Abstract
Establishment of multicellularity represents a major transition in eukaryote evolution. A subgroup of Amoebozoa, the dictyosteliids, has evolved a relatively simple aggregative multicellular stage resulting in a fruiting body supported by a stalk. Protosteloid amoeba, which are scattered throughout the amoebozoan tree, differ by producing only one or few single stalked spores. Thus, one obvious difference in the developmental cycle of protosteliids and dictyosteliids seems to be the establishment of multicellularity. To separate spore development from multicellular interactions, we compared the genome and transcriptome of a Protostelium species (Protostelium aurantium var. fungivorum) with those of social and solitary members of the Amoebozoa. During fruiting body formation nearly 4,000 genes, corresponding to specific pathways required for differentiation processes, are upregulated. A comparison with genes involved in the development of dictyosteliids revealed conservation of >500 genes, but most of them are also present in Acanthamoeba castellanii for which fruiting bodies have not been documented. Moreover, expression regulation of those genes differs between P. aurantium and Dictyostelium discoideum. Within Amoebozoa differentiation to fruiting bodies is common, but our current genome analysis suggests that protosteliids and dictyosteliids used different routes to achieve this. Most remarkable is both the large repertoire and diversity between species in genes that mediate environmental sensing and signal processing. This likely reflects an immense adaptability of the single cell stage to varying environmental conditions. We surmise that this signaling repertoire provided sufficient building blocks to accommodate the relatively simple demands for cell–cell communication in the early multicellular forms.
Keywords: multicellular development, transcriptome, Protostelium, Dictyostelia, evolution of development, signaling, Amoebozoa
Introduction
Multicellularity was independently acquired in different branches of eukaryotes. This transition from uni- to multicellular life styles may therefore have been relatively easy achieved compared with, for example, the rare event of organelle capture (Marin et al. 2005; Nowack et al. 2011). It is unlikely that in each case a new way to this transition to multicellularity was invented. Rather, building blocks already present in unicellular eukaryotes (and their last common ancestor; LCA) were likely recruited to this new task. During evolution, new layers of complexity (i.e., increase of the numbers of cell types) masked these building blocks (Hedges et al. 2004) so that we have difficulties to define them. Genomes of the unicellular ancestors of Metazoa seem to have been already complex. Exaptations of genes were later used for the evolution of multicellular development (Grau-Bove et al. 2017). Interestingly, Capsaspora owczarzaki, which can transit to an aggregative state, employs genes for this transition, the orthologs of which establish tissue architectures in Metazoa (Sebe-Pedros et al. 2013; Suga et al. 2013). Thus, aggregative multicellularity seems to have the same roots as true multicellularity at least in this evolutionary branch. Moreover, regulatory changes rather than gene innovations seemed to have played a major role in the establishment of multicellularity in Metazoa (Sebe-Pedros et al. 2016).
There seems to be no significant genomic difference between the unicellular algae Chlamydomonas and the multicellular Volvox species (Umen and Olson 2012). However, extracellular matrix proteins seem to be important for the establishment of multicellularity in this clade (Prochnik et al. 2010).
Each eukaryote branch or even closely related clades seem to follow its own route to multicellular development and have established their own toolkit for this purpose (Niklas and Newman 2013).
Multicellular organisms generally go through a unicellular spore or fertilized egg stage at least once, which then divides repeatedly to generate the multicellular form. In animals, plants, and fungi, the dividing cells remain attached to each other, representing clonal, multicellular organisms. However, in many other multicellular organisms, dispersed cells can come together when food runs out to construct a fruiting body with aerially borne spores. This type of aggregative multicellularity was invented independently by Acrasis spp. in the eukaryote division Discoba (Brown, Silberman, et al. 2012; He et al. 2014), Fonticula alba in Holomycota (Brown et al. 2009), Guttulinopsis spp. in Rhizaria (Brown, Kolisko, et al. 2012), Sorodiplophrys stercorea in Stramenophiles (Tice, Silberman, et al. 2016), Sorogena stoianovitchae in Alveolata (Lasek-Nesselquist and Katz 2001), and Copromyxa protea and Dictyostelium spp. in Amoebozoa (Baldauf et al. 2000; Brown et al. 2011).
Among these organisms, the genetic model system Dictyostelium discoideum and its close relatives display the most complex form of aggregative multicellularity with a freely migrating “slug” stage and four to five specialized cell types. Its genome and those of species representative of the five major clades of dictyosteliids have been sequenced (Heidel et al. 2011; Glöckner 2015; Glöckner et al. 2016) and within Amoebozoa, sequenced genomes are also available for the strictly unicellular Acanthamoeba castellani (Clarke et al. 2013) and the syncytial slime mold Physarum polycephalum (Schaap et al. 2016). The latter organism alternates between unicellular amoeboid or amoeboflagellate forms, which can either individually encyst or, after mating, transform into a multinuclear syncytium. This syncytium has the choice of forming a dehydrated but viable sclerotium, or fruiting bodies (fig. 1) with multiple spores and acellular stalks (Schaap et al. 2016).
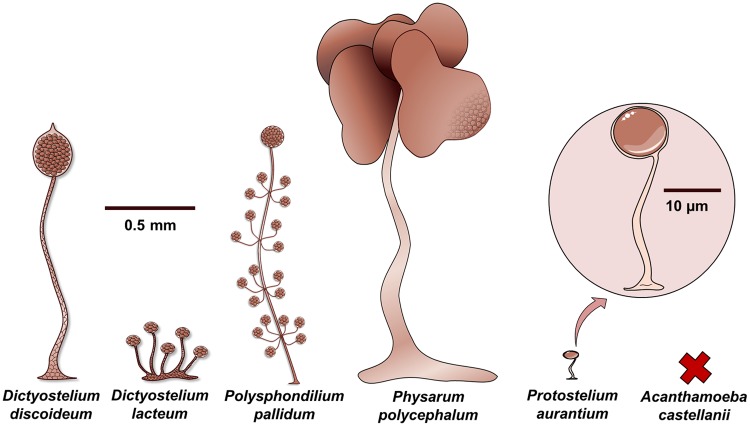
Fig. 1.
—Amoebozoa fruiting bodies. Schematic view of the different “fruiting bodies” in the Amoebozoa. Depicted are only species where a genome sequence is available including the here presented Protostelium spp. genomes. The fruiting bodies are drawn to scale, the inset shows an enlargement of the tiny Protostelium fruiting body. No fruiting body is known for Acanthamoeba castellanii, indicated by a cross. However, other Acanthamoebae do form sporocarps, as was recently discovered for A. pyriformis (Tice, Shadwick, et al. 2016).
We previously investigated the extent to which genes that are essential for multicellular development of D. discoideum are conserved throughout Dictyostelia and unicellular Amoebozoa. This study showed that genes involved in intracellular signal processing were mostly common to both multicellular and unicellular Amoebozoa, but that genes encoding membrane or secreted proteins were more unique to Dictyostelia. This suggested that novel adhesion proteins, signal, and signal sensors are prime requisites for evolving multicellularity. How these were acquired remains often elusive, but of the five known nonpeptide signals which induce cell differentiation in D. discoideum, three genes encoding their biosynthetic enzymes were likely to be acquired by Dictyostelia through lateral gene transfer (Glöckner et al. 2016). A parallel study retracing genes with conserved upregulation in multicellular development highlighted many conserved genes with likely roles in multicellular development that have not yet been functionally characterized (Schilde et al. 2016).
Multicellular development of Dictyostelia and other organisms depends not only on cell-type specialization but also on coordination of cell movement and/or cell division to generate form. This complicates identification of the mechanisms that uniquely regulate cell differentiation. Comparison of Dictyostelia with protosteloid amoebae, that lack this complication, may uniquely identify the genes involved in differentiation processes.
Fruiting bodies of diverse morphologies are widespread among the Amoebozoa, and the protosteloid type of fruiting body with a stalked spore, or very few spores, but always derived from one cell is the most common (fig. 1). A recent study presented a well resolved tree of Amoebozoa with life cycle characters mapped onto that tree (Kang et al. 2017). Species displaying protosteloid fruiting bodies were found to be scattered throughout the Amoebozoa phylogeny, but also occur within the Eumycetozoa which include the Dictyostelia and Ph. polycephalum. Recently, single cell fruiting bodies were also found to occur among the more distant Acanthamoebidae (Tice, Shadwick, et al. 2016). It is hypothesized that fruiting body formation was already present in the last common ancestor of all Amoebozoa (Kang et al. 2017), but an independent origin of fruiting body formation in different lineages is also possible (Cavalier-Smith et al. 2016).
We here compare the genome content of Protostelium aurantium var. fungivorum and Protostelium mycophagum with those of Dictyostelia, Ph. polycephalum, and Acanthamoeba castellani. We further analyze the transcriptome changes that occur in Protostelium aurantium var. fungivorum (further addressed as P. aurantium) during its transition from a feeding amoeba into a mature fruiting body. Our study suggests that fruiting body formation seemed to have evolved analogously from the similar toolbox of genes. Furthermore, despite their simple life style, Protostelium spp. already display a large variety of cell signaling genes.
