Abstract
Bacterial endosymbionts of ticks are of interest due to their close evolutionary relationships with tick-vectored pathogens. For instance, whereas many ticks contain Francisella-like endosymbionts (FLEs), others transmit the mammalian pathogen Francisella tularensis. We recently sequenced the genome of an FLE present in the hard tick Amblyomma maculatum (FLE-Am) and showed that it likely evolved from a pathogenic ancestor. In order to expand our understanding of FLEs, in the current study we sequenced the genome of an FLE in the soft tick Ornithodoros moubata and compared it to the genomes of FLE-Am, Francisella persica—an FLE in the soft tick Argus (Persicargas) arboreus, Francisella sp. MA067296—a clinical isolate responsible for an opportunistic human infection, and F. tularensis, the established human pathogen. We determined that FLEs and MA067296 belonged to a sister taxon of mammalian pathogens, and contained inactivated versions of virulence genes present in F. tularensis, indicating that the most recent common ancestor shared by FLEs and F. tularensis was a potential mammalian pathogen. Our analyses also revealed that the two soft ticks (O. moubata and A. arboreus) probably acquired their FLEs separately, suggesting that the virulence attenuation observed in FLEs are not the consequence of a single acquisition event followed by speciation, but probably due to independent transitions of pathogenic francisellae into nonpathogenic FLEs within separate tick lineages. Additionally, we show that FLEs encode intact pathways for the production of several B vitamins and cofactors, denoting that they could function as nutrient-provisioning endosymbionts in ticks.
Keywords: tick, endosymbiont, Coxiella, Francisella, Coxiella-like, Francisella-like
Introduction
Ticks (order Ixodida) are ectoparasites of reptiles, birds, and mammals. Recent molecular clock estimates indicate that they originated in the Carboniferous period (∼320 million years ago, Ma) and diverged in the early Permian (∼300 Ma) into two major families: 1) Ixodidae, or hard ticks that possess a hardened chitinous scutum, and 2) Argasidae, or soft ticks that lack scutum (Jeyaprakash and Hoy 2009; Mans et al. 2016). Ticks depend on an imbalanced diet consisting entirely of vertebrate blood. Similar to endosymbiotic bacteria in insects with specialized diets—for example, Buchnera aphidicola in aphids that feed exclusively on phloem (Baumann 2005; Bennett and Moran 2015)—endosymbiotic bacteria present within tick cells are thought to compensate for the dearth of B vitamins and cofactors in blood (Smith et al. 2015; Gottlieb et al. 2015; Gerhart et al. 2016). However, unlike most primary insect endosymbionts, tick endosymbionts are not maintained within specialized cells called bacteriocytes, but are instead found in several tissues, including salivary glands (Klyachko et al. 2007; Budachetri et al. 2014), which could explain their propensity to be horizontally transmitted between hosts (Duron et al. 2017).
Ticks carry endosymbionts mostly from the genera Coxiella, Rickettsia, or Francisella. In addition to these putative primary endosymbionts (i.e. those necessary for host survival), secondary symbionts that are not strictly necessary, including Midichloria, Arsenophonus, Rickettsiella, and Wolbachia species, are found in varying rates in hard and soft tick populations (Ahantarig et al. 2013; Duron et al. 2017). Understanding the biology and evolution of tick endosymbionts is of special interest because the three primary endosymbionts share close evolutionary relationships with tick-borne pathogens Coxiella burnetii, Rickettsia parkeri, and Francisella tularensis, respectively, that cause human and animal diseases (Petersen et al. 2009; Ahantarig et al. 2013; Duron et al. 2015). Previous studies indicate that pathogen-symbiont transitions could occur in either direction: The mammalian pathogen C. burnetii probably originated from a tick-associated nonpathogenic ancestor (Smith et al. 2015; Duron et al. 2015; Moses et al. 2017), whereas the FLE in the hard tick Amblyomma maculatum (FLE-Am) likely evolved from a mammalian pathogen (Gerhart et al. 2016).
In this study, in order to better understand the evolution and functions of FLEs in ticks, we sequenced the genome of an FLE present in the soft tick Ornithodoros moubata (FLE-Om). While this project was in progress, the genome of Francisella persica—an FLE in the soft tick Argus (Persicargas) arboreus (previously referred to as Argas persicus) was published (Hinrichs et al. 2016). By comparing the genomes of FLE-Om, FLE-Am, and F. persica, we show that FLE genomes contain intact pathways for the synthesis of several B vitamins and cofactors lacking in vertebrate blood, suggesting that FLEs function as nutrient-provisioning endosymbionts in ticks. We also found that a clinical isolate (Francisella sp. MA067296), which caused an opportunistic human infection, is more closely related to FLEs than to the established human pathogen F. tularensis. Both FLEs and MA067296 contain pseudogenized versions of virulence genes present in F. tularensis, indicating that the common ancestor of FLEs and mammalian pathogens was equipped to function as a pathogen. Furthermore, FLEs and ticks do not have corresponding phylogenies, suggesting that the FLEs in the two soft ticks were derived independently, either from environmental sources or through horizontal transfer from other hosts.
