Abstract
Clonal hematopoiesis (CH) broadly describes the expansion of a clonal population of blood cells with one or more somatic mutations. Individuals with CH are at greater risk for hematological malignancies, cardiovascular disease, and increased mortality from non-hematological cancers. Understanding the causes of CH and how these mutant cells interact with cells of other tissues will provide critical insights into preleukemic development, stem cell biology, host-immune interactions, and cancer evolution. Here we discuss the clinical manifestations of CH, mechanisms contributing to its development, the role of CH in clonal evolution toward leukemia, and the contribution of CH to non-hematological disease states.
Introduction
From the onset of an organism’s development, each cell possesses the same set of genes and template genetic sequence. How cells turn on and off these genes allows for the development of distinct tissues and functions within the organism. However, as organisms age, there is increasing evidence that cells acquire somatic mutations or encounter environmental mutagens that induce mutations (Alexandrov et al., 2013; López-Otín et al., 2013). In many cases, these mutations will have a neutral effect, landing in non-coding or non-regulatory regions of the genome. Sometimes these mutations may land in essential genes and lead to negative selection against the cells and their lineage progeny. In converse, sometimes mutations are positively selected for, allowing cells to grow at a faster rate than their neighbors and leading to clonal expansion. This process of mutation acquisition and clonal selection is a consequence of time and environmental exposure. At its extreme end, there is cancer and malignant development whereby a proliferative cell acquires a series of mutations that can lead to unrestrained growth. However, antecedent to malignant transformation, there is thought to be an expansion of cells that may pose little risk to the organism as whole, but may serve as a reservoir for future disease development. In some tissues, these premalignant cells will undergo senescence or be removed by a surveying immune system (Collado et al., 2005; Schreiber et al., 2011). How these small collections of cells develop into larger disease and what effects they have on the function of the tissue in which they inhabit are important clinical and biological questions with impacts on aging, inflammation, and malignancy.
The hematopoietic system provides an interesting lens into this premalignant process. As a whole, the hematopoietic system is responsible for producing trillions of cells a day, of which there is a multitude of cell types including erythrocytes, megakaryocytes, platelets, lymphocytes, monocytes, and granulocytes (Doulatov et al., 2012). To achieve this output, hematopoiesis employs an extensive cellular hierarchy sequestering self-renewal to a select few hematopoietic stem cells (HSCs) and offering proliferative capacity to amplifying progenitors (Orkin and Zon, 2008). Thus, the progenitors and mature cells produced from a single stem cell represent a clonal lineage. When an HSC divides, it can either do so symmetrically, producing two daughter HSCs, or asymmetrically, producing both an HSC and a daughter cell primed for differentiation and subsequent production of mature cells (Signer and Morrison, 2013). In addition, symmetric division of an HSC into two progenitors will effectively end the clonal lineage, as the progenitor cells possess limited self-renewal potential. This hierarchy allows for rapid production of cells upon infection or blood loss while safeguarding the hematopoietic system from mutation acquisition, as most cells will terminally differentiate and die.
However, with time, even slowly dividing HSCs may acquire mutations that are passed on to the next generation of cells, affecting both the HSC compartment and daughter cells. While many somatic events will have a neutral effect (Welch et al., 2012), mutations that promote increased self-renewal of the stem cell state, increased proliferation, and/or reduced cell death have the capacity to expand the HSC clone at a disproportionate rate compared to other clones. This fitness advantage relative to other clonal lineages is the defining feature of CH. Given the technological advances for detection, somatic mutations have perhaps become the most well-studied conduit for clonal expansion; however, other mechanisms include immune-mediated clonal selection and stochastic drift in clones. The spectrum of physiological consequences of CH is vast, with many instances of CH posing little to no overt effect on the individual. Whereas on the other extreme, CH cases may represent an antecedent permissive state for leukemic development, where secondary and/or tertiary mutations in HSCs or more differentiated progenitors may result in a fulminant leukemia (Bonnet and Dick, 1997; Corces-Zimmerman et al., 2014; Reya et al., 2001; Shlush et al., 2014; Welch et al., 2012). As such, understanding this spectrum of outcomes will be critical to distinguishing the technical identification of CH from a medically relevant event. The process of clonal expansion progressing toward hematological abnormality or leukemic transformation provides a critical window into identifying at-risk individuals and offering therapeutic options that might prevent the onset of leukemic development. While the clinical implementation of such a vision may be forward thinking, the easily accessible blood samples provide a window that is simply unavailable for more inaccessible solid tumors.
Furthermore, hematopoietic cells travel to different tissues of the body, affecting homeostasis among a number of different organ systems. Thus, clonal expansion of the bone marrow not only poses a risk of malignancy, but also presents an intersection point with many organ systems, whereby biological alterations within an HSC may lead to aberrant activation of pathways in terminally differentiated cells, such as macrophages, derived from that mutant HSC. Reports have demonstrated that these altered immune cells profoundly impact cardiovascular disease (Jaiswal et al., 2017), and we speculate that there might be broad impacts on diabetes, cancer, and other inflammatory syndromes.
Here we focus on reviewing the clinical identification of CH, the selective pressures associated with CH, how the mutations underlying CH inform our understanding of stem cell biology, and how these mutations impact leukemic development and non-hematological disease.
Identification of CH and Distinction from Other Clonal Hematologic Disorders
Herein, our working definition of CH is the expansion of one lineage of cells, or a clone, at a rate disproportionately greater than other clones. The first characterization of CH through modern molecular biologic techniques utilized PCR-based methods to identify non-random X inactivation in a cohort of women (Busque et al., 1996). This study found that women >60 years of age were more likely to possess skewed allelic ratios of X inactivation, while younger women showed no significant alterations. Since then, next-generation sequencing technology has allowed the detection of somatic variants with greater sensitivity for rare events. Whole-exome sequencing (WES) has offered a technical lower limit of detection for clones with a Variant Allele Frequency (VAF) at 0.07% (Jaiswal et al., 2014). Targeted sequencing to greater depth increases the sensitivity to 0.01% (Coombs et al., 2017), while newer methods utilizing targeted error correction sequencing have increased the sensitivity even further to 0.0003% (Young et al., 2016). Meanwhile, lower coverage whole-genome sequencing (WGS) studies have greatly expanded the breadth of possible variants at a cost of less sensitivity (Zink et al., 2017).
Focused sequencing assays on genes known to be implicated in hematologic malignancy—including TET2 (Busque et al., 2012) and DNMT3A (Shlush et al., 2014)—have been used to identify somatic variants in the blood of healthy adults. These studies were followed by larger unbiased sequencing studies including an analysis of 2,700 blood samples used as germline controls across 11 cancer types in the Cancer Genome Atlas (TCGA) (Xie et al., 2014). This study found age-associated recurrent mutations in DNMT3A, TET2, JAK2, ASXL1, SF3B1, PPM1D, and TP53, all of which are known to be mutated in acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), or myeloproliferative neoplasms (MPNs) (Lindsley et al., 2017; Rampal et al., 2014). Even further, the mutational spectrum within a given gene possess distinct distributions. For example, where the DNMT3A-R882H hotspot mutation is enriched in AML, it is less abundant in healthy individuals with CH (Buscarlet et al., 2017). Interestingly, other AML-related mutations in genes such as FLT3, NPM1, IDH1, and IDH2 were rarely found in the blood of TCGA patients (Xie et al., 2014). These data demonstrate that not all hematological-related mutations are equally present in the blood of cancer patients, suggesting a context-dependent selective advantage for mutations. While the TCGA cohort is unlikely to represent the general population, a similar spectrum of mutant genes were identified in a second cancer-enriched cohort (Coombs et al., 2017), as well as large cohorts of patients with cardiovascular disease (Jaiswal et al., 2017) and type 2 diabetes (Jaiswal et al., 2014), along with healthy adults (Genovese et al., 2014; McKerrell et al., 2015; Young et al., 2016). The technological differences between WGS, WES, and targeted sequencing have generated sometimes-conflicting data on the relative enrichment of hotspot mutations; however, as a whole the complimentary nature of these studies has revealed much about the mutation spectrum in CH.
Perhaps unsurprisingly, the studies above identified an increased rate of development of hematological malignancy when somatic mutations were found in the blood (Coombs et al., 2017; Genovese et al., 2014; Jaiswal et al., 2014). This indicates a preleukemic state of these cells, but also blurs the lines between what appears to be clinically benign clonal expansion and a more meaningful abnormality.
A series of terms have been used to describe these states, including CH of indeterminate potential (CHIP), age-related CH (ARCH), idiopathic cytopenias of undetermined significance (ICUS), and clonal cytopenias of undetermined significance (CCUS), which share many overlapping and similar clinical descriptions (Bejar, 2017). Generally ARCH and CHIP are technical findings, not diseases, described as a somatic variant present without a cytopenia, while ICUS is considered to present with a low-level cytopenia without a detectable somatic variant (Steensma et al., 2015). CCUS has been offered as verbiage for a patient presenting with both a cytopenia and a somatic variant, from a normal bone marrow exam (Kwok et al., 2015; Malcovati et al., 2017) (Figure 1). Here, WHO guidelines indicate thresholds for cytopenia as hemoglobin <10 g/dl, absolute neutrophil count <1.8 × 109/l, or platelet count <100 × 109/l (Arber et al., 2016). In addition to these parameters, distinguishing these states requires a VAF cutoff to demarcate whether an individual possesses a bona fide somatic mutation that is relevant to hematologic malignancy. Steensma et al. have suggested that a VAF of 2% serve as a cutoff, a value that will likely be revised as sequencing technology develops and more data become available (Steensma et al., 2015). Importantly, identification of a particular VAF cutoff is of importance if it affects clinical decision-making, but until that time it remains of technical concern.