Materials and Methods
Species Isolation and Maintenance
Protosteloid amoebae were maintained as spore suspensions in 86% [v/v] glycerol at −80°C. The type strain of Protostelium mycophaga Olive and Stoianovitch was obtained from the Culture Collection of Algae and Protozoa (CCAP, Argyll, United Kingdom) and from the American Type Culture Collection (ATCC, Manassas, VA). Protostelium aurantium var. fungivorum was isolated as described for protosteliids by Spiegel et al. (2006) with slight modifications. Briefly, beech leaves were cut to 1 cm2 and centrally placed on the surface of buffered wMY agar (0.002 g l−1 malt extract, 0.002 g l−1 yeast extract, 2 mM potassium phosphate buffer pH 6.4 [PB], and 15 g l−1 agar) in petri dishes. The basidiomycetous yeast Rhodotorula mucilaginosa DSM70404 was obtained from the Jena Microbial Resource collection and was streaked in a distance of 5 mm to the leaf pieces to promote the outgrowth of fungivorous protosteloid amoeba. Plates were sealed with parafilm and incubated at 22°C for 10–14 days with daily microscopic inspections for trophozoite growth or the formation of fruiting bodies. Isolation was achieved by four consecutive transfers of single amoeba or sporocarps (fruiting body of a single cell) to new petri dishes. Liquid cultures of Protostelium sp. were carried out in petridishes filled with 2 mM PB buffer (pH 6.4) and R. mucilaginosa as the only food source. Cell numbers for both organisms were determined either using a hemocytometer or an automatic cell counter (Casy TT Cell Counter, OLS Bio, Bremen, Germany).
Induction of the Developmental Cycle and Preparation of Samples for RNA Isolation
Protosteloid amoebae were cultivated in liquid cultures with successive additions of R. mucilaginosa as a food source. For P. aurantium, the number of trophozoites reached confluency at 48- to 60-h postinoculation. At this time point, the growth medium was aspirated and residual yeast cells were removed by two consecutive washes with PB. Adherent amoebae were scraped from the surface, resuspended in PB and transferred to a wMY-agar surface in 10 µl droplets. To obtain sufficient material for RNA isolation, cells from 40 10-µl droplets were pooled for each time point in one of the three biological replicates. Vegetative cells were harvested from plates directly after plating (timepoint 0 h). Starved cells were harvested after 1.5 h and nearly completed fruiting bodies were harvested from plates incubated for 8 h.
Nucleic Acids Isolation and Sequencing
Chromosomal DNA was isolated from liquid cultures of protosteloid amoebae. Adherent cells were washed in phosphate buffer and harvested by centrifugation at 200 x g which was sufficient to deplete nearly all residual yeast cells. Trophozoites were lysed in 50 mM Tris–HCl (pH 7.5) with 2% [w/v] of SDS and 0.5 mM Na2EDTA, followed by a 1-h incubation at 60°C with RNAase A (Sigma–Aldrich, Taufkirchen, Germany) and Proteinase K (Sigma–Aldrich) at final concentrations of 100 and 200 µg ml−1, respectively. Further purification steps like extractions with phenol-chloroform and precipitation with isopropanol were carried out as described (Sambrook and Russell 2001).
For the isolation of total RNA, cells were rapidly harvested from the agar surface, resuspended in 2 ml of phosphate buffer, and centrifuged for 1 min at 6,000×g. Pellets were shock-frozen in liquid N2 and stored −80°C. RNA was prepared by phenol extraction with subsequent additions of 500 µl of TRIsure (Bioline, Luckenwalde, Germany) and 200 µl of chloroform. Phase separation was achieved by centrifugation followed by an additional extraction with 500 µl of chloroform. Precipitation was as for DNA. RNA quantity and impurities were determined spectrophotometrically. Integrity of the isolated RNA was checked by agarose gel electrophoresis
DNA Sequencing, Assembly, and Mapping
Library preparation was done using Illumina’s TruSeq DNA PCR free library preparation kit following the manufacturer’s description for 550-bp insert libraries. Library quality check and quantification was done as described earlier. Sequencing was done in one lane of a HiSeq 2500 running in paired-end/2× 101 cycle/rapid mode. The reads derived from genomic DNA were assembled using abyss-pe with a kmer size of 45–65 with step increments of 2. The assembly sizes for the P. aurantium genome (excluding contaminants) was in a range from 39 (kmer 45) to 68 Mb (kmer 65). The kmer 45 assembly was chosen for further improvement after examination of contig lengths, coverage, and contribution of bacterial and fungal sequences to the assembly for both species. Specifically, the kmer45 assembly was the most contiguous of all assemblies and therefore represented the genomes best. The GC content of the protosteliid genomes was <50.1% while contaminating DNA from other sources (bacteria and the food fungus) was found to have a higher GC content. Thus, the GC content was a first indicator to readily discriminate between the Protostelium spp. genomes and contaminating sequences. Furthermore, all contigs were compared with fungal and bacterial genomes to exclude contaminating sequences from further analysis. Here, we were using BLAST analysis to identify fungal or bacterial sequences (BLASTn in case of bacteria and BLASTx for fungi). If a contig had a higher similarity to known bacterial or fungal sequences than to any Amoebozoa genome, it was excluded from the analysis. Further gap closure was done with PAGIT (http://www.sanger.ac.uk/science/tools/pagit; last accessed January 31, 2018) using default values. For assembly of the mitochondrial genome, we extended the contig using SSPACE (Boetzer et al. 2011). The resulting overlap between contig ends was then removed to yield a complete, closed circle mitochondrial genome.
RNA Sequencing
Each of the three time points of the RNAseq experiment was covered by three biological replicates. The quality and amount of RNA was initially checked using the Agilent Bioanalyzer 2100 in combination with a RNA 6000 nano kit (both Agilent technologies). The RIN (RNA integrity number) varies from 8.4 to 10 with an average of 9.44. Total RNA of 1 µg was used for library preparation using Illumina’s TruSeq RNA sample preparation kit v2 following the manufacturer’s description which included individual labeling by sequence barcodes. The libraries were again quantified and quality checked using the Bioanalyzer 2100 and DNA 7500 kit. For sequencing, libraries were pooled, individually labeled, and loaded in two lanes of a HiSeq 2500 running in single-end/51 cycle/high output mode. Data extraction was done in FastQ format using the tool bcl2FastQ v1.8.4 (provided by Illumina). Mapping of these reads to the genome was done with tophat2. The counts per gene were used to calculate differential expression using DEseq2 (Love et al. 2014) with the Bioconductor package of R.
Gene Predictions
For the annotation of the P. mycophagum genome, we used GeneMarkES in self-training mode, which uses no a priori knowledge on gene structures. For the annotation of the P. aurantium genome, we made use of the RNAseq data, which were mapped to the genome as described earlier. The mapped reads were used to define 52,801 splice sites. These data were used for the GeneMarkET program to enable a better gene prediction.
ncRNAs
For prediction of nonprotein-coding RNAs (ncRNAs), we used GORAP (www.rna.uni-jena.de/en/software; last accessed January 31, 2018), which searches for 2,474 known ncRNA families from Rfam version 12.1 (Nawrocki et al. 2015). GORAP uses improved homology search strategies based on sequence and secondary structures and was run with default settings. All predictions passed Rfam thresholds. All results and files in fasta, gff, and stockholm format are available at http://www.rna.uni-jena.de/supplements/protostelium; last accessed January 31, 2018.
Orthology and Protein Annotation
We defined the orthology between the two Protostelium genomes and to the dictyosteliid genomes using OrthoMCL (Li et al. 2003). The protein functions were evaluated by aligning the protein sequences to the protein sequence database obtained from NCBI (version from 5/2016) using BLAST. Domains were searched using InterproScan (Jones et al. 2014). The biochemical pathways were analyzed using the KAAS server (Moriya et al. 2007). All programs were run with default values.
Phylogenetic Analysis of Adenylate and Guanylate Cyclases
The output of an Interproscan of the translated Protostelium aurantium transcripts was scanned for proteins harboring Interpro domain IPR001054 for class III adenylate and guanylate cyclases. The P. aurantium proteins harboring this domain were aligned with the five Dictyostelium cyclases and a few signature Ph. polycephalum and A. castellanii cyclases using Clustal Omega with five combined iterations (Sievers and Higgins 2014). After deletion of segments that did not align unambiguously, alignments were subjected to Bayesian inference for phylogeny reconstruction using a mixed amino acid model in MrBayes software (Ronquist and Huelsenbeck 2003). Analyses were run until convergence, and trees were rooted at midpoint (Hess and De Moares Russo 2007). For the earliest diverging branches of each major clade, closest homologs were identified by BLASTp of all NCBI nonredundant sequences. These protein sequences were aligned and included in Bayesian inference of the final tree.