FLE-Om Shares More Genome Characteristics with FLE-Am than with F. persica
Intracellular “coccoid” bacteria in the malpighian tubules and ovaries of O. moubata were first described more than four decades ago using microscopy (Reinhardt et al. 1972). Recently, 16S rDNA PCR was used to confirm that the vertically transmitted bacterium was closely related to the human pathogen F. tularensis (Noda et al. 1997). Using Illumina high-throughput sequencing, we analyzed DNA isolated from an adult female O. moubata obtained from a laboratory colony (Rego et al. 2005). Out of the ∼180 million sequencing reads, the vast majority were of host origin, with <5% of reads contributing to the assembly of the eight FLE-Om contigs (supplementary table S1, Supplementary Material online). In addition, low quantities of reads that correspond to Clostridium spp. and Burkholderia spp. were also present (<0.5% of reads). No other bacterium was detected at significant levels, including the previously described Coxiella-like “Symbiont A” (Noda et al. 1997). By screening several male and female O. moubata adults using both bacterial and Coxiella-specific 16S rDNA PCR primers, we confirmed that FLE-Om but not the Coxiella-like “Symbiont A” has been retained in the O. moubata laboratory colony. As shown previously in insects, this observation suggests that whereas FLE-Om is likely a primary endosymbiont that provides a critical function to O. moubata, the Coxiella-like “symbiont A” is a secondary symbiont or a transient bacterium (McCutcheon and Moran 2010; Hall et al. 2016).
The ∼1.56 Mb genome of FLE-Om is similar in size to that of other FLEs (FLE-Am and F. persica), but considerably smaller than the ∼1.90 Mb genome of the human pathogen F. tularensis (table 1). Concordantly, the G + C content and number of protein-coding genes are also lower in the three FLEs in comparison to F. tularensis, signifying a more intimate host association in FLEs, which leads to small, A + T-biased genomes with fewer functional genes (McCutcheon and Moran 2010). Intriguingly, while FLE-Om and FLE-Am have similar GC%, coding density, average gene length, pseudogene content, rRNA gene content, etc., these characteristics are very distinct in F. persica, indicating that its evolutionary history is different from that of the other two FLEs.
Table 1.
Genome Features of the Human Pathogen F. tularensis and FLEs
| Feature | F. tularensis | FLE-Am | F. persica | FLE-Om |
|---|---|---|---|---|
| Length (bp) | 1,892,772 | 1,556,255 | 1,540,154 | 1,564,197 |
| G+C% | 32.30 | 31.80 | 31.39 | 31.80 |
| No. of coding genes | 1,556 | 1,001 | 1,096 | 989 |
| Coding density (%) | 92 | 57 | 68 | 56 |
| Average gene length (bp) | 937 | 886 | 964 | 881 |
| No. of pseudogenes | 227 | 484 | 205 | 543 |
| No. of single copy genesa | 106/111 | 106/111 | 106/111 | 106/111 |
| 16S rDNA | 3 | 2 | 3 | 2 |
| 23S rDNA | 3 | 2 | 3 | 2 |
| 5S rDNA | 4 | 3 | 3 | 3 |
| tRNAs | 38 | 32 | 38 | 34 |
| IS elementsb | 72 | 3 | 2 | 3 |
| Accession # | NC_006570 | LNCT00000000 | NZ_CP013022 | LVCE00000000 |
Albertsen et al. (2013).
Both coding and pseudogenized versions of IS elements have been included.
Francisella persica Has Higher Virulence Potential than Other FLEs
All three FLEs encode several genes shown to be critical to the virulence of F. tularensis (Rowe and Huntley 2015; Meibom and Charbit 2010). However, in FLE-Am and FLE-Om, as expected in nutrient-provisioning endosymbionts, most virulence genes have been inactivated or appear to have been deleted (table 2, supplementary table S2, Supplementary Material online). Francisella persica, however, contains a considerably higher number of potentially functional virulence-associated genes, suggesting that this endosymbiont could be pathogenic to mammals. In fact, F. persica (previously misidentified as Wolbachia persica) was found to cause fever and death in guinea pigs (Suitor and Weiss 1961). Similarly, MA067296, a Francisella strain that caused an opportunistic human infection (Kugeler et al. 2008; Challacombe et al. 2017) also encodes a larger complement of intact virulence genes than FLE-Om and FLE-Am, suggesting that the presence of full-length virulence genes corresponds to the ability of a bacterium to cause disease in mammals. Accordingly, while it has not yet been tested, the inactivation of most virulence genes in FLE-Om and FLE-Am is assumed to have rendered these tick endosymbionts nonpathogenic to mammals.
Table 2.