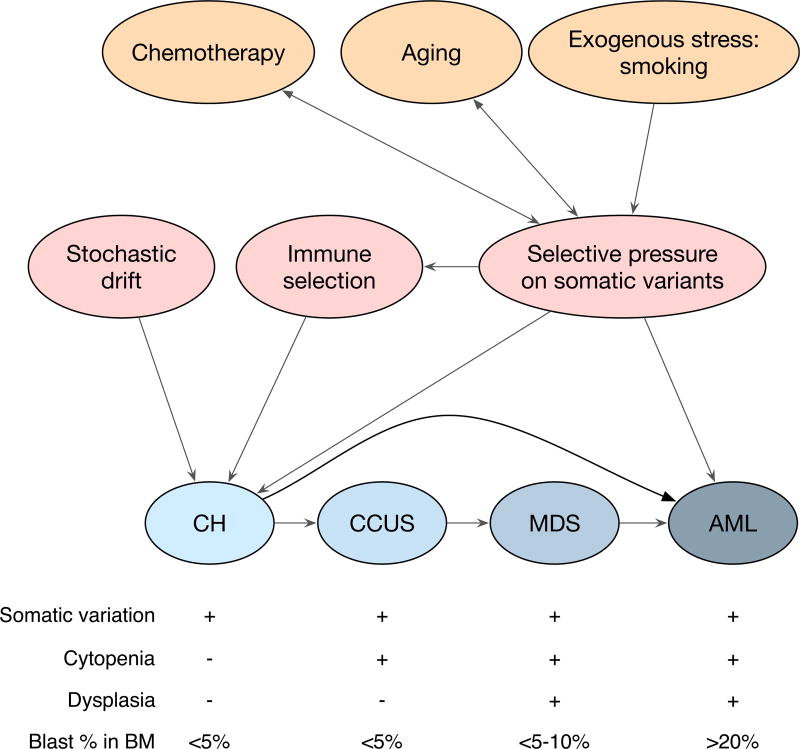
Figure 1. Clonal Hematopoiesis in Relation to Selective Pressures and Secondary Hematological Disease.
Highlighted in orange are potential sources of mutagenic stress that can generate somatic variation in CH, and these include aging-associated mutagenesis, chemotherapy-induced somatic variation, and exposure to exogenous genotoxic stresses, including smoking. Chemotherapy and aging also serve as selective pressures (indicated by double-headed arrows) whereby somatic variants may be selected for if they have better fitness. These selective pressures can lead to clonal outgrowth (as depicted in Figure 2) and the development of CH. Somatic variants can also lead directly to de novo AML or MDS formation. CH can be followed by the development of cytopenias as evident in CCUS, with additional dysplasia resulting in MDS. AML development as a result of CH can arise from an MDS intermediate, but can also likely bypass this progression. CH, clonal hematopoiesis; CCUS, clonal cytopenias of undetermined significance; MDS, myelodysplatic syndrome; AML, acute myeloid leukemia.
There has been some effort to distinguish between these relatively benign hematological states from MDS, which can present with clinically significant cytopenias and dysplasias (Arber et al., 2016). The distinction may be difficult except when MDS present with excess blasts (>5%), recurrent chromosomal abnormalities, and/or multilineage dysplasia (abnormal cellular morphology) on a bone marrow exam (Arber et al., 2016). Many of the genes associated with MDS development, and the more indolent cytopenias (CCUS), are also present in CH clones, including mutations in ASXL1 and TET2 (Cargo et al., 2015; Malcovati et al., 2017). These data suggest that both CCUS and MDS may be natural, albeit not obligatory, progressions from an antecedent CH clone. Thus, secondary AML, resultant from an MDS or MPN diagnosis, may be the emergent malignant event produced by progression of a CH clone to CCUS, MDS, and finally AML (Figure 1). This path, however, is not the obligatory outcome of all CH events, nor is it the only path toward AML formation. Herein lies the hurdle in translating the detection of somatic mutations to implementing clinical action. When is a mutation a clone, and when is that clone likely to exert a pathophysiological effect? This is likely to depend on both the mutation present in the emergent clone and the circumstances that likely generated the clone. Understanding the selective pressures that drive a mutated cell to become a clonal event will provide greater insights into the natural history of any ensuing hematological state.
Selective Pressures Associated with CH
Here we will review the selective pressures and features of the fitness landscape that contribute to CH. This includes aging-associated CH and therapy-induced selective pressures that drive the emergence of a select number of clones (Figure 2). In addition, autoimmunity and microenvironmental alterations are likely to contribute to clonal selections and are reviewed elsewhere (Medyouf, 2017). While these features are discussed discretely below, there is likely an intricate interplay between these factors at any given point in time.
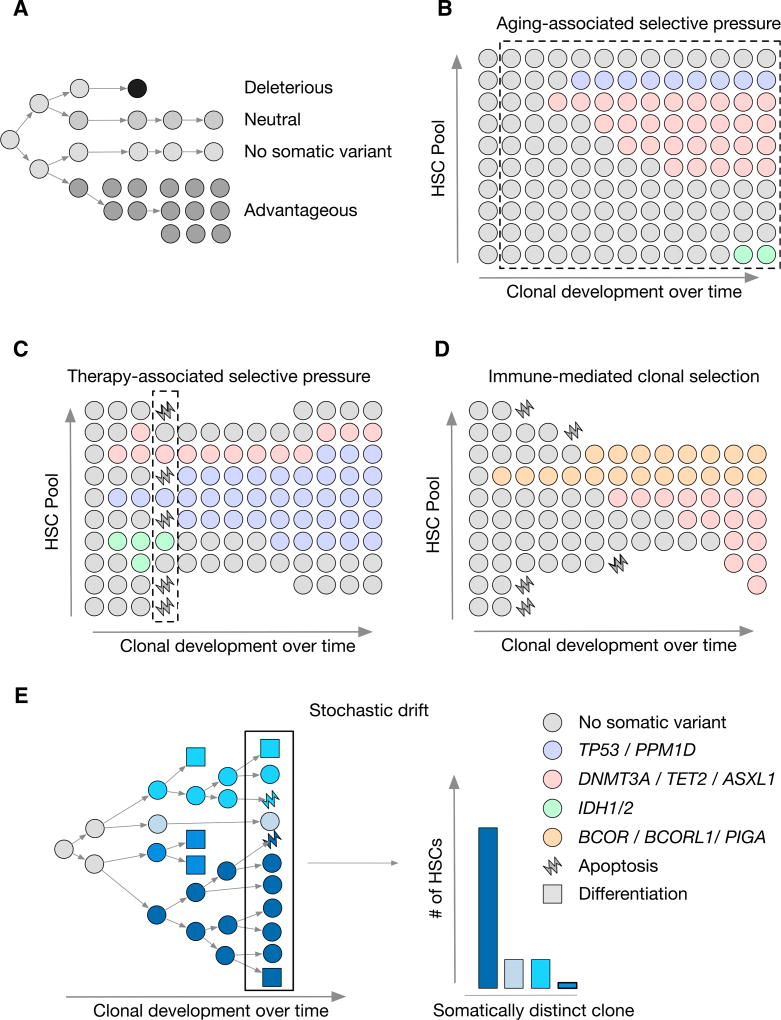
Figure 2. Clonal Expansion under Selective Pressure.
(A) Schematic depicting clonal evolution under negative selection with a deleterious event, no somatic variation, neutral selection, and positive selection with an advantageous event.
(B–D) Depiction of selective pressures: (B), aging; (C), chemotherapy; and (D), immune-mediated pressure. Clonal outgrowth is represented with time on the×axis and relative abundance of HSC clones on the y axis. The dashed line indicates the duration of time that the selective pressure is present. The color of the dot indicates the somatic variants as indicated.
(E) Stochastic drift is shown with the outgrowth of a single HSC clone yielding somatically distinct neutral daughter cells indicated by differing shades of blue. The bar plot on the right indicates abundance of HSCs following stochastic drift for each clone following three generations of HSC division.
Aging-Associated Mutation Acquisition and Selective Pressure
The most clear clinical correlate with CH development is aging, a finding that has been recapitulated in nearly every study to date (Buscarlet et al., 2017; Busque et al., 2012; Genovese et al., 2014; Gillis et al., 2017; Jaiswal et al., 2014, 2017; Takahashi et al., 2017; Xie et al., 2014). Fitting with the propensity for progression of CH to a more malignant hematological state, aging is also associated with a number of hematological complications including anemia, defects in innate and adaptive immunity, and increased prevalence in MDS, MPN, and AML (Adams et al., 2015; Geiger et al., 2013). To understand the mechanisms by which aging contributes to CH, we must consider both the cumulative effect of time on clonal distribution and the molecular and cellular properties of aged HSCs.
On a per cell basis, previous work has demonstrated that aged HSCs possess decreased engraftment capacity, despite an overall increase in the number of immuno-phenotypically defined HSCs (Kuranda et al., 2011; Pang et al., 2011). This alteration in functionality likely creates a fitness landscape such that the HSCs that best retain their self-renewal and differentiation capacity are most likely to expand. The molecular mechanisms behind this functional decline may involve age-associated increases in DNA damage (Flach et al., 2014) and oxidative stress associated DNA damage (Yahata et al., 2011). Flach and colleagues found that aged HSCs possessed increased replication-associated DNA damage response, a finding that did not extend to general cytotoxic agents. This endogenous genotoxic stress has the capacity to both trigger apoptosis and end a clonal lineage, but also potentially generate new mutations that might alter the fitness of the clone (Tubbs and Nussenzweig, 2017). Moreover, DNA damage in HSCs has also been shown to trigger differentiation of cells, providing another avenue for aging-associated genotoxic stress to affect the HSC pool (Wingert et al., 2016). Interestingly, C:T DNA base pair transversions are the most common outcome from replicative mutagenesis, a transversion that is a hallmark of aging-associated mutagenesis (Alexandrov et al., 2013).