Results and Discussion
The Species
Protostelium mycophagum, the type strain (Olive and Stoianovitch 1969) was maintained in culture together with the fungus Rhodotorula mucilaginosa. As fruiting body formation in this P. mycophagum strain occurred at lower frequencies, and growth was comparably slow, we decided to isolate a wild strain by means of food choice selection, that is, incubating environmental samples together with Rhodotorula cultures (Materials and Methods). This way we isolated a protosteloid amoeba in sensu Spiegel (Spiegel et al. 1994, 2006; Shadwick et al. 2009) from a dead aerial beech leaf in the Jena forest. The amoeba revealed striking similarities to the type strain of Protostelium mycophagum as described previously (Olive and Stoianovitch 1960). Phylogenic analysis based on its SSU rRNA (fig. 2A) as well as diagnostic morphological traits identified the isolate as a strain of Protostelium aurantium as recently described by Shadwick et al. (2017). Due to its rapid phagocytic feeding on the basidiomycetous yeast Rhodotorula mucilaginosa on solid agar surfaces (supplementary data set S1, fig. S1A, Supplementary Material online), we coined this new isolate Protostelium aurantium var. fungivorum. A detailed taxonomic description of this isolate is available within the supplementary material of this article (supplementary data set S1, Description of Protostelium aurantium, Supplementary Material online).
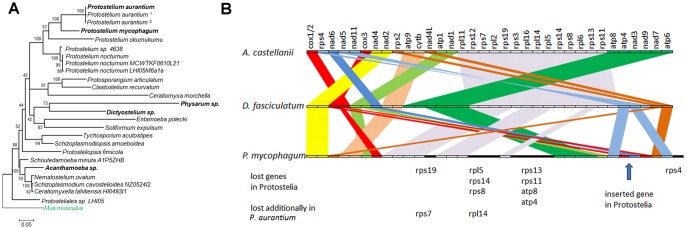
Fig. 2.
—Phylogeny and mitochondrial organization. (A) Phylogeny of selected protosteloid amoeba together with selected other amoebozoa based on 18 S RNA sequences. The species of which the genome sequences were analyzed are given in bold letters. Superscript numbers refer to recently renamed isolates of Protostelium aurantium with accession numbers 1FJ766461.1 and 2FJ766463.1. The alignment was cleaned of ambiguous positions using GBLOCKS (Talavera et al. 2007) reducing the observable distance between species and clades. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories [+G, parameter = 0.6214]). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Evolutionary analyses were conducted in MEGA6 (Tamura et al. 2013). (B) Mitochondrial synteny between selected Amoebozoa. Genes are drawn as boxes of equal length. Syntenic regions are connected by colored bands.
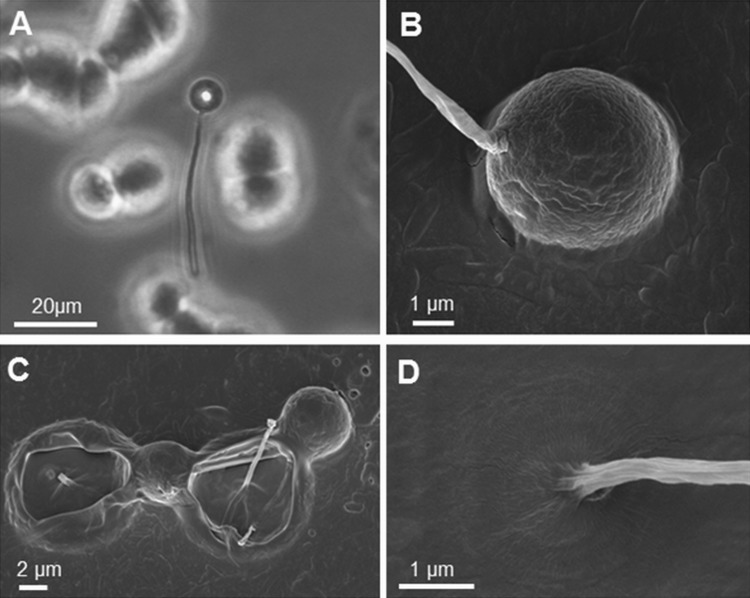
Most characteristic for its identification as a protosteloid amoeba was the abundant formation of globose fruiting bodies carrying a single spore (fig. 3A–C). The sporocarp (fruiting body of a single cell) culminated on an acellular stalk fixed on the substratum (fig. 3D). As described for P. mycophagum, asynchronous fruiting body formation occurred on solid surfaces in the absence of the fungal food source. The majority of cells initiated their full developmental cycle 2–4 h following the onset of starvation, passing through the formation of immobile discs, cellular condensation, the formation of the sporocarp, and stalk elevation, with each stage being completed in 10–15 min (supplementary data set S1, fig. S2, Supplementary Material online).
Fig. 3.
—Unicellular fruiting bodies of Protostelium aurantium. Light micrograph of a stalked fruiting body (A) and scanning electron micrographs of a tilted, but intact fruiting body (B), which will eventually open and liberate single spores (C), and the rigid base of the stalk (D).
Phylogeny of the Species and Their Genomes
For phylogenetic reconstruction, we sampled 18 S RNA sequences of Amoebozoa representatives from the NCBI database, aligned them, and performed a maximum likelihood analysis (fig. 2A). We rooted our tree with the 18 S gene from Mus musculus. The tree readily identified our environmental isolate as a member of the Protostelium aurantium clade and revealed its distant relationship to Dictyostelia and Physarum. Despite their phylogenetic distance within the Amoebozoa, all three species share the ability to construct a fruiting body. As gene expression during fruiting body formation of dictyosteliids has been particularly well studied and P. mycophagum did not sporulate readily, we focused mainly on the P. aurantium genome and expression data and its comparison to Dictyostelia.
Based on our sequencing approach, we assembled the complete mitochondrial genomes and obtained draft nuclear genomes for both Protostelium species. The mitochondrial genomes are comparable with those in dictyosteliids with respect to coding capacity and size. The order of genes differs between the two species only for one segment of genes and a few tRNA locus rearrangements (fig. 2B). Furthermore, P. aurantium lacks the genes rps7 and rpl14, located in the rearranged segment. Comparisons to other Amoebozoa mitochondrial genomes show that these additionally lost genes lie in the same region where other ribosomal subunit genes were lost in Protostelium spp. (fig. 2B). Thus, loss of such mitochondrial genes seems to be common in the Protostelium spp. analyzed.
Based on the gene prediction, both nuclear haploid genomes are comparable in size and also encode a similar number of genes (table 1). For P. aurantium, this number was further supported by RNAseq data (supplementary data set S1, fig. S4, Supplementary Material online) using GeneMarkET (Borodovsky and Lomsadze 2011). A distinguishing hallmark of both genomes is the high number of introns per gene (supplementary data set S1, fig. S5, Supplementary Material online). The median intron size is very small with 40–50 bases, but larger than the smallest intron sizes reported so far (Slabodnick et al. 2017). Overall, the Protostelium spp. genomes analyzed here harbor circa 30% more genes than Dictyostelia.
Table 1.
Genomic Features of the Two Protostelium Species
| Protostelium mycophagum | Protostelium aurantium | |
|---|---|---|
| Genome size in Mb (haploid) | 39.7 | 38.3 |
| Scaffolds | 2,578 | 822 |
| Mean GC content | 42.2 | 47.9 |
| Palindrome size (kb) | 13.4 | 16.3 |
| Mitochondrium (kb) | 48.6 | 44.5 |
| Protein coding genes | 15,691 | 17,172 |
| Mean size CDS/AA | 1,386/462 | 1,512/504 |
| tRNAs | 426 | 405 |
| Introns | 126,081 | 145,637 |
| Mean intron length | 83.2 | 63.4 |
| Median intron length | 50 | 46 |
| Introns/gene | 8 | 8.5 |
| Intron space (Mb) | 10.5 | 9.2 |
| CDS space (Mb) | 21.8 | 26 |
In line with the extensive use of introns, we found not only all small nuclear RNAs (snRNAs) of the major spliceosome but also the minor spliceosome to be present in both species. In total, a similar number of noncoding RNAs were annotated for P. mycophagum (590) and P. aurantium (525) (supplementary data set S1, table S1, Supplementary Material online).
The amount of mobile elements in P. aurantium is estimated at ∼0.13% of the genome. Although several reverse transcriptase domains of both long terminal repeat (LTR) and non-LTR retrotransposons could be determined, the corresponding elements are highly fragmented and mostly present in single copy. Only four LTR retrotransposons could be analyzed in some detail, and they belong to the Skipper and DGLT-A families of Ty3/gypsy type of LTR retrotransposons known from the dictyosteliid clade (Spaller et al. 2016).