Intact and Pseudogenized Virulence-Associate Genes in F. tularensis, Francisella sp. MA067296, and FLEs
| Genes |
F. tularensis |
MA067296 |
F. persica |
FLE-Am |
FLE-Om |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intacta | Φb | Intact | Φ | Intact | Φ | Intact | Φ | Intact | Φ | |
| LPS and capsule | 22 | 0 | 16 | 2 | 13 | 0 | 8 | 5 | 9 | 2 |
| Antigen synthesis | 11 | 0 | 10 | 0 | 7 | 0 | 5 | 2 | 5 | 2 |
| Type IV pili | 17 | 0 | 12 | 3 | 7 | 4 | 2 | 12 | 2 | 10 |
| Outer membrane | 8 | 0 | 8 | 0 | 7 | 0 | 6 | 2 | 5 | 3 |
| Inner membrane | 10 | 0 | 10 | 0 | 8 | 0 | 8 | 0 | 9 | 0 |
| Type VI sec. system | 17 | 0 | 16 | 1 | 16 | 0 | 2 | 6 | 3 | 9 |
Number of full-length genes.
Number of pseudogenes.
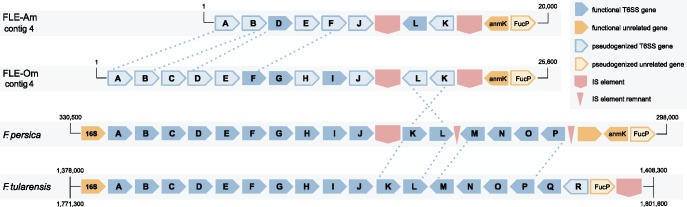
Mobile elements may have played a significant role in pathogen attenuation through the disruption and removal of virulence-associated genes. Past studies have demonstrated that association with hosts leads to a proliferation of mobile elements in bacterial genomes, followed by an extensive purge as the genome is minimized to its essentials with the development of an obligate host–bacterium relationship (Moran and Plague 2004; McCutcheon and Moran 2010). In accordance with this trend, the genome of F. tularensis, which has a less intimate relationship with its host, is replete with insertion sequences (IS), whereas the genomes of host-restricted FLEs contain very few IS elements (table 1). Type VI secretion system gene clusters in FLEs appear to have been particularly disrupted by these selfish genetic elements, as a concentration of partial and pseudogenized IS elements are found in this gene cluster. It is likely that insertion and transposition of IS elements resulted in the loss of two virulence-associated genes from F. persica and six genes from FLE-Om and FLE-Am (fig. 1).
Fig. 1.
—Mobile element invasion of Francisella Type VI secretion system gene cluster. The positions of the identified elements suggest that virulence-associated genes M-R were lost as a consequence of composite transposon excision in FLE-Am and FLE-Om. The movement of these elements also appears to have caused a 4.5 kb inversion in these species. The pictured region occurs twice in the genome of Francisella tularensis, but only once in each of the FLEs, which may also be attributable to IS duplication and transposition. Genes A-R correspond to F. tularensis FTT_1344–FTT_1361c, respectively.
FLEs May Provide B Vitamins and Cofactors to Ticks
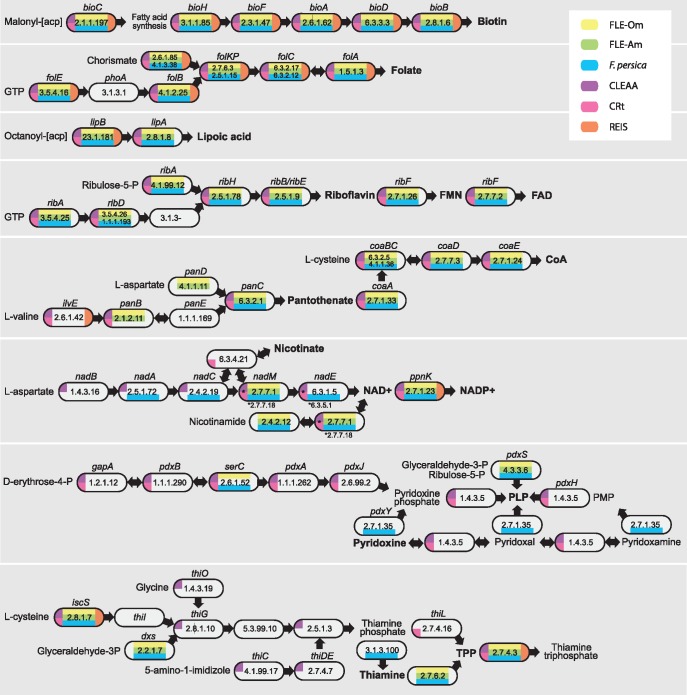
Vertebrate blood lacks several B vitamins and cofactors; therefore, vertically inherited intracellular bacteria that are present across tick lineages are thought to provide these essential nutrients to the host. We examined the metabolic pathways present in FLEs and found that they encode complete pathways for the syntheses of biotin (B7), folate (B9), lipoic acid, riboflavin (B2), and FAD (fig. 2). All lack phoA gene in the folate synthesis pathway and a phosphoric monoester hydrolase (EC 3.1.3.-) in the riboflavin pathway. However, previous studies have shown that phoA might not be required to synthesize folate (Bermingham and Derrick 2002), and these two genes have not been retained in most mutualists, suggesting that their functions could be compensated by other genes (Klein et al. 2013; Smith et al. 2015; Gottlieb et al. 2015; Manzano-Marín et al. 2015; Boyd et al. 2017). The three FLEs displayed somewhat different capabilities to synthesize the remaining B vitamins and cofactors. Whereas FLE-Am and FLE-Om could use aspartate to produce pantothenate (B5) and convert it to Coenzyme A (CoA), F. persica lacks the ability to make pantothenate but could synthesize CoA if provided with B5. Conversely, only F. persica encodes the pathway required for producing NAD+/NADP+ from aspartate (except nadB, which contains an internal stop site), whereas FLE-Am and FLE-Om could only make the two cofactors from nicotinamide. None of the FLEs has the ability to synthesize pyridoxine (B6), but they could make the cofactor PLP (pyridoxal phosphate) from glyceraldehyde-3-phosphate and ribulose-5-phosphate, which are byproducts of the pentose phosphate pathway present in the bacteria. Similarly, none of the FLEs can synthesize thiamine (B1) from either cysteine or glyceraldehyde-3-phosphate, but have the enzyme thiamine diphosphokinase to produce the cofactor TPP (thiamine pyrophosphate) from vitamin B1.