These molecular events may underlie a fitness landscape that favors HSCs capable of retaining self-renewal, blocking differentiation, and resolving DNA damage without initiating apoptosis. Mutations in the epigenetic modifiers DNMT3A, TET2, or ASXL1 would be well-suited to offer a selective advantage over non-mutated clones through maximizing self-renewal and enacting a differentiation blockade (discussed below; Figure 2). Meanwhile, mutations in DNA-damage regulators, such as TP53 and PPM1D, likely offer a similar fitness advantage in the face of genotoxic stress.
Therapy-Induced CH and Selective Pressure
In addition to aging-associated mutational processes and endogenous genotoxic stress, exogenous stress such as that elicited by radiation or chemotherapy also likely contributes to the altered fitness landscape that promotes CH. Two studies in particular were enriched for patients who had previously undergone chemotherapeutic treatment either for non-hematological conditions or as a part of a conditioning regiment for autologous stem cell transplant therapy (Coombs et al., 2017; Gibson et al., 2017). In addition to recurrent epigenetic modifiers (DNMT3A, TET2, and ASXL1), these studies identified recurrent mutations in TP53, PPM1D, ATM, and CHEK2, all of which play some role in the response to DNA damage (Kastan and Bartek, 2004; Kastenhuber and Lowe, 2017; Kleiblova et al., 2013). These mutations suggest that expansion of a DNA-damage-resistant clone may occur following genotoxic stress such as chemotherapy or irradiation. In support of this, Wong and colleagues analyzed TP53 mutation status in therapy-induced AML (t-AML); they that found TP53 mutations were present at a higher frequency in t-AML patients than in de novo AML patients (Wong et al., 2015). Critically, the total mutation number did not significantly differ between t-AML and primary AML patients, indicating a lack of mutagenic activity upon chemotherapy. Instead, in several patients, TP53 mutations were found to be present in the blood before the onset of disease, and before prior chemotherapy administration in two patients. This is the most clear evidence that TP53 mutations may provide a selective advantage upon genotoxic insult leading to clonal expansion, an expansion taken to its extreme in t-AML.
Recent genomic studies on large cohorts of patients found that TP53 and PPM1D mutations are both enriched in therapy-induced MDS compared to primary MDS (Lindsley et al., 2017), offering supporting evidence that clones harboring these mutations thrive under therapy-induced selective pressure. Likewise, mutations in PPM1D have been found to be present in the blood of patients with ovarian (Akbari et al., 2014; Ruark et al., 2013; Swisher et al., 2016), prostate (Cardoso et al., 2016), and lung (Zajkowicz et al., 2015) cancer. Genetic studies in mice found that Tp53 mutant bone marrow did not possess a clonal advantage upon transplantation, but it did exhibit clonal outgrowth following irradiation (Marusyk et al., 2010), reinforcing the context-dependent selective advantage offered by TP53 mutations. Collectively, these studies suggest that mutations in DNA damage signaling pathways are not a result of chemotherapy or prior treatment per se. Rather, the therapy-induced genotoxic stress applies increased selective pressure such that these previously neutral mutations now provide a positive selective advantage relative to the cells with intact DNA damage response.
This is in stark contrast to mutations in genes such as TET2, which provide a direct selective advantage to stem cells through increased self-renewal even in the absence of therapy-induced genotoxic insult (Moran-Crusio et al., 2011). Moreover, recent studies demonstrate that point mutations in DNMT3A-R882H increase stem cell frequency, similar to TET2 (Dai et al., 2017; Guryanova et al., 2016). However, unlike TET2 mutations, DNMT3A-R882H mutations, but not DNMT3A loss of function, led to anthracycline resistance (Guryanova et al., 2016). Thus, DNMT3A mutations may possess both a positive selective advantage in the absence of therapy and an additional selective advantage in the presence of anthracycline treatment depending upon whether or not the mutation occurs at the R882H hotspot. Interestingly, this chemoprotective property of DNMT3A mutant cells did not extend to all treatments, as DNMT3A status did not affect responsiveness to etoposide (Guryanova et al., 2016). Future studies profiling how frequent CH mutations affect responsiveness to common chemotherapeutic regimens and other therapeutic perturbations will be of critical interest in understanding how exogenous selective pressures may skew the bone marrow compartment with treatment.
Stochastic Processes Contributing to CH
Both aging-associated and therapy-induced stresses are mechanisms that may support CH either through the acquisition of mutations or through bestowing the ability to survive external genotoxic insult. In addition to these sources, CH has frequently been associated with smoking, another potential source of mutagenic stress contributing to clonal evolution (Buscarlet et al., 2017; Coombs et al., 2017; Jaiswal et al., 2014; Zink et al., 2017). However, it is also possible that CH can manifest through stochastic processes alone in the absence of these stresses. This idea was first proposed and tested using a series of stochastic mathematical models and autologous transplant data in large animal models (Abkowitz et al., 1996). In human samples, the most compelling evidence for stochastic processes in CH was based on WGS data drawn from healthy individuals (Zink et al., 2017). This approach demonstrated that the population-wide prevalence of CH is dramatically underestimated when mutations in candidate genes alone are used to classify individuals. Critically, Zink and colleagues found clonal non-coding somatic variants in a significant fraction of the cohort that were not in known candidate drivers of hematologic malignancy (DNMT3A, ASXL1, TET2, etc.) (Zink et al., 2017). Interestingly, the presence of CH remained a significant prognostic indicator of secondary hematological malignancy, even when no candidate drivers were mutated. One trivial explanation for this finding is that the catalog of known driver mutations is incomplete, and some of these seemingly silent mutations in fact provide a functional selective advantage. However, given the breadth of mutations found, it remains more likely that stochastic processes controlling division, apoptosis, and differentiation are at play (Figure 2). With this as a possibility, CH may be an inevitable emergent property of an aging system, a possibility supported by both Zink et al. and McKerrel et al. While mathematical models support stochastic CH as a possibility, the burden of proof is high. New methodologies for measuring CH will be necessary, as definitively proving the neutral fitness advantage of somatic variants will be an arduous, uncertain task given the unknown fitness landscape leading to a snapshot of clonal emergence.
Epigenetic Mutations Associated with CH and Their Role in Stem Cell Biology
To properly understand how these selective pressures result in clonal expansion of HSCs, we must better understand the biology of the genes that are mutated in these clones. The most common somatic mutations present in CH are present in genes associated with epigenetic modifications, including TET2, DNMT3A, and ASXL1, and splicing factors, including SF3B1, SRSF2, and U2AF1 (reviewed extensively elsewhere; cf. Inoue et al., 2016a). Here we will focus on mutations known to affect DNA chemical modifications and histone posttranslational modifications.
TET2 and DNMT3A Coordinate DNA Methylation in Stem Cells
DNA methylation plays a critical role in gene regulation and shaping cell identity, with methylation, particularly at promoters, generally considered a repressive mark (Smith and Meissner, 2013). Central to this regulatory process are the enzymes responsible for establishing methylation marks, DNA methyltransferases (DNMTs) and the ten-eleven translocation (TET) family, which are capable of removing these marks (Smith and Meissner, 2013) (Figure 3). DNMT1 is generally considered to have a role in maintaining established methylation marks, while DNMT3A/B are considered responsible for establishing de novo methylation marks (Smith and Meissner, 2013). The three TET family members TET1/2/3 are methylcytosine dioxygenases primarily responsible for catalyzing the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) and subsequent hydroxylation events (Ito et al., 2010, 2011; Ko et al., 2010; Tahiliani et al., 2009). Thus loss of TET2 is associated with DNA hypermethylation, while loss-of-function mutations in the DNA methyltransferase 3A (DNMT3A) is associated with hypomethylation, associations that persist into malignancy (Figure 3) (Figueroa et al., 2010; Zhang et al., 2016). While other TET and DNMT family members are capable of catalyzing these reactions, TET2 and DNMT3A remain the primary family members that are mutated in hematological malignancies (Papaemmanuil et al., 2016) and the most commonly mutated genes in CH (Buscarlet et al., 2017).
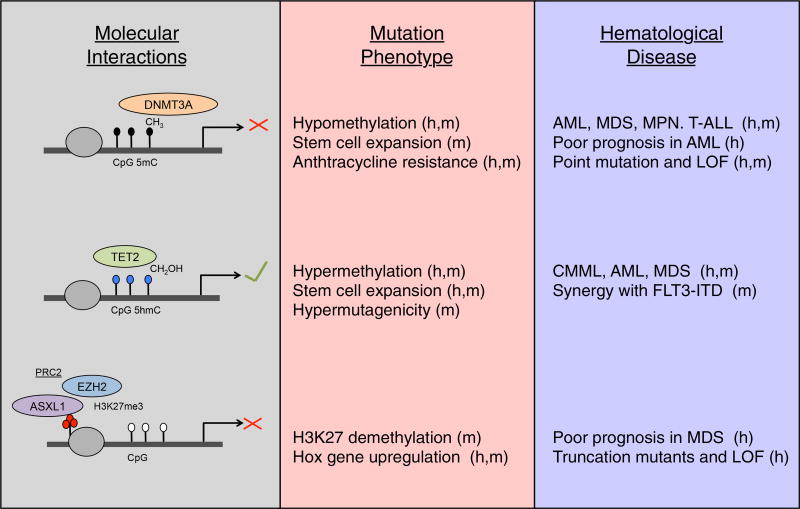
Figure 3. Epigenetic Regulators of Clonal Hematopoiesis.
Schematic of DNMT3A catalyzing CpG methylation (black circles), TET2 generating 5hmC (blue circles), and ASXL1 generating H3K27 trimethylation (red circles) by recruiting the PRC2 complex member EZH2. Associations with loss of function and hematological disease are listed to the right. Findings supported by mouse and human data are denoted with an “m” and “h,” respectively.