Gene Families and Orthology Relationships
Both Protostelium spp. share among them 6,278 1:1 orthologs and 1,708 gene families with more than one member in at least one species. Protostelium mycophagum has 6,264 genes not shared with P. aurantium, whereas P. aurantium has 5,942 such orphan genes, meaning that they were also not found in other species. These numbers are somewhat comparable to the 4,156 orphan genes reported for A. castellanii (Clarke et al. 2013). Interestingly, several of the largest gene families in both species encode proteins with Leucine-rich repeats, which are often involved in protein–protein interactions. Other large gene families comprise Ankyrin repeat containing proteins, nucleotidyl cyclases, or kinases (supplementary data set S1, table S2, Supplementary Material online).
Gene Regulation during Fruiting Body Formation
Upon starvation on a solid substratum, P. aurantium cells individually differentiate into a spore supported by an acellular stalk. As P. aurantium displayed a higher frequency and abundancy of these structures relative to P. mycophagum, we chose P. aurantium to analyze the transcriptional changes during this differentiation process by RNAseq over three time points (vegetative cells at 0 h, starvation and early differentiation at 1.5 h, and the formation of prespores and mature fruiting bodies at 8 h). Although sporocarp formation was asynchronous among the population, these time points were discernible via the predominant morphology (supplementary data set S1, figs. S2 and S6A, Supplementary Material online). We first assessed which genes are potentially differentially expressed between any of those time points. The biological replicates showed little variation (supplementary data set S1, fig. S6B and C, Supplementary Material online). The DEseq2 analysis (Love et al. 2014) implemented in the BioConductor R package revealed that there are 7,787 genes significantly downregulated and 3,980 genes significantly upregulated. An example of an upregulated gene is given in supplementary data set S1, figure S7, Supplementary Material online. A significance threshold of 0.001 reduces these numbers to 6,579 and 2,685, respectively. We next analyzed which potential functions are encoded in this smaller gene set. This analysis revealed extensive reprogramming of several functions during the developmental cycle (fig. 4). A gene ontology (GO) term enrichment analysis of all upregulated genes using Gotermfinder (Boyle et al. 2004) showed that in the GO category “molecular function” ATP and GTP binding proteins are strongly overrepresented (supplementary data set S1, fig. S8, Supplementary Material online). However, no GO terms in the category “biological process” were significantly enriched.
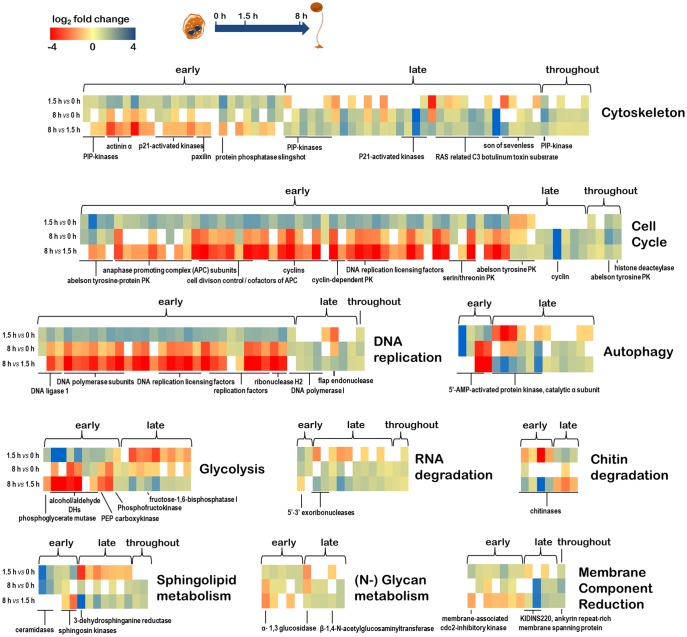
Fig. 4.
—Transcriptional reprogramming during the developmental cycle of Protostelium aurantium. Heat maps indicate higher (blue) and lower (red) expression of genes from major developmental and metabolic pathways when comparing early (0 min vs. 1.5 h, top line) or late stages (0 vs. 8 h, middle line, or 1.5 h vs. 8 h, bottom line) of development. Genes from each cluster are grouped in three temporal categories representing an upregulated expression either early, late, or throughout the developmental cycle. Heat maps illustrate the log2 expression ratio based on the mean RPKM values from three biological replicates for each time point. A more detailed table including gene accession numbers, numeric expression ratios, KEGG orthologies, and predicted protein functions are available as supplementary table S1, Supplementary Material online.
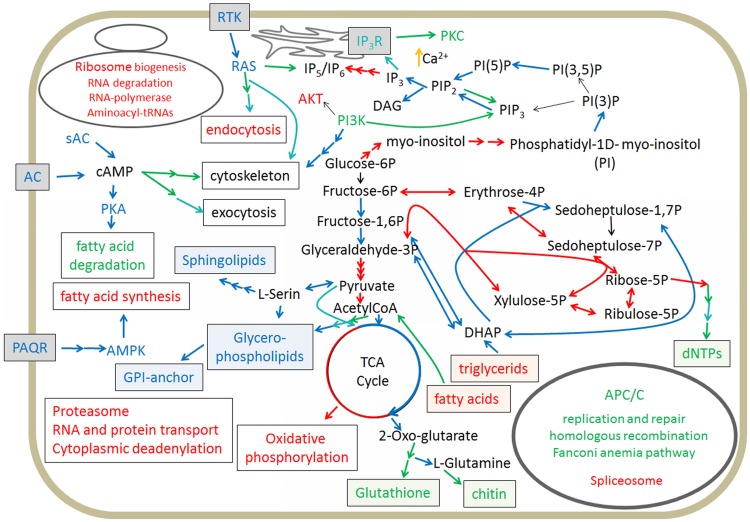
We next divided the genes into three categories (downregulation, early upregulation, and late upregulation). This only roughly approximates the real transcript dynamics since we had only three distinct developmental stages in our time course. The KEGG pathway analysis (Moriya et al. 2007) revealed that up- and downregulated genes can be part of the same pathway and concerted regulation may affect only part of a pathway (fig. 5 and supplementary table S1, Supplementary Material online). Processes related to RNA generation, fatty acid synthesis, RNA and protein transport, and oxidative phosphorylation were uniformly downregulated. This downregulation is a likely hallmark of the fact that the cells are experiencing starvation. Genes that were upregulated early tended to be involved in replication and repair, dNTP synthesis, and fatty acid degradation. These early events likely reflect the fact that the cell tries to recruit energy from fatty acids to complete the developmental cycle. In a later stage, P. aurantium devoted resources to the production of membrane components like glycerophospho- and sphingolipids, as well as chitin as a cell wall polysaccharide, indicative of the final stages of spore formation (fig. 5).
Fig. 5.
—Developmentally regulated pathways in Protostelium aurantium based on RNAseq analysis. Red indicates functions represented by genes that are downregulated upon starvation, green and blue highlight functions that are upregulated in early and late development, respectively. Presumed Calcium upregulation is depicted with a yellow arrow. Functions associated with the nucleus are listed in a circle in the right lower corner, and those with the ribosome in the upper left corner. The cytoskeleton and exocytosis are influenced by the depicted pathways, but the impact of this is not clear.
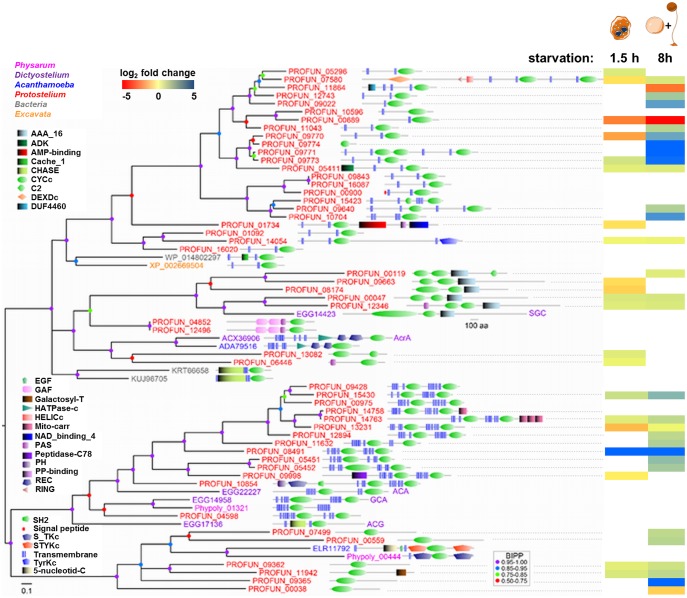
We also found that signal transduction cascades that induce cytoskeleton reorganization, survival, adhesion, and migration were upregulated early and this upregulation increased further in differentiation. The second messenger cAMP seems to play a pivotal role here since its production started early in development (supplementary data set S1, fig. S9, Supplementary Material online). Which of the numerous cyclase encoded in the genome of P. aurantium is responsible for this production or whether several cyclases are acting in a concerted way to achieve this, is currently unclear. However, one of the cyclases (PROFUN_08491) is particularly upregulated during development (fig. 6) and therefore might be responsible for the early cAMP level rise. PKA activity, which is involved in common encystation processes in Amoebozoa (Kawabe et al. 2015) is upregulated, presumably via cAMP. Throughout the differentiation process protein modification pathways (N-glycan biosynthesis) were enhanced. Interestingly, N-Glycan biosynthesis plays also a role in D. discoideum differentiation (Li et al. 2015). In P. aurantium, this pathway may be involved in production of components of the cell wall and the acellular stalk.
