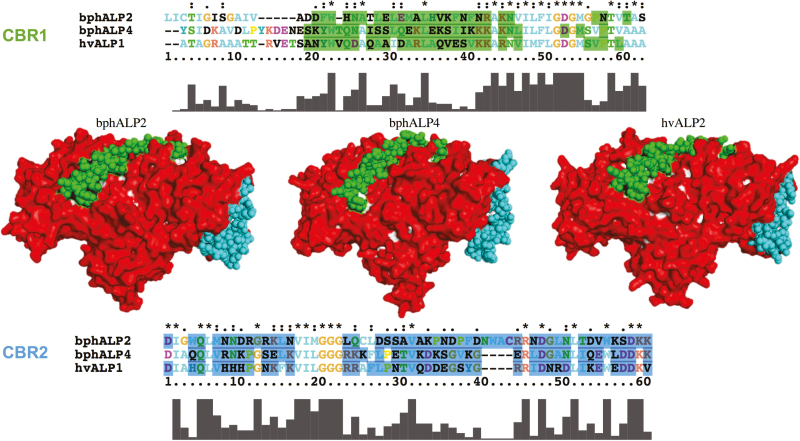
Sequence alignment of bphAPNs and bphALPs with their homologous CBPs was also conducted to identify homologous regions of previously reported CBRs. One Cry1A toxin-binding region was found from bphAPNs. Compared to the N-terminus of CBR in Cry-binding APNs, a specific motif like TQFxxTxARxAFPCxDEP at the C-terminus in bphAPN exhibited a greater similarity to Cry-binding APNs of susceptible insects (Fig. 3). Homologous modeling of the protein 3D structure showed that the structure of bphAPN1 is similar to that of the hvAPN1, which is a known receptor of Cry1Ac toxin in the midgut of H. virescens (Fig. 4). In addition, the CBR homologous region in bphAPN1 is also predicted to be exposed on the surface of the protein (Fig. 4). ALPs are important Bt receptors in the midgut of Cry-susceptible insects (Perera et al. 2009, Wang 2015). Two CBRs (95R-102G and 257N-296I) were reported to play essential roles in mediating interaction of Cry-binding ALPs with Cry toxins (Fernandez et al. 2009). Homologous sequences of the two CBRs were identified in bphALPs (Fig. 5), which showed a similar surface-exposed 3D structure to that of the Cry-binding ALP in H. virescens (Fig. 6). In addition, a highly conserved region was found in the C-terminus of CBR1 and its homologous regions of bphALPs, while the sequences of CBR2 homologs of bphALPs were closer to that of Cry-binding ALPs at the N-terminus (Fig. 5).
Fig. 3.
Schematic presentation of sequence components in bphAPNs and multiple alignments of specific motifs in bphAPNs against Cry receptor APNs. Predicted positions of signal peptide and GPI-anchored sites are boxed in red at N- and C-terminuses, respectively. Cry-binding region (CBR) and the conserved GAMEN and gluzincin sequences involved in Zn2+ binding are boxed in blue, yellow, and green, respectively. Insets show the detailed alignment of each motif between amino acid sequences of bphAPNs and Cry receptor APNs. Amino acids equal to or greater than 50% conserved are shaded in blue while those 80% conserved are shaded in red.
Fig. 4.
Three-dimensional structures of hvAPN1 and bphAPN4 were respectively constructed based on template 4wz9.1.A in the SWISS-MODEL template library. The surface structure of APN proteins was predicted by built-in Python script of the PyMOL Molecular Graphics System. CBR residues in either hvAPN1 or bphAPN4 were displayed as spheres, while the rest of the amino acids were displayed as surface structures. Surface residues in CBR were stained in green. Surface residues not included in CBR were stained in red. The top panel indicates the sequence alignment between CBRs.
Fig. 5.
Schematic presentation of sequence components in bphALPs and multiple alignments of specific motifs in bphALPs against Cry-receptor ALPs. Predicted positions of signal peptide and GPI-anchored sites are boxed in red at N- and C-terminuses, respectively. Cry-binding regions (CBRs) and the conserved alkaline phosphatase active domain (ALPAD) are boxed in blue and green, respectively. Insets show the detailed alignment of each motif between amino acid sequences of bphALPs and Cry-receptor ALPs. Amino acids equal to or greater than 50% conserved are shaded in blue while those 80% conserved are shaded in red.