Fig. 2.
—Cofactor and B vitamin biosynthetic pathways in tick endosymbionts. Pathways for the production of B vitamins and cofactors in FLE-Om, FLE-Am, Francisella persica, CLEAA (Coxiella-like endosymbiont of Amblyomma americanum), CRt (Coxiella-like symbiont in Rhipicephalus turanicus), and REIS (Rickettsia endosymbiont of Ixodes scapularis) are shown. Gene names and EC numbers of enzymes that catalyze each step in each bacterium are shown. No color indicates that no functional copy of the gene was present. Cofactors and B vitamins are in bold.
Along with FLEs, Coxiella and Rickettsia are the two most widespread tick-associated bacteria (Duron et al. 2017). Our reconstruction of biosynthetic pathways present in the Rickettsia endosymbiont of Ixodes scapularis (REIS; Gillespie et al. 2012) showed that it only has the ability to produce biotin and folate, indicating that REIS is likely a parasite and not a nutrient-providing endosymbiont. In contrast, the two Coxiella-like endosymbionts (CLEs) have complete pathways for the synthesis of several vitamins and cofactors, including for those missing in FLEs, suggestive of their roles as primary cofactor-provisioning endosymbionts in ticks. In addition to cofactor production, FLEs—but not CLEs—have retained the heme biosynthesis pathway, and have the ability to recycle nitrogen by incorporating ammonia, a metabolic waste product, into the synthesis of glutamine, as shown in several insect endosymbionts (e.g. Zientz et al. 2006; López-Sánchez et al. 2009; Sabree et al. 2009; Hansen and Moran 2011). In sum, FLEs have the potential to supply several vitamins and cofactors that are present in very low concentrations in their hosts’ blood meals, thereby improving tick fitness and providing the impetus for ticks to maintain FLEs through generations via vertical transmission. Furthermore, unlike most other arthropod endosymbionts, F. persica has been cultured in laboratory media (Hinrichs et al. 2016), and it should be feasible to develop growth media for FLE-Am and FLE-Om using our genomic data, as previously done for other fastidious bacteria (Renesto et al. 2003; Omsland et al. 2009).
FLEs Evolved from a Pathogenic Ancestor and Soft Ticks Have Acquired FLEs Multiple Times
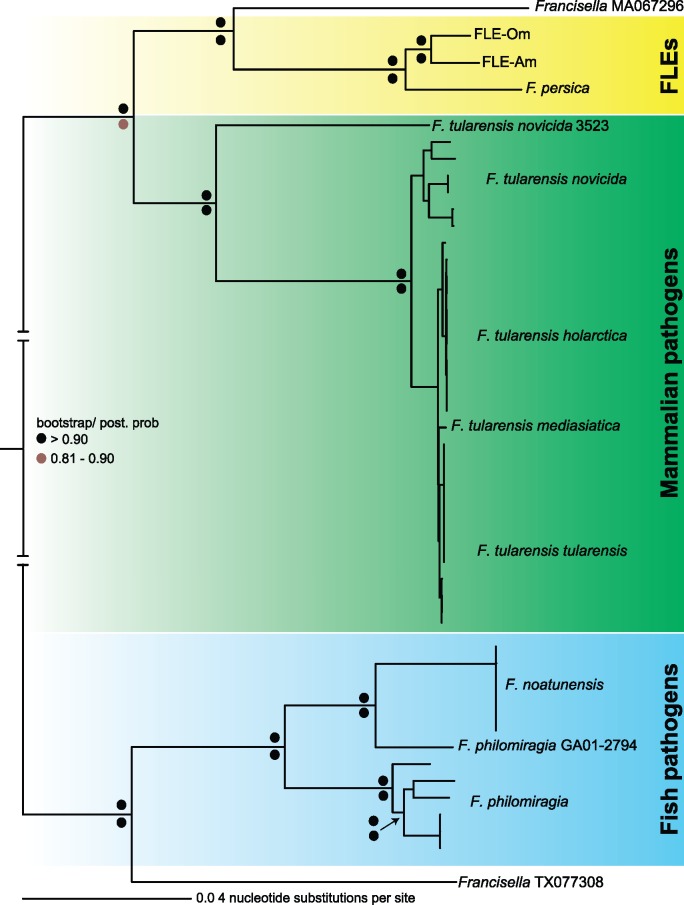
Our phylogenetic estimation using 404 genes in 49 fully sequenced Francisella genomes revealed that FLEs and the opportunistic human pathogen Francisella sp. MA067296 are closely related to each other and belong to a sister taxon of mammalian pathogens (fig. 3). This evolutionary relationship, combined with the presence of homologous virulence genes in both the FLE and mammalian-pathogen branches of the Francisella tree, indicate that the common ancestor of FLEs and F. tularensis was armed with potentially pathogenic characteristics (table 2, supplementary table S2, Supplementary Material online). The ability of MA067296 and F. persica to cause morbidity in humans and guinea pigs, respectively, further supports the conclusion that FLEs likely evolved from a pathogenic ancestor (Suitor and Weiss 1961; Kugeler et al. 2008; Challacombe et al. 2017). However, it should be noted that we cannot completely rule out the possibility that each Francisella acquired its complement of virulence genes via horizontal gene transfer or that some of the apparently pseudogenized genes in FLEs are actually functional.