Fig. 6.
—Phylogeny of adenylate and guanylate cyclases. The phylogenetic tree was calculated with Bayesian inference (see Materials and Methods).The protein identifiers are color coded to reflect species names as indicated in the figure, and are annotated with the functional domain architecture of the proteins as determined by SMART (Schultz et al. 1998). PFAM domains are represented as colored to black graded rectangles. Posterior probabilities (BIPP) of tree nodes are indicated by colored dots. The heatmap indicates relative expression levels at 1.5 h (starvation) and 8 h (fruiting body formation) compared with the vegetative state. The cartoon illustrates the predominant cell types at these time points (see supplementary fig. S7, Supplementary Material online).
Comparison between Social Amoebae and P. aurantium Developmental Genes
Social amoebae have ∼60% orthologs between them as a comparative study showed (Heidel et al. 2011). Since the evolutionary distance between dictyosteliids and the protosteliids analyzed here is even higher than within the dictyosteliids it is not surprising that only 37% of the genes in P. aurantium show similarities to genes in D. discoideum, as detected by BLAST with a bit score threshold of 200. In two previous studies, we have defined two sets of dictyosteliid genes which are involved in the developmental cycle of social amoebae (Glöckner et al. 2016; Schilde et al. 2016). The set of developmentally essential (DevEs) genes contains 374 genes that upon knock-out cause a developmental defect (Glöckner et al. 2016). The set of developmentally upregulated (DevUp) genes contains 794 genes that are consistently developmentally upregulated across the Dictyostelium taxon groups (Schilde et al. 2016). In total, these two sets comprise 1,168 genes (table 2 and supplementary data set S1, fig. S11, Supplementary Material online). A higher percentage of DevEs than DevUp genes has identifiable counterparts in the P. aurantium genome (76% vs. 48%). However, of the in total 669 D. discoideum proteins with homologs in P. aurantium, 172 have the same homolog, indicating gene family expansions in D. discoideum. Strikingly, the directionality of developmental regulation of P. aurantium counterparts is often opposite (table 2). For example, statA with a major role in D. discoideum chemotaxis and stalk formation (Kawata 2011) is upregulated in D. discoideum, whereas its P. aurantium ortholog (PROFUN_03920) is slightly downregulated.
Table 2.
Gene Regulation Differences between Social Amoebae, Protostelium aurantium, and Other Amoebozoa
| Gene Set | Dictyostelium sp. |
Protostelium aurantium |
Developmental Expression in P. aurantium |
Physarum polycephalum | Acanthamoeba castellanii | ||||
|---|---|---|---|---|---|---|---|---|---|
| Development | Genes with Similarity (score >200) | % of Dictyostelium sp. Genes | Multiple Hits | Up | Down | Neutral | Genes with Similarity (score >200) | Genes with Similarity (score >200) | |
| Upregulated during development | 783 | 377 | 48 | 96 | 128 | 199 | 50 | 370 | 310 |
| Essential for development | 309 | 234 | 76 | 62 | 55 | 138 | 41 | 233 | 193 |
| Upregulated and essential | 75 | 57 | 78 | 13 | 21 | 21 | 15 | 57 | 47 |
| All | 1,167 | 668 | 57 | 171 | 204 | 358 | 106 | 660 | 550 |
Note.—About 1,167 genes of D. discoideum were identified previously as being either developmentally upregulated (Schilde et al. 2016), developmentally essential (Glöckner et al. 2016), or both (commonly identified in both studies). These genes were searched in the genomes of P. aurantium, Ph. polycephalum, and A. castellanii. To reduce spurious hits a BLAST similarity bit score of 200 was applied when comparing the encoded amino acid sequences. The column “Multiple hits” counts the occasions where different D. discoideum genes yielded the same P. aurantium hit.
Overall, not more than 75 genes were consistently identified as important for development in D. discoideum by being upregulated and essential during the developmental process (Glöckner et al. 2016; Schilde et al. 2016). However, for most of these, orthologous genes were found in the genomes of P. aurantium (57), A. castellanii (47), and Ph. polycephalum (57) (table 2). It is therefore conceivable that these genes could belong to the basic toolkit for the evolution of all differentiated forms of development among the Amoebozoa.
The set of in total 233 D. discoideum DevUp and DevEs genes with no hits in either the P. aurantium, Ph. polycephalum, and A. castellanii genomes is enriched in several GO terms, for example, “extracellular matrix organization” and “multicellular organismal process” (supplementary data set S1, fig. S10, Supplementary Material online). We next searched for genes in the 1,168 gene set, which are shared exclusively between D. discoideum and P. aurantium and are not present in A. castellanii and Ph. polycephalum. This search revealed only 13 genes with a range of putative functions (supplementary data set S1, table S3, Supplementary Material online).
Three of the four genes of Dictyostelia (chlA, dgcA, dokA, iptA), which are likely derived from horizontal gene transfer from bacteria (Glöckner et al. 2016) and which either sense (dokA) or synthesize developmental signals, are not present in the P. aurantium genome. However, two histidine kinases (PROFUN_01260 and PROFUN_15316) have the same domain structure as dokA (supplementary data set S1, fig. S13, Supplementary Material online) and therefore might encode similar functions even if they are not orthologs.
We also examined, whether transcription factors (TFs) important for the development in Dictyostelia are conserved in Protostelium spp. (table 3). Of a total of twelve TFs, orthologous proteins in P. aurantium were found for only seven and none of them were highly expressed during development. Only two TFs which were previously identified in Dictyostelia were slightly conserved only in P. aurantium (crtf and stkA) but not found in Ph. polycephalum or A. castellanii. This indicates a common root for developmental regulation for only few if any TFs and a likely loss in some lineages. Further genome sequences in the Amoebozoa are needed to trace the origin of these TFs to the LCA.
Table 3.
Developmental TFs from Dictyostelium discoideum and Presence of Orthologous Proteins in Protostelium aurantium
| Gene ID | TF | Best Hit in P. aurantium | Log2 Change in Expression at 1.5 h, 8.5 h | Comment |
|---|---|---|---|---|
| DDB_G0278077 | crtf | PROFUN_06017 | −0.42, 0.45 | Expression of aggregation genes; Dictyostelia specific |
| DDB_G0281387 | srfA | PROFUN_09032 | −1.13, n.d. | Spore differentiation; MADS box; only box similar |
| DDB_G0277589 | gtaC | PROFUN_10218 | 0.41, n.d. | pstB cell sorting and basal disc formation; GATA zinc finger; also in Acanthamoeba |
| DDB_G0278971 | dimA | n.d. | – | Development; bZIP transcription factor |
| DDB_G0291372 | dimB | n.d. | – | Development; bZIP transcription factor |
| DDB_G0279529 | bzpF | PROFUN_06600 | n.d., 0.58 | Spore viability; bZIP transcription factor; common TF |
| DDB_G0281381 | dstA | PROFUN_03920 | n.d., −0.29 | Culmination, cudA expression; STAT transcription factor; also in A. castellanii |
| PROFUN_04291 | n.d., n.d. | |||
| PROFUN_12834 | 0.64, n.d. | |||
| PROFUN_01113 | n.d., n.d. | |||
| DDB_G0277147 | stkA | PROFUN_04849 | n.d., n.d. | Spore formation; GATA zinc finger; dictyosteliid specific motif C terminal of zinc finger conserved |
| PROFUN_12502 | n.d., n.d. | |||
| DDB_G0284465 | cudA | PROFUN_14255 | n.d., −0.82 | Many prespore genes and for culmination; also in A. castellanii |
| PROFUN_08418 | n.d., n.d. | |||
| DDB_G0281563 | mybC | n.d. | – | Culmination; homology to myb domains only |
| DDB_G0275445 | mybB | n.d. | – | ACA expression; homology to myb domains only |
| DDB_G0281969 | mybE | n.d. | – | Basal disc formation; homology to myb domains only |
Note.—n.d., orthologous protein or gene expression was not detected.
Overall, and somewhat contrary to expectations based on morphological similarities, the conservation of developmental genes between Dictyostelia and P. aurantium is low and possible not more than with other unicellular amoebozoan genomes. Evolution of fruiting body formation could thus have well been independent in the two species. However, analysis of further, phylogenetically more distant, protosteloid amoeba will help to resolve this question.