Fig. 6.
Three-dimensional structures of hvALP1, bphALP2, and bphALP4 were respectively constructed based on template 1zef.1.A in the SWISS-MODEL template library. The surface structure of ALP proteins was predicted by built-in Python script of the PyMOL Molecular Graphics System. CBR residues in bphALP2, bphALP4, and hvALP1 were displayed as spheres, respectively, while the rest of the amino acids were displayed as surface structures. Surface residues in CBR1 and CBR2 were stained in green and cyan, respectively. Surface residues not included in CBR were stained in red. The panels on the top and bottom indicate the sequence alignment of CBR1 and CBR2, respectively.
In addition to the CBRs, specific motifs were also predicted from BPH APNs or ALPs. A gluzincin aminopeptidase motif (GAMEN) and a zinc-binding/gluzincin motif (HEXXHX18E), which are highly conserved in APNs, were detected from the sequence of five deduced bphAPNs (Supp Fig. 6). Both motifs show high sequence conservation compared to the reported APNs of lepidopteran, coleopteran, dipteran insects, and aphids (hemipterans) (Fig. 3). Similarly, as typical characteristics of ALPs, phosphatase-active sites and a phosphatase-active domain (Perera et al. 2009), which showed high sequence similarity to the ALPs of Bt-susceptible insects and the soybean aphid, were predicted in all six bphALPs (Fig. 5 and Supp Fig. 7).
Relative Transcript Abundance of Putative Cry Receptor Homologous Genes in the BPH Midgut
Relative expression of bphCadherins, bphAPNs, and bphALPs in the midgut of BPH were estimated by per kilobase of exon model per million mapped reads (RPKM), which showed that transcript abundance of bphAPN1 and bphAPN4 was significantly greater than that of other APNs in the midgut of BPH. The highest RPKM of ALP in the BPH midgut was bphALP6 and bphALP4. To verify the relative transcript abundance determined by transcriptome sequencing, RT-qPCR was conducted to examine selected BPH APN and ALP expressions, which confirmed the results calculated from the transcript reads (Table 2). Compared to APNs and ALPs, bphCadherins were expressed at a low to moderate level in the midgut (Table 2).
Table 2.
Relative transcript abundance of selected homologs of Cry receptor proteins in the midgut of BPH
| Deduced gene | RPKM | Relative abundance (△CT±SD) |
|---|---|---|
| bphcadherin1 | 5.59 | 6.68 ± 0.86 |
| bphcadherin2 | 20.39 | 7.41 ± 0.94 |
| bphcadherin3 | 3.7 | 5.45 ± 0.75 |
| bphcadherin4 | 0.69 | 2.08 ± 0.89 |
| bphAPN1 | 1182.63 | 10.88 ± 0.23 |
| bphAPN2 | 5.05 | 3.19 ± 0.37 |
| bphAPN3 | 2.00 | 3.18 ± 0.45 |
| bphAPN4 | 1263.03 | 10.25 ± 0.24 |
| bphAPN5 | 10.39 | 5.39 ± 0.50 |
| bphALP1 | 0.29 | 1.08 ± 0.34 |
| bphALP2 | 9.59 | 1.45 ± 0.41 |
| bphALP3 | 0.59 | 0.76 ± 0.52 |
| bphALP4 | 302.37 | 8.67 ± 0.29 |
| bphALP5 | 2.06 | 1.70 ± 0.41 |
| bphALP6 | 2020.66 | 11.67 ± 0.22 |
Reads Per Kilo base per million mapped reads (RPKM) of was calculated to predict transcript abundance of putative Bt receptor genes. The relative abundance (△Ct) of selected APNs, ALPs, and cadherins in the midgut was normalized using the BPH 18s rRNA threshold cycle (Ct) values that were obtained from reactions run on the same plate.