Fig. 3.
—Phylogeny estimation of FLEs. A phylogenetic tree using 404 orthologous genes in 49 fully sequenced Francisella genomes is shown. Bootstrap (Maximum Likelihood) and posterior probability (Bayesian) values are shown on top and bottom, respectively, at each node as colored dots. Francisella sp. MA067296 is an opportunistic human pathogen; Francisella sp. TX077308 was isolated from seawater (Challacombe et al. 2017).
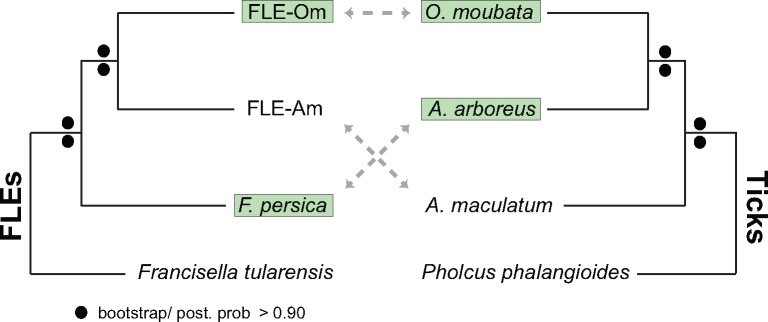
Although soft ticks O. moubata and A. arboreus host FLE-Om and F. persica, respectively, FLE-Om is more closely related to FLE-Am in the hard tick A. maculatum than to F. persica (fig. 4). This incongruence between tick and FLE phylogenies is not entirely surprising because host–endosymbiont coevolution is rare in ticks, possibly due to horizontal transfer of symbionts between unrelated tick species (Duron et al. 2015). While a few CLE and FLE strains have coevolved with their tick hosts (Duron et al. 2017; Azagi et al. 2017), a comprehensive survey of 81 species of hard and soft ticks showed that FLEs in several soft ticks were more closely related to FLEs in hard ticks than to each other, suggesting that FLEs have been exchanged between hard and soft ticks (Duron et al. 2017). On the basis of these observations, the two FLEs in soft ticks were probably acquired from independent sources: FLE-Om from a hard tick, and F. persica from an as yet unknown (possibly environmental) source. Horizontal transfer of endosymbionts is more prevalent among ticks than among insects possibly because tick endosymbionts are not restricted to specialized host cells, but instead are usually present in several tissues including in the salivary glands (Klyachko et al. 2007; Budachetri et al. 2014). Since tick-borne pathogens are typically found in salivary glands and are spread via tick saliva while the arthropod is feeding on hosts, the presence of FLEs in salivary glands is likely a holdover from their previous pathogenic lifestyle, and could facilitate their horizontal transfer to other ticks or even to other blood-feeding arthropods such as keds, while cofeeding on the same vertebrate host (Wright et al. 2015; Lee et al. 2016).
Fig. 4.
—FLEs and hosts have incongruent evolutionary histories. FLE phylogeny (left; same as in fig. 3) does not correspond to the host phylogeny (right; based on an 18S rDNA sequences). Branch lengths are not representative of evolutionary distances. Soft ticks and their FLEs are highlighted in green.