Spore and Stalk Genes
The analysis described earlier comprised all known developmentally essential and conserved developmentally upregulated genes, but many of these genes are typically involved in multicellular morphogenesis which has never been observed for Protostelium sp. In the following analysis, we focused entirely on the morphogenetic process that protosteliids and dictyosteliids do have in common, the differentiation of spore(s), supported by a stalk. Although the stalk differs from that of most Dictyostelia by not containing any cells, there is also a Dictyostelium clade, the acytosteliids, which lack cells in their stalk, but nevertheless have the same stalk-specifying genes as the species with stalk cells (Urushihara et al. 2015).
About 92 genes with roles in spore and stalk formation have been identified in Dictyostelium discoideum (Glöckner et al. 2016) and we investigated whether orthologs of these genes were also present in P. aurantium, Ph. polycephalum, and A. castellanii. Genes in the three species were validated as D. discoideum orthologs when 1) the D. discoideum and species homologs were each other’s best bidirectional BLASTp hit (BBH), 2) they had the same or very similar functional domain architectures, and 3) the species genes clustered with the dictyosteliid orthologs in phylogenetic trees that also included other close homologs (see supplementary data set S2, Supplementary Material online). All D. discoideum spore and stalk genes and their best P. aurantium, Ph. polycephalum, and A. castellanii hits are listed in supplementary data set S1, table S4, Supplementary Material online, with likely orthologs indicated in bold text. A summary of the analysis is listed in table 4.
Table 4.
Conserved Dictyostelium discoideum Spore and Stalk Genes in Three Amoebozoa
| Required for | Dictyostelium discoideum | Protostelium aurantium | Physarum polycephalum | Acanthamoeba castellani |
|---|---|---|---|---|
| Sporulation | 72 | 26 | 40 | 32 |
| Stalk formation | 20 | 2 | 6 | 2 |
Of the 92 Dictyostelium genes, 28 had orthologs in P. aurantium, but a much larger number (46) had orthologs in Ph. polycephalum. Even the nonsporulating A. castellanii has slightly more (34) spore and stalk genes in common with Dictyostelium. Largely the same genes are shared between all four species, which suggests that they have a common role independent of sporulation. Their role in normal spore and stalk formation in Dictyostelium may either result from a dictyosteliid specific recruitment into such a role, or reflect a pleiotropic effect of their loss on spore or stalk formation.
Cell Signaling
Without undue emphasis on its development into fruiting bodies, we also considered the cell signaling potential of P. aurantium in its own right. Similar to most protists, P. aurantium amoebae need to sense a range of external stimuli in order to find prey, evade predators, and adapt to environmental change. To assess its sensory potential, we investigated the presence of well-known categories of cell signaling genes. A more detailed description of such genes is provided in supplementary data set S1, pages 20–30, Supplementary Material online.
G-Protein Coupled Receptors
Transmembrane receptors that interact with heterotrimeric G-proteins are the most common sensors for external signals in eukaryotes. They are subdivided into six families with family 4 only being found in fungi. Protostelium aurantium has only 17 G-protein coupled receptors (GPCRs), mostly belonging to the family 1 rhodopsin-like receptors, and completely lack the family 3 metabotropic glutamate-like receptors (supplementary data set S1, fig. S12, Supplementary Material online). This compares rather poorly with Dictyostelium, A. castellanii, and Ph. polycephalum, which have 55, 35, and 146 GPCRs, respectively (table 5). Protostelium aurantium does have 9 heterotrimeric G-proteins, more similar to Dictyostelium, which has 12 (supplementary data set S1, fig. S12, Supplementary Material online).
Table 5.
Cell Signaling Proteins in Amoebozoa Genomes
| Category | Protostelium aurantium | Dictyostelium discoideum | Physarum polycephalum | Acanthamoeba castellanii |
|---|---|---|---|---|
| G-protein coupled receptors | 17 | 55 | 146 | 35 |
| Heterotrimeric G-proteins | ||||
| Alpha | 9 | 12 | 26 | 6 |
| Beta | 1 | 1 | 1 | n.d. |
| Gamma | 1 | 1 | 1 | n.d. |
| Histidine kinases/phosphatases | 71 | 16 | 51 | 48 |
| Cyclic nucleotide signaling | ||||
| Adenylate/guanylate cyclases | 52 | 5 | 64 | 67 |
| cNMP binding domains | 27 | 5 | 28 | 7 |
| cNMP phosphodiesterases | 16 | 7 | 11 | 10 |
| Protein kinases | ||||
| All (S/T, S/T/Y, Y) | 827 | 295 | 447 | 377 |
| Sensor tyrosine kinases (Y) | 167 | 0 | 4 | 21 |
| SH2 domain proteins | 85 | 15 | 18 | 48 |
Sensor Histidine Kinases/Phosphatases
Histidine kinases/phosphatases with an attached sensor domain are used for detecting and processing a broad range of chemical and physical stimuli in both pro- and eukaryotes. The stimulus activates either phosphorylation or dephoshorylation of a conserved histidine residue in the histidine kinase domain, which respectively triggers forward or reverse histidine–aspartate–histidine relay of the phosphate to/from an aspartate in a receiver domain. This then results in activation or inactivation of an attached effector, often an enzyme or TF (Zschiedrich et al. 2016). Protostelium aurantium appears to make up for its low number of GPCRs by having an extremely large number of 71 sensor histidine kinases/phosphatases (SHKPs) (supplementary data set S1, fig. S13, Supplementary Material online). This is over four times more than D. discoideum and also exceeding Ph. polycephalum and A. castellanii (table 5). The P. aurantium SHKPs contain a variety of sensor domains, among which is also a phytochrome domain. This domain was also found in two SHKPs of Ph. polycephalum, where a phytochrome is known to mediate light-induced sporulation (Starostzik and Marwan 1995).
Cyclic Nucleotide Signaling
Many external stimuli exert their effect by modifying intracellular levels of the cyclic nucleotides (cNMPs) cAMP and cGMP. By binding to the conserved cyclic nucleotide binding domains of protein kinases, ion channels, and other effector proteins, both molecules control a broad range of cellular responses. cAMP is particularly important in the life cycle of D.discoideum where it acts both as an extracellular chemoattractant and as an intracellular messenger to regulate aggregation, morphogenesis, spore and stalk cell differentiation, and spore dormancy. In this organism, cAMP and cGMP are synthesized by three adenylate- and two guanylate cyclases, respectively, intracellularly detected by five cNMP binding proteins and hydrolyzed by seven cNMP phosphodiesterases (Saran et al. 2002; Bader et al. 2007). Query of the P. aurantium proteome revealed a much larger repertoire of cyclases, binding proteins, and phosphodiesterases.
We detected 52 nucleotidyl cyclases in the P. aurantium genome (fig. 5), which is somewhat less than the 64 and 67 cyclases detected in the Ph. polycephalum and A. castellanii genomes, respectively (table 5). However, 66 of the A. castellanii cyclases resulted from extensive duplication of a single gene that harbors a cyclase domain flanked by two protein kinase domains (identifier ELR11792 in fig. 5), whereas the remaining cyclase is a homolog of D. discoideum AcrA. The kinase-flanked cyclases also represent about half of the Ph. polycephalum cyclases, but are not present in P. aurantium. Instead, P. aurantium has many mammalian-type cyclases with two cyclase domains and two sets of six transmembrane domains. These cyclases are usually regulated by heterotrimeric G-proteins. Dictyostelium discoideum ACA and GCA also belong to this category. There are five homologs of the soluble D. discoideum guanylate cyclase SGC with two cyclase and one AAA-ATPase domain, which is implicated in chemotaxis (Saran et al. 2002). Although there are no obvious AcrA or ACG representatives, there is a large clade of 23 cyclases with mostly two transmembrane domains and a single cyclase domain. These enzymes have closest homologs in Excavates and Prokaryotes with a similar domain configuration. The two transmembrane domains may, as is the case for ACG (Saran et al. 2002), provide the cyclases with an external sensor domain.
With a total of 27 cyclic nucleotide binding proteins (cNBPs) (supplementary data set S1, fig. S14, Supplementary Material online), P. aurantium by far surpasses the 5 and 7 cNBPs of Dictyostelium and A. castellanii, respectively (table 5). Only Ph. polycephalum has one more cNBP, but has less variety in additional functional domains, which are the likely targets for regulation by either cAMP or cGMP (Schaap et al. 2016). The number of cyclic nucleotide phosphodiesterases (PDEs) is also higher in P. aurantium than in the other Amoebozoa (supplementary data set S1, fig. S15, Supplementary Material online).