Discussion
Cry-receptor interaction is necessary for Cry toxin insecticidal activities to occur. It was observed that the deletion of putative Cry-receptor binding sites or low expression of specific receptor protein could significantly reduce the susceptibility of susceptible insects to the related Cry toxins (Wang 2015). In addition, reinstallation of the dysfunctional receptor-binding domains of the resistant insect strains could restore the toxin insecticidal activity against the previously resistant insects (Badran et al. 2016). Studies on replacements of Bt toxin (Cry or Cyt) functional domains with gut-binding peptide (GBP) of aphids and BPH revealed that GBP improved the toxin-midgut binding affinity and resulted in a significant increase in toxin activity against the aphids and BPH (Chougule et al. 2013, Shao et al. 2016). These results demonstrated that the insecticidal activity of Cry toxins was firmly related to toxin-receptor binding. Improvement of binding affinity between Bt toxins and hemipteran insect guts could possibly retarget the toxins against hemipteran insects. Hence, identification of Bt-toxin receptor homologous proteins in BPH midguts and comparison of the differences with previously reported Bt-toxin receptors may provide valuable information in understanding the mode of action of Bt toxins in BPH.
In this report, we have investigated the homologous genes of Cry receptors (cadherin, apn, and alp) from the transcriptome of N. lugens nymphs and then analyzed the protein sequence, functional domains, protein structures, and transcript abundance of the related genes. We have identified at least one cadherin, one APN, and two ALP proteins that showed similar characteristics to the previously reported CBPs in lepidopterans, coleopterans, and dipterans.
Cry-binding APNs and ALPs were reported to have a high expression level in the midgut of susceptible insects, while cadherin showed a relatively low abundance level (Chen et al. 2005, 2009). Results of RPKM calculation and RT-qPCR assay showed that bphAPN1, bphAPN4, and bphALP6 were highly abundant in the midgut of BPH, while a low to moderate expression level of all cadherins was detected in the transcription level (Table 2). The transcription abundance of bphCadherin, bphAPN, and bphALP was similar to that of their homologs in the midgut of Bt-susceptible insects (Tiewsiri and Wang 2011, Zhang et al. 2012, Chen et al. 2015). Cry-binding cadherins, APNs, and ALPs were known as membrane-bound proteins (Wang 2015). Prediction of transmembrane helix and GPI-anchored signal showed that bphAPN4, bphALP2, bphALP4, bphALP6, and all bphCadherins are potentially membrane-bound proteins with either GPI-anchor signal or transmembrane helix (Table 1). Hence, it is possible that the bphCadherins, bphAPNs, and bphALPs have features similar to their homologs in Bt-susceptible insects.
The results of sequence alignment indicated that bphAPN4, bphALP2, and bphALP4 shared moderate to high protein identity with their CBP homologs (Supp Tables 7–9), although the protein sequences of bphCadherins, bphAPNs, and bphALPs were highly divergent compared to those of the CBPs in Cry-susceptible insects (Fig. 1). In addition, bphCadherin1, bphAPN4, bphALP2, and bphALP4 contained domains similar to the Cry-binding domains of the CBPs. However, it is not surprising to observe that the majorities of the amino acids in these domains differed from CBPs (Figs. 2, 3, 5). Studies on the Cry-cadherin interaction indicated that CBRs 865NITIHITDTNN875 and 1331IPLPASILTVTV1342 in the Cry-binding cadherin of M. sexta and repeat 12 in the Cry-binding cadherin of H. virescens could interact with the domain II loop 2, loop α8, and loop 3 of Cry toxins, respectively (Bravo et al. 2007). Sequence alignment showed that nearly no sequence similarity between the corresponding regions in bphCadherins and the three CBRs of the lepidopteran Cry-binding cadherins. Cry receptor-related APN and ALP usually contain a GPI site and numbers of N- or O-glycosylation sites. Glycosyl-mediated interaction between Cry toxins and GPI-anchored APN or ALP was reported, suggesting that these glycosyl sites could be important in the mechanism of Cry insecticidal activity (Luo et al. 1999, Shinkawa et al. 1999, Fernandez et al. 2009, Lin et al. 2014). The bphAPN4, bphALP2, and bphALP4 were predicted to contain numbers of N- and O-glycosylation sites and a GPI-anchored site, suggesting that bphAPN4, bphALP2, and bphALP4 are potential to interact with Cry toxins. However, CBRs of bphAPN and bphALP are divergent to the Cry-binding APNs and ALPs. Consequently, high divergence of Cry-binding cadherins, APNs, and ALPs with their homologs in BPH may be part of the reason why BPH is not susceptible to Cry toxins.