In conclusion, we show that while all three FLEs likely originated from pathogenic francisellae and could function as nutrient-provisioning endosymbionts, the FLEs in two soft ticks have dissimilar genome characteristics, virulence potentials, and evolutionary histories, indicative of independent acquisition events. The soft ticks either gained preexisting FLEs from other hosts, or acquired pathogenic Francisella strains that evolved in parallel into FLE-Om and F. persica with varying degrees of virulence loss within their respective hosts. Similar to FLEs, previous studies suggest that Sodalis-allied endosymbionts of insects probably evolved from a pathogenic progenitor, indicating that arthropods could gain new symbionts by domesticating potentially pathogenic bacteria (Clayton et al. 2012). Because of the potential to utilize insights gained from studying FLEs to control tularemia and other tick-borne diseases, it is critical that further studies are conducted to 1) delineate the evolutionary histories of all FLEs in hard and soft ticks, 2) define the evolutionary relationships between FLEs and environmental Francisella (Barns et al. 2005; Keim et al. 2007), 3) understand the functional consequences of the genetic differences observed between FLEs, and 4) validate the nutrients potentially provided by FLEs to ticks.
Materials and Methods
Sequencing, Genome Assembly, and Bacterial Identification
A female O. moubata from a laboratory colony maintained at the Institute of Parasitology, Czech Republic was used to sequence the FLE-Om genome. The tick’s outer surface was sterilized using 70% ethanol, and midgut tissue was extracted and subjected to Protease K digestion, followed by DNA extraction using a DNeasy kit and protocol (Qiagen). Purified DNA was sequenced in a single lane of Illumina Hi-Seq 2500 (100 bp, paired-end) at Oregon Health and Science University’s Massively Parallel Sequencing Shared Resource, yielding ∼180 million read pairs. Low confidence reads were removed using Trimmomatic (Bolger et al. 2014) and assembled into contigs with IDBA (Peng et al. 2012), using a k-mer range of 49:121, a step size of two, and a minimum contig length of 1 kb. All contigs were searched using BlastN against a library of Francisella genome sequences obtained from NCBI to identify putative FLE contigs. All trimmed reads were mapped to these contigs using bowtie2 and Samtools (Langmead and Salzberg 2012; Li et al. 2009). Reads that mapped to the preliminary FLE contigs along with 20% of original reads were reassembled into a final set of eight FLE-Om contigs, which have been submitted to NCBI (accession: LVCE00000000.1). The completeness of the assembled genome was examined using a single-copy gene database (Albertsen et al. 2013), yielding identical results for FLE-Om, FLE-Am, F. persica, and F. tularensis (106/111 genes). The identities of bacteria present in the tick microbiome were determined by binning BlastN results using MEGAN5’s taxonomy browser (Huson et al. 2007). To verify the absence of the Coxiella-like “Symbiont A” (Noda et al. 1997), DNA was extracted from five adult male and female ticks and PCR was performed using both bacterial and Coxiella-specific 16S rDNA primers (Klyachko et al. 2007; Klindworth et al. 2013).
Genome Annotation and Assessment of Metabolic and Virulence Potential
FLE-Om contigs were annotated using NCBI’s prokaryotic genome annotation pipeline. Cofactor synthesis pathways in FLEs, CLEs, and REIS were identified with BlastKOALA (Kanehisa et al. 2016), which maps protein sequences to the Kyoto Encyclopedia of Genes and Genomes (Manyam et al. 2015). In addition, biosynthesis genes present in F. tularensis were searched against FLE genomes using tBlastN and verified with a reciprocal BlastP of the resultant top hits against F. tularensis coding sequences. Pathways in CLEs and REIS were verified using previously published studies (Gillespie et al. 2012; Smith et al. 2015; Gottlieb et al. 2015). A list of genes critical to the pathogenicity of F. tularensis was used to identify functional and pseudogenized versions of virulence genes present in FLEs (Rowe and Huntley 2015; Meibom and Charbit 2010). Presence of IS elements was determined using ISfinder (Siguier et al. 2006).
Phylogenetic Analyses
In addition to FLE-Om, FLE-Am and F. persica, we included 46 fully sequenced Francisella genomes to generate a robust phylogenetic tree (supplementary Data set S1, Supplementary Material online). Using reciprocal BlastP, a subset of 404 orthologous genes were identified. Concatenated sequences were aligned using Clustal Omega (Sievers et al. 2014) and trimmed using Gblocks (Talavera and Castresana 2007). Tick phylogeny was built using 18 S rDNA sequences (NCBI accessions: L76344.1, L76353.1, KJ133645.1, KY016614.1). jModelTest2 was used to select the GTR + I + G evolution model for the FLE tree and HKY85 + G model for the tick tree (Darriba et al. 2012). Maximum Likelihood trees with 1,000 bootstrap replicates were constructed using RAxML (Stamatakis 2014). Bayesian trees with a chain length of 500,000 and a burn-in fraction of 25% and sampling every 100 trees were constructed using MrBayes (Huelsenbeck and Ronquist 2001). The Gammaproteobacteria cladogram (supplementary fig. S1, Supplementary Material online) is based on a previously published phylogenetic tree (Williams et al. 2010).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Colleen Campbell, Jess Millar, Todd Smith, and Jim Archuleta for technical assistance and helpful discussions. This work was supported in part by National Institutes of Health grant AI126385 to R.R.
