Abstract.
Secondary syphilis (SS) has always been puzzling for the clinicians because of the similarity of the appearance of skin rashes with other dermatoses. Serological assays are useful, but less sensitive at an early stage of SS or when patients are immunodeficient. Therefore, there is an urgent need to develop a rapid and effective tool for the diagnosis of SS, which may play an important role in the control of epidemic syphilis outbreaks. In this study, we evaluated a loop-mediated isothermal amplification (LAMP) assay, targeting gene encoding the basic membrane protein of Treponema pallidum, to detect the presence of circulating T. pallidum DNA in the blood of SS patients. The specificity of LAMP was validated using three strains of Spirochaetales and six common clinical bacteria. The clinical applicability of LAMP assay was assessed using 642 blood samples from clinically suspected SS patients and 80 samples from healthy blood donors, showing a sensitivity of 82.1% and a specificity of 100.0% in the diagnosis of SS. Thus, our results indicate that the LAMP can be used as a supplementary method for the diagnosis of SS.
INTRODUCTION
Syphilis, a reemerging sexually transmitted disease, is caused by the spirochete bacterium Treponema pallidum. The nature history of the disease consists of multistages, including primary, secondary, and latent syphilis. Among these stages, the clinical manifestations of secondary syphilis (SS), as a consequence of widespread vascular and lymphatic dissemination of the spirochete, are the most variable. Thus, syphilis has been called “the great imitator” for its uncharacteristic symptoms as it is very hard to distinguish syphilis from other dermatoses.1
Rapid and accurate diagnosis, followed by prompt and complete treatment, is essential for the effective control of syphilis. Currently, the definitive diagnosis of SS is largely based on clinical findings with support by serological methods, which can detect the specific and nonspecific antibodies. Nevertheless, the joint application of two categories of antibodies in the clinical setting, according to the reverse algorithm released by the International Union Against Sexually Transmitted Infections, has greatly enhanced the diagnosis of patients infected with T. pallidum.2 Unfortunately, serological assays cannot distinguish between treated and untreated infected patients. Another disadvantage of these antibody-based serological assays is the inability to detect the corresponding antibody in immunocompromised or immunodeficient patients. Therefore, another detection method is needed to circumvent the limitations of serological assays for syphilis. Nucleic acid amplification testing (NAAT) can directly detect the presence of pathogens and is a useful diagnostic tool. Over the past decade, it has been reported that T. pallidum DNA can be found in the blood of syphilis patients at various stages of the disease.3 Therefore, polymerase chain reaction (PCR)-based methods have been mostly developed for detecting pathogenic T. pallidum in blood samples,4–6 resulting, to a certain extent, in a progress in the diagnosis of syphilis. However, the sensitivity of PCR detection was unsatisfactory because of the low blood bacteria load, highlighting the need to increase the positive rate of NAAT.
A promising NAAT technology, called loop-mediated isothermal amplification (LAMP), may be a good approach to overcome the limitations of current molecular techniques because of its rapidity, simplicity, high sensitivity and specificity, and low cost.7 The reaction uses a DNA polymerase with strand displacement activity and multiple primers recognizing different sequences in the target DNA. The whole reaction of LAMP is usually completed within a short period of time, which is far less than that of PCR-based test. More importantly, LAMP can amplify a negligible amount of DNA to more than 109 copies.7,8 Currently, similar methods have been developed for the detection of a range of pathogens, such as Plasmodium,9 Schistosome,10 Aspergillus fumigatus,11 Mycobacterium ulcerans,12 Dengue virus,13 Didymella bryoniae,14 and Rickettsiae.15 However, the application of LAMP technology to the diagnosis of syphilis has not yet been reported.
In this study, we developed a LAMP assay targeting the basic membrane protein (bmp) gene to detect the presence of T. pallidum DNA in the peripheral blood of SS patients and preliminarily evaluated the feasibility of using this assay for the diagnosis of SS.
MATERIALS AND METHODS
Clinical samples.