Improper binding of Cry toxins in the midgut of BPH and pea aphid has been known to be a possible reason for the low toxicity of these toxins (Li et al. 2011, Shao et al. 2013, Niu et al. 2017). Our results provide not only sequence characteristics but also structural characteristics of the CBP-like proteins of BPH. Information from this study may also enrich our knowledge of the mode of action of Bt toxins in the midgut of hemipteran insects.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Acknowledgments
We thank Connie Allison for critical reading and editing of the manuscript. We also thank Institute of Plant Virology, FAFU, for providing us the wild-type N. lugens strain. This study was supported by Project of Fujian-Taiwan Joint Center for Ecological Control of Crop Pests (grant number Minjiaoke 2013-51); National Natural Science Foundation of China (grant number 31401802 and 31772539); Science and Technology Project in Fujian Province Department of Education (grant number JA15161); Research development fund of Fujian Agricultural and Forestry University (grant number KF2015041) and Science and Technology Major Project of Fujian Province (grant number 2016NZ0001-1).
References Cited
- Altschul S. F., T. L. Madden A. A. Schäffer J. Zhang Z. Zhang W. Miller, and Lipman D. J.. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., L. Bordoli J. Kopp, and Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22: 195–201. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., and Eppig J. T.. 2000. Gene ontology: tool for the unification of biology. Nature Genetics. 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badran A. H., V. M., Guzov Q., Huai M. M., Kemp P., Vishwanath W., Kain A. M., Nance A., Evdokimov F., Moshiri K. H., Turner et al. 2016. Continuous evolution of Bacillus thuringiensis toxins overcomes insect resistance. Nature. 533: 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Y. Y., Y. Wang W. J. Wu D. Zhao J. Xue B. Q. Zhang Z. C. Shen, and Zhang C. X.. 2012. De novo intestine-specific transcriptome of the brown planthopper Nilaparvata lugens revealed potential functions in digestion, detoxification and immune response. Genomics. 99: 256–264. [DOI] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T. G., Bertoni M., and Bordoli L.. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N., T. Sicheritz-Pontén R. Gupta S. Gammeltoft, and Brunak S.. 2004. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 4: 1633–1649. [DOI] [PubMed] [Google Scholar]
- Bravo A., S. S. Gill, and Soberón M.. 2007. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon. 49: 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., S. Likitvivatanavong S. S. Gill, and Soberón M.. 2011. Bacillus thuringiensis: a story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41: 423–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A., I. Gómez H. Porta B. I. García-Gómez C. Rodriguez-Almazan L. Pardo, and Soberón M.. 2013. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microb. Biotechnol. 6: 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budatha M., G. Meur, and Dutta-Gupta A.. 2007. A novel aminopeptidase in the fat body of the moth Achaea janata as a receptor for Bacillus thuringiensis Cry toxins and its comparison with midgut aminopeptidase. Biochem. j. 405: 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chen J., M. R. Brown G. Hua, and Adang M. J.. 2005. Comparison of the localization of Bacillus thuringiensis Cry1A delta-endotoxins and their binding proteins in larval midgut of tobacco hornworm, Manduca sexta. Cell Tissue Res. 321: 123–129. [DOI] [PubMed] [Google Scholar]
- Chen J., K. G. Aimanova L. E. Fernandez A. Bravo M. Soberon, and Gill S. S.. 2009. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem. j. 424: 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., C. Liu Y. Xiao D. Zhang Y. Zhang X. Li B. E. Tabashnik, and Wu K.. 2015. A toxin-binding alkaline phosphatase fragment synergizes Bt toxin Cry1Ac against susceptible and resistant Helicoverpa armigera. PLoS One. 10: e0126288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chougule N. P. and Bonning B. C.. 2012. Toxins for transgenic resistance to hemipteran pests. Toxins (Basel). 4: 405–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chougule N. P., Li H., Liu S., Linz L. B., Narva K. E., Meade T., and Bonning B. C.. 2013. Retargeting of the Bacillus thuringiensis toxin Cyt2Aa against hemipteran insect pests. Proc. Natl. Acad. Sci. USA. 110: 8465–8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 39: 783–791. [DOI] [PubMed] [Google Scholar]