Literature Cited
- Ahantarig A, Trinachartvanit W, Baimai V, Grubhoffer L.. 2013. Hard ticks and their bacterial endosymbionts (or would be pathogens). Folia Microbiol (Praha). 58(5):419–428. [DOI] [PubMed] [Google Scholar]
- Albertsen M, et al. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 31(6):533–538.http://dx.doi.org/10.1038/nbt.2579 [DOI] [PubMed] [Google Scholar]
- Azagi T, et al. 2017. Francisella like endosymbionts and Rickettsia species in local and imported Hyalomma ticks. Appl Environ Microbiol. 83(18): pii:AEM.01302-17. doi:10.1128/AEM.01302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns SM, Grow CC, Okinaka RT, Keim P, Kuske CR.. 2005. Detection of diverse new Francisella-like bacteria in environmental samples. Appl Environ Microbiol. 71(9):5494–5500.http://dx.doi.org/10.1128/AEM.71.9.5494-5500.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann P. 2005. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 59:155–189.http://dx.doi.org/10.1146/annurev.micro.59.030804.121041 [DOI] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 112(33):10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Derrick JP.. 2002. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays 24(7):637–648.http://dx.doi.org/10.1002/bies.10114 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120.http://dx.doi.org/10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd BM, et al. 2017. Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Mol Biol Evol. 34(7):1743–1757.http://dx.doi.org/10.1093/molbev/msx117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budachetri K, et al. 2014. An insight into the microbiome of the Amblyomma maculatum (Acari: ixodidae). J Med Entomol. 51(1):119–129.http://dx.doi.org/10.1603/ME12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challacombe JF, et al. 2017. Whole-genome relationships among Francisella bacteria of diverse origins define new species and provide specific regions for detection. Appl Environ Microbiol. 83(6):e00174-17–e00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, et al. 2012. A Novel Human-infection-derived bacterium provides insights into the evolutionary origins of mutualistic insect–bacterial symbioses. PLOS Genet. 8(11):e1002990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D.. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772..http://dx.doi.org/10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, et al. 2017. Evolutionary changes in symbiont community structure in ticks. Mol Ecol. 38:42–49. [DOI] [PubMed] [Google Scholar]
- Duron O, et al. 2015. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLOS Pathog. 11(5):e1004892.http://dx.doi.org/10.1371/journal.ppat.1004892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart JG, Moses AS, Raghavan R.. 2016. A Francisella-like endosymbiont in the Gulf Coast tick evolved from a mammalian pathogen. Sci Rep. 6:33670.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb Y, Lalzar I, Klasson L.. 2015. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome Biol Evol. 7(6):1779–1796.http://dx.doi.org/10.1093/gbe/evv108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, et al. 2012. A Rickettsia genome overrun by mobile genetic elements provides insight into the acquisition of genes characteristic of an obligate intracellular lifestyle. J Bacteriol. 194(2):376–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AAG, et al. 2016. Codivergence of the primary bacterial endosymbiont of psyllids versus host switches and replacement of their secondary bacterial endosymbionts. Environ Microbiol. 18(8):2591–2603.http://dx.doi.org/10.1111/1462-2920.13351 [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA.. 2011. Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 108(7):2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs SH, et al. 2016. Reclassification of Wolbachia persica as Francisella persica comb. nov. and emended description of the family Francisellaceae. Int J Syst Evol Microbiol. 66(3):1200–1205.http://dx.doi.org/10.1099/ijsem.0.000855 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F.. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8):754–755.http://dx.doi.org/10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC.. 2007. MEGAN analysis of metagenomic data. Genome Res. 17(3):377–386.http://dx.doi.org/10.1101/gr.5969107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash A, Hoy MA.. 2009. First divergence time estimate of spiders, scorpions, mites and ticks (subphylum: chelicerata) inferred from mitochondrial phylogeny. Exp Appl Acarol. 47(1):1–18.http://dx.doi.org/10.1007/s10493-008-9203-5 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Morishima K.. 2016. BlastKOALA and GhostKOALA: kEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 428(4):726–731. [DOI] [PubMed] [Google Scholar]
- Keim P, Johansson A, Wagner DM.. 2007. Molecular epidemiology, evolution, and ecology of Francisella. Ann NY Acad Sci. 1105:30–66.http://dx.doi.org/10.1196/annals.1409.011 [DOI] [PubMed] [Google Scholar]