Serine/Threonine and Tyrosine Protein Kinases
Protein kinases that phosphorylate other proteins on either serine/threonine (S/T) or tyrosine residues (Y) or both (S/T/Y) represent the major group of intracellular signal processing intermediates in eukaryotes with ∼518 members in humans (Hanks 2003). Protostelium aurantium has no less than 827 proteins with S/T, Y, or S/T/Y kinase domains, vastly surpassing the other publically available amoebozoan genomes, as well as humans (table 5). Eukaryotes have in general many S/T or S/T/Y kinases, but the Y specific kinases were previously considered to be present only in animals (Lim and Pawson 2010). Here, particularly the receptor tyrosine kinases play crucial developmental roles as sensors for secreted and exposed peptides that act as growth factors, controlling cell division, or differentiation inducing signals that control cell-type specialization (McDonell et al. 2015). More recent sequencing of protozoan genomes revealed that tyrosine kinases with and without intrinsic receptor domains are more widespread (Manning et al. 2008; Suga et al. 2012; Clarke et al. 2013; Schaap et al. 2016). Around 167 tyrosine kinases were detected in P. aurantium (supplementary data set S1, fig. S16, Supplementary Material online). Most are likely receptor tyrosine kinases with either protein–protein interaction or polysaccharide binding domains in their extracellular regions, suggesting roles in cell–cell recognition or adhesion. The phosphorylated tyrosines in target proteins typically act as binding sites for SH2 domains, causing proteins with SH2 domains to directly interact with the target protein. We detected 85 proteins with SH2 domains in the P. aurantium genome, also a 2- to 5-fold increase in numbers compared with the other Amoebozoa (table 5).
Conclusions
We sequenced the genomes of Protostelium aurantium and Protostelium mycophagum as the first representatives of the morphologically similar but genetically diverse protosteliids, which are characterized by forming a fruiting structure from a single amoeba consisting of a single spore or few spores on an acellular stalk.
With a size of 38 Mb and 17,000 genes, the P. aurantium genome is similar to that of A. castellanii (42 Mb, 15,000 genes), markedly smaller than the Ph. polycephalum genome (189 Mb, 29,000 genes) and larger than the Dictyostelium genomes, which range from 34 Mb, 13,000 genes (D. discoideum) to 23 Mb and 10,000 genes (D. lacteum). Although Ph. polycephalum’s large genome may reflect that this organism displays many alternative morphologies and life cycle stages (amoeba, flagellate, cyst, sporulating syncytium, or sclerotic syncytium), it is quite remarkable that the dictyosteliids with their complex multicellular life cycle and alternative abilities to form either haploid or zygotic cysts have the smallest genomes of currently sequenced amoebozoa. Apparently, the multicellular life style did not require numerically more genes. This is in agreement with analyses of the evolution of multicellularity in other eukaryote lineages (Niklas and Newman 2013; Nguyen et al. 2017). It is even conceivable that social amoebae were freed from an unknown selection pressure due to the evolution of their multicellularity. Thus, they might have been able to jettison surplus genes associated with signaling cascades and reception.
Because the single-celled P. aurantium fruiting body might be considered as a prototype for the multicellular Dictyostelium fruiting body, we had expected that a significant number of Dictyostelium developmental genes originated in the LCA of both lineages as was shown for developmental genes in Metazoa (Sebe-Pedros et al. 2013). However, contrary to this expectation, the conservation of developmentally essential Dictyostelium genes in Protostelium spp. is limited and actually not higher than for the evolutionary more distant A. castellanii. Our data and the recent finding of a sporulating Acanthamoeba species (Tice, Shadwick, et al. 2016), makes it conceivable that the entire genus Acanthamoeba could comprise at least the genomic capacity for protosteloid fruiting. It is well possible that the lack of apparent conservation in Dictyostelium discoideum could be due to the evolutionary distance between these taxa and that Dictyostelia have invented their own toolbox for multicellularity.
Using RNAseq data from discernable time points during development, we were able to dissect early and late events in fruiting body formation. The same processes might be at work in many other Amoebozoa, even if not true orthologous but analogous proteins are being utilized.
All free-living amoeba sequenced thus far do show a large repertoire of sensor histidine kinases, which in Dictyostelium regulate the activity of the intracellular cAMP phosphodiesterase RegA and thereby the activity of PKA (Loomis 2014). All amoebae genomes also contain several to many adenylate cyclases for cAMP production to activate PKA (Clarke et al. 2013; Schaap et al. 2016). In Dictyostelium, cAMP acting on PKA critically regulates the transition from growth to development, the encapsulation of spore and stalk cells and the dormancy of spores (Loomis 2014). In both Dictyostelia, which have retained the unicellular survival strategy of encystation, and in A. castellanii, cAMP acting on PKA also mediate stress-induced encystation, with RegA antagonizing this process and favoring the trophozoite stage (Du et al. 2014; Kawabe et al. 2015). Since similar to Dictyostelium (Brenner 1978), cAMP levels were also found to increase during P. aurantium development into fruiting bodies (supplementary data set S1, fig. S9, Supplementary Material online), it is likely that this is one of the core processes that is conserved between Amoebozoa, to mediate their transition into a walled dormant stage, when experiencing stress.
Another conclusion of our analyses is that despite its simple life cycle compared with Dictyostelium and Ph. polycephalum, and having a five times smaller genome than Ph. polycephalum, P. aurantium has an extremely large repertoire of proteins for the detection and processing of external stimuli. This implies that P. aurantium has many more interactions with other organisms and/or with members of its own species than is currently being realized. It is also remarkable that the multicellular Dictyostelium with its complex life cycle has both a smaller number and less variety in its signal detection and processing proteins than the unicellular Amoebozoa. Could this mean that the Dictyostelia evolved multicellularity, because they were less adaptable than other Amoebozoa? Comparisons between genomes of related uni- and multicellular organisms in other eukaryote divisions might reveal whether this is a general trend.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported in part by the Leibniz association and a grant of the European Social Fund ESF—“Europe for Thuringia” (to F.H.). S.N. was supported by the DFG-funded excellence graduate school Jena School of Microbial Communication (JSMC). We thank Ivonne Görlich and Cornelia Luge for their skillful laboratory assistance and Emanuel Barth for his assistance in file conversion and data processing. We also thank the Cologne Center for Genomics for sequencing data on P. aurantium.
Author Contributions
The study was conceptualized by F.H., T.W., and G.G. The formal analysis of data was done by T.S., M.M., G.G., P.S., G.F., F.H., M.G., S.N., M.W., I.F., and K.R. All project parts were supervised by F.H. and G.G. The original draft preparation was done by G.G. and the final manuscript review and editing by T.W., F.H., G.G., and P.S.