The consequences of DNMT3A loss of function have been informed through mechanistic studies in the mouse. Complete knockout of Dnmt3a and Dnmt3b in mice, and thus elimination of de novo DNA methylation, led to a reduction in HSC self-renewal, but without any skewing in lineage commitment (Tadokoro et al., 2007). Subsequent studies revealed that, given enough time, loss of Dnmt3a alone was sufficient to drive expansion of stem cells, inevitably leading to the development of both lymphoid and myeloid leukemias (Challen et al., 2014). These leukemias possessed global hypomethylation with lineage-specific alterations, with a striking hypermethylation in lymphoid malignancies at promoters (Challen et al., 2014). While similar methylation patterns have been identified in AML patient studies, understanding the role and potential kinetics of methylation in CH has been limited due to a lack of studies on the methylation status of DNMT3A mutant non-leukemic cells. One study, however, profiled the blood of a 9-year-old patient who possessed a germline DNMT3A-R882H mutation and displayed characteristics of Tatton-Brown-Rahman Syndrome (Spencer et al., 2017). Here the authors found hypomethylation similar to that of AML patients when compared to a healthy sibling. Despite the caveats of the germline nature of the mutation and the age of the patient, these data suggest that the hypomethylation pattern found in DNMT3A-mutant AML precedes leukemic development and may play a role in CH progression.
In contrast to DNMT3A loss, TET2 loss-of-function mutations are associated with hypermethylation. Mouse studies using a conditional Tet2 loss-of-function mutant revealed an expansion of Lineage−Sca1+cKit+ (LSK) cells concomitant with decreased 5hmC levels (Moran-Crusio et al., 2011; Quivoron et al., 2011). In healthy individuals with CH, the ratio of 5hmC to 5mC has been shown to track with TET2 VAF, such that higher mutant VAF leads to lower 5hmC, supporting the prevalence of these biochemical processes in human biology (Buscarlet et al., 2017). The exact mechanisms by which TET2 loss of function results in increased stem cell expansion remains an unsolved issue, though the self-renewal and differentiation phenotypes likely involve complex pleiotropic regulation of multiple loci. One study focused on erythropoiesis identified dynamic changes in 5hmC during differentiation with increases in methylation at genomic loci associated with binding of the erythroid enriched transcription factors GATA1, GATA2, and KLF1 (Ge et al., 2014). These studies suggest that loss of 5mC and increases in 5hmC are associated with differentiation, revealing that perhaps TET2 activity permits stem cells to engage in signal-dependent differentiation processes.
If TET2 loss of function leads to a differentiation blockade, what role, if any does TET2 provide under homeostasis in HSCs? The answer to this may lie in the cofactors necessary for TET2 enzymatic activity. TET2 is an Fe(II), alpha-ketoglutarate, O2-dependent dioxgenase whose activity has been shown to be induced by vitamin C (Blaschke et al., 2013). Recent studies in mice have demonstrated that vitamin C is abundant within HSCs and its withdrawal mimics TET2 loss of function leading to HSC expansion (Agathocleous et al., 2017). In addition, HSCs have been shown to reside in a hypoxic perivascular niche (Boulais and Frenette, 2015; Chen et al., 2016). Combined, these factors may contribute to finely tuned control of TET2 enzymatic activity in HSCs. Future studies modulating TET2 activity with vitamin C in the setting of CH may offer insights into the potential reversibility of clonal expansion.
One curiosity among the genes associated with CH is the relative absence of mutations in IDH1 and IDH2, since frequent early hits in AML (Papaemmanuil et al., 2016) are exceedingly rare in CH (Coombs et al., 2017; Jaiswal et al., 2014, 2017). Mutations in IDH1 and IDH2 lead to the production of the neometabolite 2-hydroxyglutarate (2-HG), a potent inhibitor of alpha-ketoglutarate-dependent dioxygenases, including TET2. Mutations in IDH1/2 and TET2 are largely mutually exclusive in myeloid malignancies, supporting the idea that mutant IDH1/2 function in part through the downstream inhibition of TET2. Understanding why IDH1/2 mutations are not as equally enriched in CH as TET2 mutations will likely lend insights into whether different mechanisms are responsible for CH development and AML initiation without CH. One answer might lay in the pleiotropic effects of 2-HG function; in addition to inhibiting TET2, 2-HG inhibits, among a number of enzymes, a series of histone demethylases leading to increases in methylation of H3K27, H3K9, and H3K4 (Inoue et al., 2016b; Xu et al., 2011). This histone methylation led to condensation of chromatin around the DNA damage regulator Atm, decreasing expression levels particularly in aged mice. Subsequently IDH1/2 mutant cells were found to have increased 53BP1 foci and increased sensitivity to daunorubicin (Inoue et al., 2016b). Thus, IDH1/2 mutants may present the opposite scenario from DNMT3A-R882H mutant clones, where IDH1/2 mutations lead to a negative selective pressure in the presence of genotoxic stress. IDH1/2 mutant cells further diverge from TET2 mutants upon aging, where IDH1 mutants possess a decreased frequency of long-term HSCs, while aging exacerbates the expansion of these cells in TET2 mutants (Inoue et al., 2016b). Additional explanations may include potential non-cell-autonomous effects on 2-HG on the surrounding bone marrow niche, generating a supportive environment for AML development.
ASXL1 Mutations and Regulation of the Polycomb Repressive Complex
In addition to the regulation of DNA methylation described above, epigenetic regulation of histones through posttranslational modifications is another major regulator of cellular state. Among the many readers and writers of histones reviewed extensively elsewhere (Allis and Jenuwein, 2016), the polycomb repressive complex (PRC) is of particular interest given its role in stem cell biology (Avgustinova and Benitah, 2016) and Hox gene regulation. ASXL1 is one of three mammalian homologs of the Drosophila gene Asx, a member of the PRC (Fisher et al., 2003), and exhibits its effects through two different polycomb repressive complexes. The PRC2 complex catalyzes methylation of lysine 27 of the histone H3 tail (H3K27me3), a reaction that is executed by the methyltransferases EZH1 and EZH2. Meanwhile the PRC1 complex binds H3K27me3 by the protein CBX and catalyzes the ubiquitination of lysine 119 on Histone H2A (H2AK119ub). Both of these marks lead to a repression of transcription and the formation of a heterochromatin state at multiple loci across the genome including Hox genes (Schwartz and Pirrotta, 2013). In mammals, ASXL1 loss has been shown to alter the activity of the PRC2 complex with global downregulation of H3K27me3 and general narrowing of repressive domains, a phenomenon attributed to decreased EZH2 association with chromatin (Figure 3). Interestingly, while loss of Drosophila Asx leads to a reduction in H2AK119Ub levels, the evidence for a similar event for ASXL1 loss in mammals is unclear. Mouse models of hematopoietic-specific Asxl1 loss revealed progressive anemia and leukopenia in aged animals as well as an expansion of the stem cell compartment. This expansion of stem cells was associated with increased Hox gene expression and decreased H3K27me3 at this locus. These models mirror what is seen in patients, with recurrent frameshift and nonsense mutations in exon 11 and exon 12 of ASXL1 in MDS, MPN, and secondary AML patients. Whether these truncation mutants lead to stable protein products remains an area of active study, with some reports finding no stable ASXL1 truncation products and others reporting truncated products in cell lines.
Clonal Evolution to Hematological Abnormalities and Leukemic Transformation
In multiple studies, the presence of CH mutations has been associated with an increased propensity toward developing hematological malignancies (Coombs et al., 2017; Genovese et al., 2014; Jaiswal et al., 2014; Jaiswal et al., 2017). While the selective pressures associated with the previously discussed mutations are obvious explanations for this predisposition, other answers might lay in the biology associated with the mutated genes, and how they synergize with secondary and tertiary mutations. Given the baseline rate of mutation in normal, nonleukemic HSCs, there are many mutations that are present in leukemic samples that are likely to be passengers, acquired before leukemic transformation (Welch et al., 2012). Genomic studies have aided the identification of recurrent, potentially synergistic mutations; yet, causality and distinction between driver mutations and passenger events requires verification in model systems.
CH in Aplastic Anemia
The connection between CH and leukemic development involves evolutionary progression, where clones may acquire additional mutations conferring increased fitness to the clone. Similar principles apply to bone marrow failure syndromes, including aplastic anemia where immune processes impose a selective pressure limiting the HSC pool (Young et al., 2006). Recent genomic studies on the blood of aplastic anemia patients identified recurrent mutations in BCOR, BCORL1, and PIGA, as well as the often-mutated DNMT3A and ASXL1 identified in other CH-related studies (Yoshizato et al., 2015). The authors found that while the incidence of DNMT3A and ASXL1 mutations increased with age, PIGA and BCOR/BCORL1 mutations were equally represented across all age groups. Moreover, PIGA/BCOR/BCORL1 mutations were associated with increased responsiveness to immunotherapeutic intervention, while patients with the AML/MDS-related mutations were less responsive. These data suggest at least two distinct modes for aplastic anemia development: (1) immune-evasion-mediated clonal selection and (2) HSC self-renewal-mediated clonal selection (Ogawa, 2016). While the role of BCOR/BCORL1 in aplastic anemia remains unsolved, some evidence exists for the other commonly mutated genes. PIGA mutations are well known in Paroxysmal nocturnal hemoglobinuria (PNH), where PIGA mutant cells are destroyed in a complement-mediated manner. Despite this killing mechanism, PIGA mutant clones expand, a finding likely explained by the decreased production of glycosylphosphatidylinositol, a potential target of CD1D+ restricted T cells (Gargiulo et al., 2013). The evidence for DNMT3A and ASXL1 mediating stem cell self-renewal is described above; however, if and how these mutations specifically interact with the immune system remains to be seen.