- Fernandez L. E., C. Martinez-Anaya E. Lira J. Chen A. Evans S. Hernández-Martínez H. Lanz-Mendoza A. Bravo S. S. Gill, and Soberón M.. 2009. Cloning and epitope mapping of Cry11Aa-binding sites in the Cry11Aa-receptor alkaline phosphatase from Aedes aegypti. Biochemistry. 48: 8899–8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez I., L. Pardo-López C. Muñoz-Garay L. E. Fernandez C. Pérez J. Sánchez M. Soberón, and Bravo A.. 2007. Role of receptor interaction in the mode of action of insecticidal Cry and Cyt toxins produced by Bacillus thuringiensis. Peptides. 28: 169–173. [DOI] [PubMed] [Google Scholar]
- Gómez J. E., S. A. López-Pazos, and Cerón J.. 2012. Determination of Cry toxin activity and identification of an aminopeptidase N receptor-like gene in Asymmathetes vulcanorum (Coleoptera: Curculionidae). j. Invertebr. Pathol. 111: 94–98. [DOI] [PubMed] [Google Scholar]
- Grabherr M. G., B. J., Haas M., Yassour J. Z., Levin D. A., Thompson I., Amit X., Adiconis L., Fan R., Raychowdhury Q., Zeng et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N., M. C. Peitsch, and Schwede T.. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 30: S162–S173. [DOI] [PubMed] [Google Scholar]
- Hibino H. 1996. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 34: 249–274. [DOI] [PubMed] [Google Scholar]
- Jia D., N. Guo H. Chen F. Akita L. Xie T. Omura, and Wei T.. 2012. Assembly of the viroplasm by viral non-structural protein Pns10 is essential for persistent infection of rice ragged stunt virus in its insect vector. J. Gen. Virol. 93: 2299–2309. [DOI] [PubMed] [Google Scholar]
- Julenius K., A. Mølgaard R. Gupta, and Brunak S.. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology. 15: 153–164. [DOI] [PubMed] [Google Scholar]
- Kaur R., Sharma A., Gupta D., Kalita M., and Bhatnagar R. K.. 2014. Bacillus thuringiensis toxin, Cry1C interacts with 128HLHFHLP134 region of aminopeptidase N of agricultural pest, Spodoptera litura. Process Biochem. 49: 688–696. [Google Scholar]
- Kiefer F., K. Arnold M. Künzli L. Bordoli, and Schwede T.. 2009. The SWISS-MODEL repository and associated resources. Nucleic Acids Res. 37: D387–D392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. doi: 10.1002/0471250953.bi1107s32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S. Q. and Gascuel O.. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25: 1307–1320. [DOI] [PubMed] [Google Scholar]
- Li B., and Dewey C. N.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 12: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., N. P. Chougule, and Bonning B. C.. 2011. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris). J. Invertebr. Pathol. 107: 69–78. [DOI] [PubMed] [Google Scholar]
- Lin P., T. Cheng S. Jin L. Jiang C. Wang, and Xia Q.. 2014. Structural, evolutionary and functional analysis of APN genes in the Lepidoptera Bombyx mori. Gene. 535: 303–311. [DOI] [PubMed] [Google Scholar]
- Liu S., N. P. Chougule D. Vijayendran, and Bonning B. C.. 2012. Deep sequencing of the transcriptomes of soybean aphid and associated endosymbionts. Plos One. 7: e45161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Zhu P., Gurr G. M., Zheng X., Chen G., and Heong K. L.. 2015. Rice pest management by ecological engineering: a pioneering attempt in China, pp. 161–178, Rice Planthoppers. Springer. [Google Scholar]
- Luo K., J. R. McLachlin M. R. Brown, and Adang M. J.. 1999. Expression of a glycosylphosphatidylinositol-linked Manduca sexta aminopeptidase N in insect cells. Protein Expr. Purif. 17: 113–122. [DOI] [PubMed] [Google Scholar]
- Min X. J., G. Butler R. Storms, and Tsang A.. 2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33: W677–W680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K., K. Yaoi Y. Nagino H. Hara M. Kitami S. Atsumi N. Miura, and Sato R.. 2002. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella – their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 519: 215–220. [DOI] [PubMed] [Google Scholar]