Literature Cited
- Bader S, Kortholt A, Van Haastert PJM.. 2007. Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP. Biochem J. 402(1):153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF.. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290(5493):972–977. [DOI] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W.. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27(4):578–579. [DOI] [PubMed] [Google Scholar]
- Borodovsky M, Lomsadze A.. 2011. Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinformatics Chapter 4:Unit 4 6 1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, et al. 2004. GO:: TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20(18):3710–3715.http://dx.doi.org/10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner M. 1978. Cyclic AMP levels and turnover during development of the cellular slime mold Dictyostelium discoideum. Dev Biol. 64(2):210–223.http://dx.doi.org/10.1016/0012-1606(78)90073-8 [DOI] [PubMed] [Google Scholar]
- Brown MW, Kolisko M, Silberman JD, Roger AJ.. 2012. Aggregative multicellularity evolved independently in the eukaryotic supergroup Rhizaria. Curr Biol. 22(12):1123–1127.http://dx.doi.org/10.1016/j.cub.2012.04.021 [DOI] [PubMed] [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW.. 2011. “Slime molds” among the Tubulinea (Amoebozoa): molecular systematics and taxonomy of Copromyxa. Protist 162(2):277–287. [DOI] [PubMed] [Google Scholar]
- Brown MW, Silberman JD, Spiegel FW.. 2012. A contemporary evaluation of the acrasids (Acrasidae, Heterolobosea, Excavata). Eur J Protistol 48(2):103–123.http://dx.doi.org/10.1016/j.ejop.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Brown MW, Spiegel FW, Silberman JD.. 2009. Phylogeny of the “forgotten” cellular slime mold, Fonticula alba, reveals a key evolutionary branch within Opisthokonta. Mol Biol Evol. 26(12):2699–2709. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE, Lewis R.. 2016. 187-gene phylogeny of protozoan phylum Amoebozoa reveals a new class (Cutosea) of deep-branching, ultrastructurally unique, enveloped marine Lobosa and clarifies amoeba evolution. Mol Phylogenet Evol. 99:275–296.http://dx.doi.org/10.1016/j.ympev.2016.03.023 [DOI] [PubMed] [Google Scholar]
- Clarke M, et al. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol. 14(2):R11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, et al. 2014. The cyclic AMP phosphodiesterase RegA critically regulates encystation in social and pathogenic amoebas. Cell Signal. 26(2):453–459.http://dx.doi.org/10.1016/j.cellsig.2013.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G. 2015. Social amoebae and their genomes: on the brink to true multicellularity In: Ruiz-Trillo I, Nedelcu AM, editors. Evolutionary transitions to multicellular life. Stuttgart: Springer. p; 363–378. [Google Scholar]
- Glöckner G, et al. 2016. The multicellularity genes of dictyostelid social amoebas. Nat Commun. 7:12085.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grau-Bove X, et al. 2017. Dynamics of genomic innovation in the unicellular ancestry of animals. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK. 2003. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 4:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D, et al. 2014. An alternative root for the eukaryote tree of life. Curr Biol. 24(4):465–470.http://dx.doi.org/10.1016/j.cub.2014.01.036 [DOI] [PubMed] [Google Scholar]
- Hedges SB, Blair JE, Venturi ML, Shoe JL.. 2004. A molecular timescale of eukaryote evolution and the rise of complex multicellular life. BMC Evol Biol. 4(1):2..http://dx.doi.org/10.1186/1471-2148-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel AJ, et al. 2011. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Res. 21(11):1882–1891.http://dx.doi.org/10.1101/gr.121137.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess PN, De Moares Russo CA.. 2007. An empirical test of the midpoint rooting method. Biol J Linn Soc. 92(4):669–674.http://dx.doi.org/10.1111/j.1095-8312.2007.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, et al. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30(9):1236–1240.http://dx.doi.org/10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, et al. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol. 34(9):2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabe Y, Schilde C, Du Q, Schaap P.. 2015. A conserved signalling pathway for amoebozoan encystation that was co-opted for multicellular development. Sci Rep. 5(1):9644..http://dx.doi.org/10.1038/srep09644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawata T. 2011. STAT signaling in Dictyostelium development. Dev Growth Differ. 53(4):548–557.http://dx.doi.org/10.1111/j.1440-169X.2010.01243.x [DOI] [PubMed] [Google Scholar]
- Lasek-Nesselquist E, Katz LA.. 2001. Phylogenetic position of Sorogena stoianovitchae and relationships within the class Colpodea (Ciliophora) based on SSU rDNA sequences. J Eukaryot Microbiol. 48(5):604–607.http://dx.doi.org/10.1111/j.1550-7408.2001.tb00197.x [DOI] [PubMed] [Google Scholar]
- Li CL, Chen G, Webb AN, Shaulsky G.. 2015. Altered N-glycosylation modulates TgrB1- and TgrC1-mediated development but not allorecognition in Dictyostelium. J Cell Sci. 128(21):3990–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS.. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189.http://dx.doi.org/10.1101/gr.1224503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Pawson T.. 2010. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 142(5):661–667.http://dx.doi.org/10.1016/j.cell.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF. 2014. Cell signaling during development of Dictyostelium. Dev Biol. 391(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550..http://dx.doi.org/10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Young SL, Miller WT, Zhai Y.. 2008. The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A. 105(28):9674–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin B, Nowack EC, Melkonian M.. 2005. A plastid in the making: evidence for a second primary endosymbiosis. Protist 156(4):425–432.http://dx.doi.org/10.1016/j.protis.2005.09.001 [DOI] [PubMed] [Google Scholar]
- McDonell LM, Kernohan KD, Boycott KM, Sawyer SL.. 2015. Receptor tyrosine kinase mutations in developmental syndromes and cancer: two sides of the same coin. Hum Mol Genet. 24(R1):R60–R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server):W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43(D1):D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, et al. 2017. Innovation and constraint leading to complex multicellularity in the Ascomycota. Nat Commun. 8:14444.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niklas KJ, Newman SA.. 2013. The origins of multicellular organisms. Evol Dev. 15(1):41–52.http://dx.doi.org/10.1111/ede.12013 [DOI] [PubMed] [Google Scholar]
- Nowack EC, et al. 2011. Endosymbiotic gene transfer and transcriptional regulation of transferred genes in Paulinella chromatophora. Mol Biol Evol. 28(1):407–422.http://dx.doi.org/10.1093/molbev/msq209 [DOI] [PubMed] [Google Scholar]
- Olive LS, Stoianovitch C.. 1960. Two new members of the Acrasiales. Bull Torrey Bot Club 87(1):1–20.http://dx.doi.org/10.2307/2483057 [Google Scholar]
- Olive SL, Stoianovitch C.. 1969. Monograph of the genus Protostelium. Am J Bot. 56(9):979–988.http://dx.doi.org/10.2307/2440919 [Google Scholar]
- Prochnik SE, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329(5988):223–226.http://dx.doi.org/10.1126/science.1188800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574.http://dx.doi.org/10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW.. 2001. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Saran S, et al. 2002. cAMP signaling in Dictyostelium – complexity of cAMP synthesis, degradation and detection. J Musc Res Cell Motil. 23(7/8):793–802. [DOI] [PubMed] [Google Scholar]
- Schaap P, et al. 2016. The Physarum polycephalum genome reveals extensive use of prokaryotic two-component and metazoan-type tyrosine kinase signaling. Genome Biol Evol. 8(1):109–125.http://dx.doi.org/10.1093/gbe/evv237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C, et al. 2016. A set of genes conserved in sequence and expression traces back the establishment of multicellularity in social amoebae. BMC Genomics 17(1):871..http://dx.doi.org/10.1186/s12864-016-3223-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP.. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 95(11):5857–5864.http://dx.doi.org/10.1073/pnas.95.11.5857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, et al. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165(5):1224–1237.http://dx.doi.org/10.1016/j.cell.2016.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, et al. 2013. Regulated aggregative multicellularity in a close unicellular relative of metazoa. Elife 2:e01287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadwick JDL, Silberman JD, Spiegel FW.. 2017. Variation in the SSUrDNA of the genus Protostelium leads to a new phylogenetic understanding of the genus and of the species concept for Protostelium mycophaga (Protosteliida, Amoebozoa). [DOI] [PubMed]
- Shadwick LL, et al. 2009. Eumycetozoa = Amoebozoa?: SSUrDNA phylogeny of protosteloid slime molds and its significance for the amoebozoan supergroup. PLoS One 4(8):0006754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Higgins DG.. 2014. Clustal omega, accurate alignment of very large numbers of sequences. Methods Mol Biol. 1079:105–116. [DOI] [PubMed] [Google Scholar]
- Slabodnick MM, et al. 2017. The macronuclear genome of stentor coeruleus reveals tiny introns in a giant cell. Curr Biol. 27(4):569–575.http://dx.doi.org/10.1016/j.cub.2016.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaller T, Kling E, Glöckner G, Hillmann F, Winckler T.. 2016. Convergent evolution of tRNA gene targeting preferences in compact genomes. Mob DNA 7(1):17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel FW, Gecks SC, Feldman J.. 1994. Revision of the genus Protostelium (Eumycetozoa) I: the Protostelium mycophaga group and the P. irregularis group. J Eukaryot Microbiol. 41(5):511–515.http://dx.doi.org/10.1111/j.1550-7408.1994.tb06051.x [Google Scholar]
- Spiegel FW, Shadwick JD, Hemmes DE.. 2006. A new ballistosporous species of Protostelium. Mycologia 98(1):144–148.http://dx.doi.org/10.1080/15572536.2006.11832721 [DOI] [PubMed] [Google Scholar]
- Starostzik C, Marwan W.. 1995. A photoreceptor with characteristics of phytochrome triggers sporulation in the true slime mould Physarum polycephalum. FEBS Lett. 370(1–2):146–148. [DOI] [PubMed] [Google Scholar]
- Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 4:2325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, et al. 2012. Genomic survey of premetazoans shows deep conservation of cytoplasmic tyrosine kinases and multiple radiations of receptor tyrosine kinases. Sci Signal. 5(222):ra35.. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice AK, et al. 2016. Expansion of the molecular and morphological diversity of Acanthamoebidae (Centramoebida, Amoebozoa) and identification of a novel life cycle type within the group. Biol Direct 11(1):69..http://dx.doi.org/10.1186/s13062-016-0171-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice AK, et al. 2016. Sorodiplophrys stercorea: another novel lineage of sorocarpic multicellularity. J Eukaryot Microbiol. 63(5):623–628.http://dx.doi.org/10.1111/jeu.12311 [DOI] [PubMed] [Google Scholar]
- Umen JG, Olson BJ.. 2012. Genomics of volvocine Algae. Adv Bot Res. 64:185–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urushihara H, et al. 2015. Comparative genome and transcriptome analyses of the social amoeba Acytostelium subglobosum that accomplishes multicellular development without germ-soma differentiation. BMC Genomics 16(1):80..http://dx.doi.org/10.1186/s12864-015-1278-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich CP, Keidel V, Szurmant H.. 2016. Molecular mechanisms of two-component signal transduction. J Mol Biol. 428:3752–3775.http://dx.doi.org/10.1016/j.jmb.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.