Mutations in DNMT3A Synergize with Oncogenic Mutations to Promote Malignant Transformation
DNMT3A is one of the most commonly mutated genes in CH among both healthy cohorts of patients and those with other hematological and non-hematological malignancies. Furthermore, DNMT3A is one of the most commonly mutated genes in adult AML and T cell acute lymphoid leukemia (ALL) with mutated frequency averaging between 22% and 35% of patients depending upon the cohort (Ley et al., 2010; Yang et al., 2015). In stark contrast, DNMT3A mutations are exceedingly rare in pediatric disease (Bolouri et al., 2018). These findings lend insight into the role of CH as a precursor to adult disease, but suggest that other mechanisms might be at play in pediatric leukemias. Large adult AML cohorts have identified co-mutations in the receptor tyrosine kinase FLT3; RAS signaling pathway members NRAS, KRAS, and PTPN11; and mutations in the multifunctional nuclear protein NPM1 (Ley et al., 2013; Papaemmanuil et al., 2016). Experimental studies in mice have confirmed these findings; with conditional loss-of-function Dnmt3a mice developing both myeloid and lymphoid malignancy with age (Mayle et al., 2015). While few recurrent mutations outside of Notch1 were found in these aged mice, Dnmt3a mutations were found to synergize with mutant Nras expression, leading to the rapid development of leukemia. Similar studies were performed with mutant Flt3 overexpression, leading to the development of both myeloid and lymphoid leukemias (Yang et al., 2016). In contrast, a genetic mouse model of the Dnmt3a-R882H (R878H in mouse) combined with Flt3-ITD, as well as Npm1 mutations, led to robust AML formation (Guryanova et al., 2016), a combination that is a poor prognostic mutational genotype in patients (Papaemmanuil et al., 2016).
This triple combination of mutations appears to be particularly enriched in younger patients, suggesting against its enrichment in patients with antecedent CH. Indeed, further evidence beyond the presence of a DNMT3A somatic mutation is necessary to declare that a patient’s AML proceeded from a CH clone. One approach to retroactively identify AML patients with antecedent CH clones is to determine the VAF of the presumed initiating mutation in both myeloid and lymphoid cells. If a mutation is present in both AML blasts and T cells, it suggests a cell of origin in a stem cell compartment capable of mutlilineage differentiation, consistent with CH. It is worth noting that the converse is not necessarily true, that the absence of a mutation in T cells formally excludes an antecedent CH clone, especially given the myeloid bias in aged stem cells (Pang et al., 2011). Regardless, Shlush and colleagues identified recurrent DNMT3A mutations that appeared in both AML blasts and sorted T cells, while the AML-restricted NPM1c mutation was only found in AML blasts (Shlush et al., 2014). This study demonstrated that in some cases, mutated HSCs capable of multilineage differentiation might serve as preleukemic clones. More recently, Thol and colleagues found that DNMT3A mutant AML patients that possessed mutations in both myeloid and lymphoid cells (lympho-myeloid CH or LM-CH) had a distinct mutational spectrum from those that bore no DNMT3A mutations in the lymphoid lineage (Thol et al., 2017). LM-CH patients were enriched for additional mutations in TET2, RUNX1, and EZH2, while patients without LM-CH were enriched for the aforementioned NPM1 and FLT3 mutations.
In this cohort of patients, the DNMT3A-R882H hotspot mutation was enriched in patients with LM-CH compared to those without LM-CH. This was a surprising finding, given that this hotspot mutation is underrepresented in individuals with CH compared to de novo AML patients. One explanation is that CH clones with DNMT3A hotspot mutations may be more likely to go on to form AML compared to those with other DNMT3A mutations. In such a setting, it will be of interest to determine if a patient ever had CH per se, or if they were already in the early stages of leukemic development. The distinction between these states might depend upon the latency between second or third mutations that are acquired following the initial DNMT3A mutation. Whether this is an important feature of disease development remains to be seen. In addition, this highlights the possibility that some individuals with CH who do not possess hematological abnormalities at the time of sample collection may rapidly progress to AML or other myeloid malignancies. Such a scenario complicates the interpretation of studies assessing the rate at which patients progress to secondary hematological malignancy.
TET2 Mutations Cooperate with JAK2 and FLT3 Mutations to Promote Myeloid Transformation
Like Dnmt3a, Tet2 mutant mice have been shown to synergize with Flt3 mutant mice, leading to the development of AML with full penetrance (Shih et al., 2015). As an expected consequence of Tet2 loss, many loci were hypermethylated. Interestingly, these sites changed dramatically with the combined Flt3 mutation, demonstrating a synergy between the two mutations. While the methylation state of many loci changed, methylation and transcription-repression at the Gata2 locus in particular seemed to play a causative role in disease development. Indeed, Gata2 re-expression blunted leukemia formation and resulted in differentiation. This locus may serve as an interesting biomarker for monitoring TET2 mutant CH and its progression toward a more aggressive condition.
In addition to FLT3, mutations in TET2 have been shown to cooperate with expression of mutant Kit in mast cells (De Vita et al., 2014), AML-ETO (Rasmussen et al., 2015), and loss of Notch signaling (Lobry et al., 2013). However, in each case, the specific disease phenotype differs, suggesting that the subsequent mutations acquired by the CH clone play an instructive role in dictating disease phenotype. A recent study identified a hypermutagenicity phenotype in aged Tet2 knockout mice compared to wild-type littermates (Pan et al., 2017a), providing an additional potential source for oncogenic allele generation. These mutations were enriched at sites of 5hmC accumulation in wild-type mice, suggesting a potential site-specific protective role of genotoxic stress for TET2. Importantly, these mutations were largely enriched for G:C and A:T transversions, suggestive of a mutational process distinct from that of aging-associated mutational acquisition, which is characterized by C:T transitions.
While clearly associated with CH, TET2 mutations are not always present as the first, or dominant, clonal mutation in leukemias. In TET2 mutant AML, a study inferring clonality and temporal mutation acquisition through variant allele frequency has proposed that patients’ disease may arise as either TET2-first or JAK2-first (Ortmann et al., 2015). In this setting, patients with JAK2-first disease appeared to have more aggressive disease with increased risk of thrombosis and polycythemia vera. These studies lend insight into the situation during which two CH mutations intertwine, demonstrating that the natural history of the ensuing disease can be altered based on the order of event acquisition. Similar cooperation between JAK2 and TP53 mutations has been shown to result in aggressive leukemia formation in the mouse (Rampal et al., 2014), a combination prevalent among secondary AML patients. Given the selective pressures that influence TP53 mutant clonal emergence, this provides a second interesting scenario where mutation order may reflect distinct courses of disease development.
CH Mutations in Cardiovascular Disease
CH mutations are associated with an expansion of the stem cell pool likely serving as antecedent clones for potential future hematological disease; however, the differentiation blockade is not completely penetrant. Indeed, CH mutant stem cells give rise to mature, differentiated cells that contribute to homeostasis and host defense. Thus understanding how TET2 loss, for instance, affects mature myeloid development and function may provide insights into what role CH may play in homeostasis and aging-associated disease.
One study by Zhang et al. identified a role for Tet2 in inflammation resolution through the suppression of IL-6 (Zhang et al., 2015). Tet2 knockout mice displayed greater tissue damage to both the lung upon exposure to LPS and the gut following dextran sodium sulfate (DSS) treatment. This disruption of tissue architecture following inflammatory insult poses the hypothesis where individuals with CH may be at increased risk in inflammatory disease. Indeed, one study found that the presence of TET2 mutant clones was associated with the development of COPD/asthma (Buscarlet et al., 2017), while another study demonstrated an increased association with smoking behavior, pulmonary disease, and psychiatric treatment (Zink et al., 2017) (Figure 4). Another study found that elderly individuals (age >70) with CH mutations had a greater risk of all-cause mortality (Jaiswal et al., 2014), specifically associated with an increased risk in developing type 2 diabetes and coronary heart disease. Follow-up studies on a larger patient cohort revealed that individuals with CH possessing mutations in DNMT3A, JAK2, ASXL1, and TET2 were at greater risk for cardiovascular disease (Jaiswal et al., 2017). Collectively these data demonstrate that CH is associated with inflammatory conditions; however, with the exception of cardiovascular disease described below, there remains limited understanding into whether these associations are causal. For instance, future studies will need to determine whether the association of TET2 mutation status with COPD is a direct association with smoking, or whether TET2 mutant cells might play a mechanistic role in exacerbating COPD.
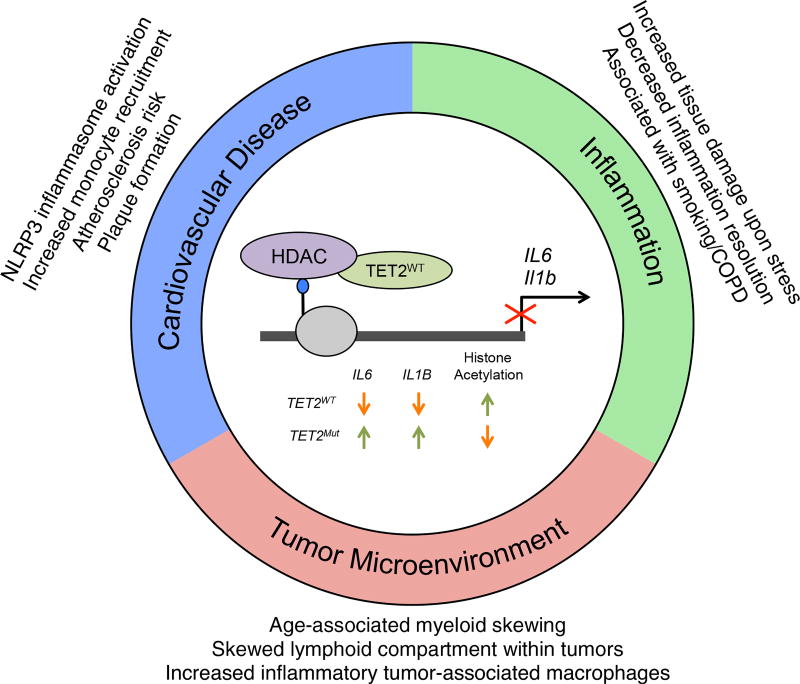
Figure 4. CH-Associated TET2 Mutations and Interaction with Nonhematologic Disease.