- Klein CC, et al. 2013. Biosynthesis of vitamins and cofactors in bacterium-harbouring trypanosomatids depends on the symbiotic association as revealed by genomic analyses. PLoS ONE. 8(11):e79786.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, et al. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41(1):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyachko O, Stein BD, Grindle N, Clay K, Fuqua C.. 2007. Localization and visualization of a Coxiella-type symbiont within the lone star tick, Amblyomma americanum. Appl Environ Microbiol. 73(20):6584–6594.http://dx.doi.org/10.1128/AEM.00537-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugeler KJ, et al. 2008. Isolation and characterization of a novel Francisella sp. from human cerebrospinal fluid and blood. J Clin Microbiol. 46(7):2428–2431.http://dx.doi.org/10.1128/JCM.00698-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.http://dx.doi.org/10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-H, et al. 2016. Novel detection of Coxiella spp., Theileria luwenshuni, and T. ovis endosymbionts in deer keds (Lipoptena fortisetosa). PLOS ONE. 11(5):e0156727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, et al. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25(16):2078–2079.http://dx.doi.org/10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Sánchez MJ, et al. 2009. Evolutionary convergence and nitrogen metabolism in Blattabacterium strain Bge, primary endosymbiont of the cockroach Blattella germanica. PLoS Genet. 5(11):e1000721.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, et al. 2016. Ancestral reconstruction of tick lineages. Ticks Tick Borne Dis. 7(4):509–535.http://dx.doi.org/10.1016/j.ttbdis.2016.02.002 [DOI] [PubMed] [Google Scholar]
- Manzano-Marín A, Oceguera-Figueroa A, Latorre A, Jiménez-García LF, Moya A.. 2015. Solving a bloody mess: b-vitamin independent metabolic convergence among gammaproteobacterial obligate endosymbionts from blood-feeding arthropods and the leech Haementeria officinalis. Genome Biol Evol. 7(10):2871–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manyam G, Birerdinc A, Baranova A.. 2015. KPP: kEGG pathway painter. BMC Syst Biol. 9(Suppl 2):S3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 million years of evolution. Genome Biol Evol. 2:708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meibom KL, Charbit A.. 2010. The unraveling panoply of Francisella tularensis virulence attributes. Curr Opin Microbiol. 13(1):11–17.http://dx.doi.org/10.1016/j.mib.2009.11.007 [DOI] [PubMed] [Google Scholar]
- Moran NA, Plague GR.. 2004. Genomic changes following host restriction in bacteria. Curr Opin Gen Dev. 14(6):627–633.http://dx.doi.org/10.1016/j.gde.2004.09.003 [DOI] [PubMed] [Google Scholar]
- Moses AS, Millar JA, Bonazzi M, Beare PA, Raghavan R.. 2017. Horizontally acquired biosynthesis genes boost Coxiella burnetii’s physiology. Front Cell Infect Microbiol. 7:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda H, Munderloh UG, Kurtti TJ.. 1997. Endosymbionts of ticks and their relationship to Wolbachia spp. and tick-borne pathogens of humans and animals. Appl Environ Microbiol. 63(10):3926–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omsland A, et al. 2009. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci USA. 106(11):4430–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y, Leung HCM, Yiu SM, Chin FYL.. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28(11):1420–1428.http://dx.doi.org/10.1093/bioinformatics/bts174 [DOI] [PubMed] [Google Scholar]
- Petersen JM, Mead PS, Schriefer ME.. 2009. Francisella tularensis : an arthropod-borne pathogen. Vet Res. 40(2):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rego ROM, et al. 2005. Molecular cloning and comparative analysis of fibrinogen-related proteins from the soft tick Ornithodoros moubata and the hard tick Ixodes ricinus. Insect Biochem Mol Biol. 35(9):991–1004.http://dx.doi.org/10.1016/j.ibmb.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Reinhardt C, Aeschlimann A, Hecker H.. 1972. Distribution of Rickettsia-like microorganisms in various organs of an Ornithodorus moubata laboratory strain (Ixodoidea, Argasidae) as revealed by electron microscopy. Zeitschrift Fur Parasitenkd. 39(3):201–209.http://dx.doi.org/10.1007/BF00329456 [DOI] [PubMed] [Google Scholar]
- Renesto P, et al. 2003. Genome-based design of a cell-free culture medium for Tropheryma whipplei. Lancet 362(9382):447–449.http://dx.doi.org/10.1016/S0140-6736(03)14071-8 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Huntley JF.. 2015. From the Outside-In: the Francisella tularensis envelope and virulence. Front Cell Infect Microbiol. 5:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabree ZL, Degnan PH, Moran NA.. 2009. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci USA. 106(46):19521–19516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2014. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539.http://dx.doi.org/10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M.. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(90001):D32–D36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TA, Driscoll T, Gillespie JJ, Raghavan R.. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the lone star tick. Genome Biol Evol. 7(3):831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313.http://dx.doi.org/10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suitor EC, Weiss E.. 1961. Isolation of a Rickettsialike microorganism (Wolbachia persica, n. sp.) from Argas persicus (Oken). J Infect Dis. 108(1):95–106. [Google Scholar]
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577.http://dx.doi.org/10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Williams KP, et al. 2010. Phylogeny of gammaproteobacteria. J Bacteriol. 192(9):2305–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CL, Sonenshine DE, Gaff HD, Hynes WL.. 2015. Rickettsia parkeri transmission to Amblyomma americanum by cofeeding with Amblyomma maculatum (Acari: ixodidae) and potential for spillover. J Med Entomol. 52(5):1090–1095.http://dx.doi.org/10.1093/jme/tjv086 [DOI] [PubMed] [Google Scholar]
- Zientz E, Beyaert I, Gross R, Feldhaar H.. 2006. Relevance of the endosymbiosis of Blochmannia floridanus and carpenter ants at different stages of the life cycle of the host. Appl Environ Microbiol. 72(9):6027–6033.http://dx.doi.org/10.1128/AEM.00933-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.