- Niu L., Mannakkara A., Qiu L., Wang X., Hua H., Lei C., Jurat-Fuentes J. L., and Ma W.. 2017. Transgenic Bt rice lines producing Cry1Ac, Cry2Aa or Cry1Ca have no detrimental effects on Brown Planthopper and Pond Wolf Spider. Sci. Rep. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-López L., M. Soberón, and Bravo A.. 2013. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol. Rev. 37: 3–22. [DOI] [PubMed] [Google Scholar]
- Peng X., W. Zha R. He T. Lu L. Zhu B. Han, and He G.. 2011. Pyrosequencing the midgut transcriptome of the brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 20: 745–762. [DOI] [PubMed] [Google Scholar]
- Perera O. P., J. D. Willis M. J. Adang, and Jurat-Fuentes J. L.. 2009. Cloning and characterization of the Cry1Ac-binding alkaline phosphatase (HvALP) from Heliothis virescens. Insect Biochem. Mol. Biol. 39: 294–302. [DOI] [PubMed] [Google Scholar]
- Petersen T. N., S. Brunak G. von Heijne, and Nielsen H.. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 8: 785–786. [DOI] [PubMed] [Google Scholar]
- Shao E., S. Liu L. Lin, and Guan X.. 2013. Proteolytic processing of Bacillus thuringiensis toxin Cry1Ab in rice brown planthopper, Nilaparvata lugens (Stål). J. Invertebr. Pathol. 114: 255–257. [DOI] [PubMed] [Google Scholar]
- Shao E., L. Lin C. Chen H. Chen H. Zhuang S. Wu L. Sha X. Guan, and Huang Z.. 2016. Loop replacements with gut-binding peptides in Cry1Ab domain II enhanced toxicity against the brown planthopper, Nilaparvata lugens (Stål). Sci. Rep. 6: 20106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkawa A., K. Yaoi T. Kadotani M. Imamura N. Koizumi H. Iwahana, and Sato R.. 1999. Binding of phylogenetically distant Bacillus thuringiensis cry toxins to a Bombyx mori aminopeptidase N suggests importance of Cry toxin’s conserved structure in receptor binding. Curr. Microbiol. 39: 14–20. [DOI] [PubMed] [Google Scholar]
- Shu C., S. Tan J. Yin M. Soberón A. Bravo C. Liu L. Geng F. Song K. Li, and Zhang J.. 2015. Assembling of Holotrichia parallela (dark black chafer) midgut tissue transcriptome and identification of midgut proteins that bind to Cry8Ea toxin from Bacillus thuringiensis. Appl. Microbiol. Biotechnol. 99: 7209–7218. [DOI] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., and Zdobnov E. M.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. doi: 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Talavera G. and Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiewsiri K., and Wang P.. 2011. Differential alteration of two aminopeptidases N associated with resistance to Bacillus thuringiensis toxin Cry1Ac in cabbage looper. Proc. Natl. Acad. Sci. 108: 14037–14042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenborre G., G. Smagghe, and Van Damme E. J.. 2011. Plant lectins as defense proteins against phytophagous insects. Phytochemistry. 72: 1538–1550. [DOI] [PubMed] [Google Scholar]
- Wang P. 2015. Mechanism of Cry1Ac resistance in cabbage loopers–a resistance mechanism selected in insect populations in an agricultural environment. Bt Resistance: Characterization and Strategies for GM Crops Producing Bacillus thuringiensis Toxins 4: 87. [Google Scholar]
- Xie F., Xiao P., Chen D., Xu L., and Zhang B.. 2012. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80: 75–84. [DOI] [PubMed] [Google Scholar]
- Xie R., M. Zhuang L. S. Ross I. Gomez D. I. Oltean A. Bravo M. Soberon, and Gill S. S.. 2005. Single amino acid mutations in the cadherin receptor from Heliothis virescens affect its toxin binding ability to Cry1A toxins. J. Biol. Chem. 280: 8416–8425. [DOI] [PubMed] [Google Scholar]
- Zhang X., W. Kain, and Wang P.. 2013. Sequence variation and differential splicing of the midgut cadherin gene in Trichoplusia ni. Insect Biochem. Mol. Biol. 43: 712–723. [DOI] [PubMed] [Google Scholar]
- Zhang X., K. Tiewsiri W. Kain L. Huang, and Wang P.. 2012. Resistance of Trichoplusia ni to Bacillus thuringiensis toxin Cry1Ac is independent of alteration of the cadherin-like receptor for Cry toxins. Plos One. 7: e35991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.