Schematic of TET2-dependent repression of IL-6 and IL-1beta by recruitment of HDAC proteins. TET2 loss of function leads to de-repression of Il6 and Il1b cytokine expression. This deregulation has impacts on atherosclerotic plaque formation, inflammation resolution, and the tumor microenvironment. Age-related myeloid skewing and mutation-dependent lymphoid department skewing alters the tumor microenvironment immune landscape.
Recent studies utilizing genetic mouse models of atherosclerosis development support a causative role for subclonal TET2 loss of function in atherosclerosis development (Fuster et al., 2017). Using an Ldlr-deficient model of atherosclerotic plaque formation, the authors transplanted either WT or Tet2-deficient bone marrow at a subclonal ratio, such that mutant cells only made up 10% of the hematopoietic compartment. This led to an increase in plaque size, a finding that was recapitulated with a macrophage-specific knockout of Tet2. In line with previous studies, the authors identified an increase in IL-6 expression, as well as an inability to suppress IL-1beta expression following the resolution of an inflammatory challenge with LPS (Figure 4). The increase in expression was complemented by increased secretion and activation of the NLRP3 inflammasome, a necessary component in IL-1beta activity. Functionally, the authors found that inhibition of the NLRP3 inflammasome, and thus IL-1beta, led to reduced plaque size in the Tet2 mutant mice, with little activity in wild-type controls. Similar results were reported in the aforementioned study that highlighted the key role of CH mutations in cardiovascular risk (Jaiswal et al., 2017). Further insights into how CH mutations impact cardiovascular disease development may provide a framework for integrating personalized CH-relevant therapy into risk reduction strategies.
This study provides striking mechanistic insight into how a CH mutation may alter non-hematologic disease progression through specific alterations of differentiated cell biology. Critically the authors found that suppression of IL-1beta did not depend upon the catalytic activity of Tet2 (Fuster et al., 2017). These data suggest that that not all TET2 mutations are the same, and perhaps only those that lead to truncated or nonsense protein products may pose a risk to cardiovascular disease development, while mutations leading to reduced catalytic function may offer a preleukemic risk. This non-enzymatic loss of function also suggests that boosting enzymatic activity with increased vitamin C availability, an approach that appears to provide value in the setting of leukemia (Cimmino et al., 2017), might not be applicable in the setting of TET2 mutant-associated cardiovascular disease.
CH Mutations in Non-hematological Malignancy
Recently, a large cohort of cancer patients was analyzed for the prevalence of CH mutations in the blood (Coombs et al., 2017). Among the 8,800 patients, over 25% acquired a mutation in a gene commonly mutated in CH. Mutations in DNMT3A and TET2 were the most common among all patients, with mutant TP53 and PPM1D being specifically enriched in patients that were previously exposed to chemo and radiotherapy. Perhaps unsurprisingly, the patients with CH mutations were found to be significantly older than non-CH patients, were more likely to have undergone prior chemo or radiotherapy, and were more likely to be current or former smokers. These findings suggest that aging and exposure to mutagenic environments likely contribute to the acquisition and/or selective expansion of CH mutations. Indeed, the most commonly mutated genes can be divided into those that have been shown to be related to aging and therapy-naive hematologic malignancy (DNMT3A, TET2, ASXL1, JAK2, and SF3B1) or are related to DNA damage repair (TP53, PPM1D, ATM, and CHEK2). These mutations were further classified based on their association as potential drivers (PDs) in hematological malignancy. Furthermore, while CH status was found to not be an independent prognostic indicator, CH-PD status was found to be associated with worse prognosis independent of age, gender, and smoking history, largely due to progression of the primary non-hematologic tumor.
These findings suggest that it is not simply clonal skewing of the hematopoietic compartment that alters disease course, but also molecular functions of the mutant alleles that elicit the CH phenotype. DNMT3A was the most commonly mutated gene in this cohort and included truncating alleles, a broad spectrum of missense mutations, and the mutational hotspot R882H that is highly prevalent in AML (Papaemmanuil et al., 2016). The third most commonly mutated gene was PPM1D, which appeared to be overrepresented here relative to studies of CH in the general population. This is likely due to the selection bias in this cohort of patients who were either currently or had previously undergone chemo or radiotherapy. Collectively these findings suggest that CH in cancer patients may be either derived from an antecedent clonal expansion or induced by chemotherapeutic treatment and/or selection. It will be of future interest to determine whether these two paths are indeed distinct and perhaps elicit different effects systemically or locally in the tumor microenvironment (TME).
In addition to the studies that focused on the prevalence of CH mutations in the blood of patients, other studies have identified somatic mutations in the stroma of breast (Kurose et al., 2002; Patocs et al., 2007) and bladder (Paterson et al., 2003) cancer patients. In an analysis targeted to hematopoietic cells, Kleppe et al. performed targeted sequencing and WES on CD45+ tumor-initiating lymphocytes (TILs) as well as CD45− epithelial cells isolated from the primary breast cancer samples (Kleppe et al., 2015). Through both flow cytometry assisted cell sorting and laser microdissection, these analyses identified mutations in NOTCH2, EZH2, NF1, TET2, BCOR, DNMT3A, and JAK1, all of which were selectively found in TILs, but not the tumor epithelium, indicating the hematopoietic origin of these mutants. TET2 and DNMT3A mutations are clearly abundant in CH; however, the remaining mutations, while relevant to hematological malignancy as a whole, are not particularly abundant or enriched in individuals with CH. A finer understanding of the origin of these mutant cells may offer insights into whether the CH-associated mutations represent a distinct biological entity from the remaining mutant cells. These studies also identified mutations in the blood that were present in the TILs, albeit at a lower VAF. While the sample size was limited, this suggests that certain CH mutations may lead to greater accumulation of TILs within the TME. Future studies will be critical to determine if this expansion in the TME is due to increased recruitment from the periphery or expansion within the TME.
Beyond changes in cellular abundance, it remains to be seen what, if any, functional effects these CH mutations may elicit on the cells of the TME. Recent reports by Pan et al. have demonstrated that Tet2 expression and 5hmC accumulation was increased in tumor-associated macrophages (TAMs) as disease progressed (Pan et al., 2017b). Importantly, loss of Tet2 in myeloid cells led to a reduction in tumor growth accompanied by a skewing of CD4+ and CD8+ lymphocytes. While this offers an informative look into the role of Tet2 in TAMs, it will be of further interest to determine the consequences of subclonal Tet2 loss of function in these models, given that most patients with CH possess a VAF substantially lower than the recombination efficiency of the LysM:Cre line.
Importantly, future analyses on tumor samples will be necessary to determine whether mutations are enriched in lymphocytes, myeloid cells, or both in order to properly contextualize the potential role these mutations play in disease. Previous studies on AML patients identified an accumulation of DNMT3A mutations across mature lymphocyte populations as well as myeloid cells (Shlush et al., 2014). Additional studies found that secondary AML patients were enriched for this lymphomyeloid mutational distribution, while primary AML patients tended to possess mutations restricted to the myeloid lineage (Thol et al., 2017). These studies suggest that there is at least the potential for mutations to manifest in multiple lineages; however, future studies are necessary to determine whether this remains true in patients without AML and how these findings in the periphery relate to the tumor.
Perspective on Clonal Evolution and Somatic Mosaicism
CH presents an opportunity to view the early stages of cancer development. While many solid tumors are nearly undetectable until the later, inoperable malignant state of disease, leukemias and other hematological malignancies may be readily detectable as sequencing technology allows even greater sensitivity in detecting mutant alleles in the blood. However, unlike solitary solid tumors that can be surgically removed, it remains unclear how to best eradicate these premalignant threats in blood, or whether any actions are necessary. As clinical data continues to build, and mechanistic studies reveal the role of hematopoietic-derived somatic mutations on homeostasis, one might imagine a scenario where CH mutations offer a biomarker for nonhematologic disease intervention. In this setting, eradication of a clone, or attempts at doing so, must be taken with care so as to not provide a selective pressure leading to the emergence of a more aggressive clone capable of hematological malignancy.
Easy access to blood samples has provided a logical opportunity to study somatic mosaicism in homeostasis in disease development; however, it is unlikely to be the only major organ system to attain somatic mutations leading to clonal dominance. Indeed, somatic alterations in copy number and retrotransposon mobility have been well studied in neuron differentiation and skin (Abyzov et al., 2012; Muotri et al., 2005). Interestingly, the somatic mosaicism here is predominately established during development (McConnell et al., 2017); as neurons are largely post mitotic, they offer a biological example of the distinction between somatic mosaicism with and without clonal expansion. Another example involves patients with hereditary tyrosinemia type I, who possess loss-of-function mutations in fumarylacteoacetate hydrolase (Kvittingen et al., 1994). Reversion of mutations in this gene has been identified in clusters of hepatocytes within the livers of these patients, indicating clonal expansion under selective pressure. Given the rate of cancer development across such a wide variety of tissues, it is likely that somatic variations are abundant across multiple organ systems (O’Huallachain et al., 2012). The nature of the hematopoietic system and its accessibility to nearly every tissue befits an opportunity for these somatic mutations to impact both homeostasis and pathology in a widespread manner.
Future Clinical Perspective on CH
In the past few years, CH has emerged from an intriguing observation (Busque et al., 1996) to the focus of intense research. The studies discussed here demonstrate that CH is a common, age-dependent state, and that CH is associated with an increased risk of subsequent hematologic malignancies, an increased cardiovascular risk, and adverse outcomes in patients with advanced malignancies. From the clinical context, this leads to several important considerations. The first and foremost of these relates to diagnosis. Should we screen for CH on a population basis, and if so, to what depth and for which genetic alterations? Should patients have CH screening as part of their risk assessment for cardiovascular disease? To address this critical question, it will take careful assessment of the ever-expanding data being assembled from population-based studies.
The broader issue relates to whether age-dependent CH represents our first glimpse into the role of somatic mutations in tissue-resident stem cells as a pathogenic factor in human disease. It is likely that CH will have an impact on a broad spectrum of age-associated diseases, in part through mutant-derived inflammatory signaling and likely through yet-to-be defined additional mechanisms. Such investigations will lead to novel insights into the relationship between CH and additional disease states, and more importantly, will elucidate additional risk factors that justify screening for CH in the general population. More broadly, it is likely that somatic mutations will be identified in stem cells in non-hematopoietic lineages and that this contributes to a broad suite of human diseases. We would contend that CH is likely our first glimpse into a broader phenomenon.
Most importantly, it is incumbent upon the field to determine if there are therapeutic strategies that can be used to target CH and reduce the fitness advantage of mutant hematopoietic cells. Such approaches would have the potential to reduce the risk of hematopoietic malignancies, cardiovascular disease, and likely additional age-dependent disease states. We would expect such approaches to be tailored to specific mutations such that a “one size fits all” mindset to targeting CH will not be the optimal approach, and discovery studies will be required to identify mutant-specific dependencies in different CH genotypes. Such an approach would represent the beginning of a new era of “precision” prevention, in which we can tailor intervention studies to genetically defined at-risk populations to prevent subsequent adverse outcomes. Until precise risk factors are identified, the clinical significance of CH in individuals with normal blood counts is undetermined.
Acknowledgments
We are grateful to the members of the Levine lab for insight and discussion. R.L.B. is supported by the Damon Runyon Cancer Research Foundation. This study was supported in part by NIH National Cancer Institute (NCI) grant P30 CA008748 (Cancer Center Support Grant).
Footnotes
DECLARATION OF INTERESTS
R.L.L. is a consultant for Novartis, is on the Supervisory Board of QIAGEN, and is on the Scientific Advisory Board of Loxo.
References
- Abkowitz JL, Catlin SN, Guttorp P. Evidence that hematopoiesis may be a stochastic process in vivo. Nat. Med. 1996;2:190–197. doi: 10.1038/nm0296-190. [DOI] [PubMed] [Google Scholar]
- Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino AF, Rosenberg Belmaker LA, Szekely A, Wilson M, et al. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012;492:438–442. doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Jasper H, Rudolph KL. Aging-Induced Stem Cell Mutations as Drivers for Disease and Cancer. Cell Stem Cell. 2015;16:601–612. doi: 10.1016/j.stem.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, Cowin BL, Bruner E, Murphy MM, Chen W, et al. Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature. 2017;549:476–481. doi: 10.1038/nature23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari MR, Lepage P, Rosen B, McLaughlin J, Risch H, Minden M, Narod SA. PPM1D mutations in circulating white blood cells and the risk for ovarian cancer. J. Natl. Cancer Inst. 2014;106:djt323. doi: 10.1093/jnci/djt323. [DOI] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- Avgustinova A, Benitah SA. Epigenetic control of adult stem cell function. Nat. Rev. Mol. Cell Biol. 2016;17:643–658. doi: 10.1038/nrm.2016.76. [DOI] [PubMed] [Google Scholar]
- Bejar R. CHIP, ICUS, CCUS and other four-letter words. Leukemia. 2017;31:1869–1871. doi: 10.1038/leu.2017.181. [DOI] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Farrar JE, Triche T, Jr, Ries RE, Lim EL, Alonzo TA, Ma Y, Moore R, Mungall AJ, Marra MA, et al. The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med. 2018;24:103–112. doi: 10.1038/nm.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Boulais PE, Frenette PS. Making sense of hematopoietic stem cell niches. Blood. 2015;125:2621–2629. doi: 10.1182/blood-2014-09-570192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscarlet M, Provost S, Zada YF, Barhdadi A, Bourgoin V, Lépine G, Mollica L, Szuber N, Dubé MP, Busque L. DNMT3A and TET2 dominate clonal hematopoiesis and demonstrate benign phenotypes and different genetic predispositions. Blood. 2017;130:753–762. doi: 10.1182/blood-2017-04-777029. [DOI] [PubMed] [Google Scholar]
- Busque L, Mio R, Mattioli J, Brais E, Blais N, Lalonde Y, Maragh M, Gilliland DG. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood. 1996;88:59–65. [PubMed] [Google Scholar]
- Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso M, Paulo P, Maia S, Teixeira MR. Truncating and missense PPM1D mutations in early-onset and/or familial/hereditary prostate cancer patients. Genes Chromosomes Cancer. 2016;55:954–961. doi: 10.1002/gcc.22393. [DOI] [PubMed] [Google Scholar]
- Cargo CA, Rowbotham N, Evans PA, Barrans SL, Bowen DT, Crouch S, Jack AS. Targeted sequencing identifies patients with preclinical MDS at high risk of disease progression. Blood. 2015;126:2362–2365. doi: 10.1182/blood-2015-08-663237. [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, Mallaney C, Celik H, Yang L, Xia Z, et al. Dnmt3a and Dnmt3b have overlapping and distinct functions in hematopoietic stem cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Miyanishi M, Wang SK, Yamazaki S, Sinha R, Kao KS, Seita J, Sahoo D, Nakauchi H, Weissman IL. Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature. 2016;530:223–227. doi: 10.1038/nature16943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, Ng V, Xia B, Witkowski MT, Mitchell-Flack M, et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell. 2017;170:1079–1095. e1020. doi: 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M, Benguría A, Zaballos A, Flores JM, Barbacid M, et al. Tumour biology: senescence in premalignant tumours. Nature. 2005;436:642. doi: 10.1038/436642a. [DOI] [PubMed] [Google Scholar]
- Coombs CC, Zehir A, Devlin SM, Kishtagari A, Syed A, Jonsson P, Hyman DM, Solit DB, Robson ME, Baselga J, et al. Therapy-Related Clonal Hematopoiesis in Patients with Non-hematologic Cancers Is Common and Associated with Adverse Clinical Outcomes. Cell Stem Cell. 2017;21:374–382.e4. doi: 10.1016/j.stem.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai YJ, Wang YY, Huang JY, Xia L, Shi XD, Xu J, Lu J, Su XB, Yang Y, Zhang WN, et al. Conditional knockin of Dnmt3a R878H initiates acute myeloid leukemia with mTOR pathway involvement. Proc. Natl. Acad. Sci. USA. 2017;114:5237–5242. doi: 10.1073/pnas.1703476114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita S, Schneider RK, Garcia M, Wood J, Gavillet M, Ebert BL, Gerbaulet A, Roers A, Levine RL, Mullally A, Williams DA. Loss of function of TET2 cooperates with constitutively active KIT in murine and human models of mastocytosis. PLoS ONE. 2014;9:e96209. doi: 10.1371/journal.pone.0096209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CL, Berger J, Randazzo F, Brock HW. A human homolog of Additional sex combs, ADDITIONAL SEX COMBS-LIKE 1, maps to chromosome 20q11. Gene. 2003;306:115–126. doi: 10.1016/s0378-1119(03)00430-x. [DOI] [PubMed] [Google Scholar]
- Flach J, Bakker ST, Mohrin M, Conroy PC, Pietras EM, Reynaud D, Alvarez S, Diolaiti ME, Ugarte F, Forsberg EC, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuster JJ, MacLauchlan S, Zuriaga MA, Polackal MN, Ostriker AC, Chakraborty R, Wu CL, Sano S, Muralidharan S, Rius C, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355:842–847. doi: 10.1126/science.aag1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargiulo L, Papaioannou M, Sica M, Talini G, Chaidos A, Richichi B, Nikolaev AV, Nativi C, Layton M, de la Fuente J, et al. Glycosylphosphatidylinositol-specific, CD1d-restricted T cells in paroxysmal nocturnal hemoglobinuria. Blood. 2013;121:2753–2761. doi: 10.1182/blood-2012-11-469353. [DOI] [PubMed] [Google Scholar]
- Ge L, Zhang RP, Wan F, Guo DY, Wang P, Xiang LX, Shao JZ. TET2 plays an essential role in erythropoiesis by regulating lineage-specific genes via DNA oxidative demethylation in a zebrafish model. Mol. Cell. Biol. 2014;34:989–1002. doi: 10.1128/MCB.01061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger H, de Haan G, Florian MC. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 2013;13:376–389. doi: 10.1038/nri3433. [DOI] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CJ, Lindsley RC, Tchekmedyian V, Mar BG, Shi J, Jaiswal S, Bosworth A, Francisco L, He J, Bansal A, et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J. Clin. Oncol. 2017;35:1598–1605. doi: 10.1200/JCO.2016.71.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis NK, Ball M, Zhang Q, Ma Z, Zhao Y, Yoder SJ, Balasis ME, Mesa TE, Sallman DA, Lancet JE, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18:112–121. doi: 10.1016/S1470-2045(16)30627-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guryanova OA, Shank K, Spitzer B, Luciani L, Koche RP, Garrett-Bakelman FE, Ganzel C, Durham BH, Mohanty A, Hoermann G, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat. Med. 2016;22:1488–1495. doi: 10.1038/nm.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Bradley RK, Abdel-Wahab O. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016a;30:989–1001. doi: 10.1101/gad.278424.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Li WY, Tseng A, Beerman I, Elia AJ, Bendall SC, Lemonnier F, Kron KJ, Cescon DW, Hao Z, et al. Mutant IDH1 Downregulates ATM and Alters DNA Repair and Sensitivity to DNA Damage Independent of TET2. Cancer Cell. 2016b;30:337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, McConkey M, Gupta N, Gabriel S, Ardissino D, et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017;377:111–121. doi: 10.1056/NEJMoa1701719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiblova P, Shaltiel IA, Benada J, Ševčík J, Pecháčková S, Pohlreich P, Voest EE, Dundr P, Bartek J, Kleibl Z, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. J. Cell Biol. 2013;201:511–521. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppe M, Comen E, Wen HY, Bastian L, Blum B, Rapaport FT, Keller M, Granot Z, Socci N, Viale A, et al. Somatic mutations in leukocytes infiltrating primary breast cancers. NPJ Breast Cancer. 2015;1:15005. doi: 10.1038/npjbcancer.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuranda K, Vargaftig J, de la Rochere P, Dosquet C, Charron D, Bardin F, Tonnelle C, Bonnet D, Goodhardt M. Age-related changes in human hematopoietic stem/progenitor cells. Aging Cell. 2011;10:542–546. doi: 10.1111/j.1474-9726.2011.00675.x. [DOI] [PubMed] [Google Scholar]
- Kurose K, Gilley K, Matsumoto S, Watson PH, Zhou XP, Eng C. Frequent somatic mutations in PTEN and TP53 are mutually exclusive in the stroma of breast carcinomas. Nat. Genet. 2002;32:355–357. doi: 10.1038/ng1013. [DOI] [PubMed] [Google Scholar]
- Kvittingen EA, Rootwelt H, Berger R, Brandtzaeg P. Self-induced correction of the genetic defect in tyrosinemia type I. J. Clin. Invest. 1994;94:1657–1661. doi: 10.1172/JCI117509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, Flamholz R, Dabbas B, Yung A, Al-Hafidh J, et al. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood. 2015;126:2355–2361. doi: 10.1182/blood-2015-08-667063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ, Jr, Laird PW, Baty JD, et al. Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Saber W, Mar BG, Redd R, Wang T, Haagenson MD, Grauman PV, Hu ZH, Spellman SR, Lee SJ, et al. Prognostic Mutations in Myelodysplastic Syndrome after Stem-Cell Transplantation. N. Engl. J. Med. 2017;376:536–547. doi: 10.1056/NEJMoa1611604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J. Exp. Med. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcovati L, Gallì A, Travaglino E, Ambaglio I, Rizzo E, Molteni E, Elena C, Ferretti VV, Catricalà S, Bono E, et al. Clinical significance of somatic mutation in unexplained blood cytopenia. Blood. 2017;129:3371–3378. doi: 10.1182/blood-2017-01-763425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Porter CC, Zaberezhnyy V, DeGregori J. Irradiation selects for p53-deficient hematopoietic progenitors. PLoS Biol. 2010;8:e1000324. doi: 10.1371/journal.pbio.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, Goodell MA. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell MJ, Moran JV, Abyzov A, Akbarian S, Bae T, Cortes-Ciriano I, Erwin JA, Fasching L, Flasch DA, Freed D, et al. Brain Somatic Mosaicism Network. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science. 2017;356:356. doi: 10.1126/science.aal1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, Crawley C, Craig J, Scott MA, Hodkinson C, et al. Understanding Society Scientific Group. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medyouf H. The microenvironment in human myeloid malignancies: emerging concepts and therapeutic implications. Blood. 2017;129:1617–1626. doi: 10.1182/blood-2016-11-696070. [DOI] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- O’Huallachain M, Karczewski KJ, Weissman SM, Urban AE, Snyder MP. Extensive genetic variation in somatic human tissues. Proc. Natl. Acad. Sci. USA. 2012;109:18018–18023. doi: 10.1073/pnas.1213736109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S. Clonal hematopoiesis in acquired aplastic anemia. Blood. 2016;128:337–347. doi: 10.1182/blood-2016-01-636381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortmann CA, Kent DG, Nangalia J, Silber Y, Wedge DC, Grinfeld J, Baxter EJ, Massie CE, Papaemmanuil E, Menon S, et al. Effect of mutation order on myeloproliferative neoplasms. N. Engl. J. Med. 2015;372:601–612. doi: 10.1056/NEJMoa1412098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Wingo TS, Zhao Z, Gao R, Makishima H, Qu G, Lin L, Yu M, Ortega JR, Wang J, et al. Tet2 loss leads to hypermutagenicity in haematopoietic stem/progenitor cells. Nat. Commun. 2017a;8:15102. doi: 10.1038/ncomms15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W, Zhu S, Qu K, Meeth K, Cheng J, He K, Ma H, Liao Y, Wen X, Roden C, et al. The DNA Methylcytosine Dioxygenase Tet2 Sustains Immunosuppressive Function of Tumor-Infiltrating Myeloid Cells to Promote Melanoma Progression. Immunity. 2017b;47:284–297. e285. doi: 10.1016/j.immuni.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter NE, Heuser M, Thol F, Bolli N, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N. Engl. J. Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson RF, Ulbright TM, MacLennan GT, Zhang S, Pan CX, Sweeney CJ, Moore CR, Foster RS, Koch MO, Eble JN, Cheng L. Molecular genetic alterations in the laser-capture-microdissected stroma adjacent to bladder carcinoma. Cancer. 2003;98:1830–1836. doi: 10.1002/cncr.11747. [DOI] [PubMed] [Google Scholar]
- Patocs A, Zhang L, Xu Y, Weber F, Caldes T, Mutter GL, Platzer P, Eng C. Breast-cancer stromal cells with TP53 mutations and nodal metastases. N. Engl. J. Med. 2007;357:2543–2551. doi: 10.1056/NEJMoa071825. [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- Rampal R, Ahn J, Abdel-Wahab O, Nahas M, Wang K, Lipson D, Otto GA, Yelensky R, Hricik T, McKenney AS, et al. Genomic and functional analysis of leukemic transformation of myeloproliferative neoplasms. Proc. Natl. Acad. Sci. USA. 2014;111:E5401–E5410. doi: 10.1073/pnas.1407792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev. 2015;29:910–922. doi: 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, et al. Breast and Ovarian Cancer Susceptibility Collaboration; Wellcome Trust Case Control Consortium. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. A new world of Polycombs: unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, Antony-Debre I, Viale A, Socci N, Sun Y, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell. 2015;27:502–515. doi: 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, et al. HALT Pan-Leukemia Gene Panel Consortium. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RA, Morrison SJ. Mechanisms that regulate stem cell aging and life span. Cell Stem Cell. 2013;12:152–165. doi: 10.1016/j.stem.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS, Fronick CC, O’Laughlin M, Heath SE, Shinawi M, et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell. 2017;168:801–816. e813. doi: 10.1016/j.cell.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swisher EM, Harrell MI, Norquist BM, Walsh T, Brady M, Lee M, Hershberg R, Kalli KR, Lankes H, Konnick EQ, et al. Somatic Mosaic Mutations in PPM1D and TP53 in the Blood of Women With Ovarian Carcinoma. JAMA Oncol. 2016;2:370–372. doi: 10.1001/jamaoncol.2015.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Wang F, Kantarjian H, Doss D, Khanna K, Thompson E, Zhao L, Patel K, Neelapu S, Gumbs C, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18:100–111. doi: 10.1016/S1470-2045(16)30626-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Klesse S, Köhler L, Gabdoulline R, Kloos A, Liebich A, Wichmann M, Chaturvedi A, Fabisch J, Gaidzik VI, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017;31:1286–1295. doi: 10.1038/leu.2016.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A, Nussenzweig A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingert S, Thalheimer FB, Haetscher N, Rehage M, Schroeder T, Rieger MA. DNA-damage response gene GADD45A induces differentiation in hematopoietic stem cells without inhibiting cell cycle or survival. Stem Cells. 2016;34:699–710. doi: 10.1002/stem.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong TN, Ramsingh G, Young AL, Miller CA, Touma W, Welch JS, Lamprecht TL, Shen D, Hundal J, Fulton RS, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518:552–555. doi: 10.1038/nature13968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat. Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata T, Takanashi T, Muguruma Y, Ibrahim AA, Matsuzawa H, Uno T, Sheng Y, Onizuka M, Ito M, Kato S, Ando K. Accumulation of oxidative DNA damage restricts the self-renewal capacity of human hematopoietic stem cells. Blood. 2011;118:2941–2950. doi: 10.1182/blood-2011-01-330050. [DOI] [PubMed] [Google Scholar]
- Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat. Rev. Cancer. 2015;15:152–165. doi: 10.1038/nrc3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, Jeong M, Curry CV, Kim SB, Ruau D, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29:922–934. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizato T, Dumitriu B, Hosokawa K, Makishima H, Yoshida K, Townsley D, Sato-Otsubo A, Sato Y, Liu D, Suzuki H, et al. Somatic Mutations and Clonal Hematopoiesis in Aplastic Anemia. N. Engl. J. Med. 2015;373:35–47. doi: 10.1056/NEJMoa1414799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NS, Calado RT, Scheinberg P. Current concepts in the pathophysiology and treatment of aplastic anemia. Blood. 2006;108:2509–2519. doi: 10.1182/blood-2006-03-010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 2016;7:12484. doi: 10.1038/ncomms12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajkowicz A, Butkiewicz D, Drosik A, Giglok M, Suwiński R, Rusin M. Truncating mutations of PPM1D are found in blood DNA samples of lung cancer patients. Br. J. Cancer. 2015;112:1114–1120. doi: 10.1038/bjc.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, Zhao D, Liu Y, Wang C, Zhang X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–393. doi: 10.1038/nature15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Su J, Jeong M, Ko M, Huang Y, Park HJ, Guzman A, Lei Y, Huang YH, Rao A, et al. DNMT3A and TET2 compete and cooperate to repress lineage-specific transcription factors in hematopoietic stem cells. Nat. Genet. 2016;48:1014–1023. doi: 10.1038/ng.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink F, Stacey SN, Norddahl GL, Frigge ML, Magnusson OT, Jonsdottir I, Thorgeirsson TE, Sigurdsson A, Gudjonsson SA, Gudmundsson J, et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood. 2017;130:742–752. doi: 10.1182/blood-2017-02-769869. [DOI] [PMC free article] [PubMed] [Google Scholar]