Between March 2013 and June 2015, anticoagulated blood specimens (5 mL) were collected consecutively from patients suspected of having SS from hospitals and clinics throughout Hunan province, China, including Hengyang, Changsha, Chenzhou, Yueyang, Changde, and Yongzhou, to evaluate the performance of the LAMP assay for SS diagnosis. In the laboratory, the blood samples were also submitted for routine screening by T. pallidum particle agglutination (TPPA), rapid plasma reagin, and HIV test. The diagnostic criteria of SS were based on the European Center for Disease Prevention and Control guidelines.2 Those patients who had dermal manifestations and unsafe sexual contact(s) were suspected to be infected by T. pallidum. Study subjects were excluded if they had been treated with antibiotics within 1 month before collecting the blood. In addition, a panel of 80 blood samples from healthy blood donors served as the negative control. This study was approved by the Institutional Ethics Committee of the University of South China, and informed consent was obtained before enrollment.
Control strains.
Treponema pallidum Nichols strain was bestowed by Medical College of Xiamen University. DNA templates of Borrelia burgdorferi, Treponema denticola, and Leptospira interrogans were provided by Chinese Center for Disease Control and Prevention, Hunan Provincial Center for Disease Control and Prevention, and the First Affiliated Hospital, Xinjiang Medical University, respectively.
The specificity of the LAMP was evaluated by using B. burgdorferi, T. denticola, and L. interrogans (each of them is phylogenetically closed to T. pallidum) and six clinical isolates, that are commonly seen in septicemia, namely, Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. To mimic the clinical samples, all the control strains, except B. burgdorferi, T. denticola, and L. interrogans, were cultured at 37°C for 6 hours in a broth and centrifuged for 10 minutes. The pellet was suspended in sterile saline solution and vortexed. After adjusting the absorbance of the suspension to 0.1 at 450 nm, a 20-μL aliquot of suspension of each strain was added to 2 mL blood sample obtained from Hengyang Central Blood Bank (Hengyang, China), for use in LAMP and simple PCR assays.
Preparation of DNA template.
DNA was extracted from 1 mL anticoagulated blood using the Wizard Genomic DNA Purification Kit (Promega, Beijing, China) within 24 hours of collection following the manufacturer’s instructions, eluted in 50 μL deionized water, and stored at −80°C until needed. Each sample was handled separately under stringent PCR-clean conditions to avoid cross-contamination. DNA templates of the control strains were used to evaluate the specificity of the LAMP. To determine the assay sensitivity, the T. pallidum DNA concentration was measured spectrophotometrically by OD260 (DU-640, Beckman USA) and subjected to 10-fold serial dilution in sterile deionized water to yield concentrations ranging from 5.4 × 10−1 to 5.4 × 10−7 ng/μL.
LAMP primer design.
Based on our previous study,16 the T. pallidum bmp gene (GenBank accession number: M17716.1) was chosen for LAMP assay. The primer sets targeting the conserved regions of bmp were designed using Primer Explorer V4 (http://primerexplorer.jp/e/). The specificity of the chosen primers was evaluated using the Mega 4 software (available in the GenBank-EMBL database, http://www.ebi.ac.uk/) as well as the Basic Local Alignment Search Tool (available at http://blast.ncbi.nlm.nih.gov/Blast.cgi). The LAMP primers (Table 1) were synthesized by Sangon (Shanghai, China).
Table 1.
Primers for the loop-mediated isothermal amplification assay targeting the bmp gene
| Primer name | Sequence (5′→3′) |
|---|---|
| F3 | ACGCCTCCATCGTCAGAC |
| B3 | CCGAAGGGTTCAGGTCCT |
| FIP | TGCACAGGCGGGTTACTCTG-GTGGCAGTAACCGCAGTC |
| BIP | AATGTCAGCCGTGGCTTTGACA-GCGAAAGCGCAAGAGTTTG |
LAMP and simple PCR for bmp gene.
All LAMP assays were set up as 25-μL reaction mixtures using Loopamp DNA amplification kit (Eiken Chemical Co. Ltd., Tokyo, Japan). The method is based on the use of four types of primers (two inner and two outer) that recognize six distinct regions of a target gene in the presence of Bst DNA polymerase with strand-displacement activity (http://www.loopamp.eiken.co.jp/e/lamp/index.html). Briefly, 25 μL of the reaction mixture containing 4 μL of each FIB and BIP primer (equivalent to 40 pmol of each), 0.5 μL of each F3 and B3 primer (equivalent to 5 pmol of each), 12.5 μL of 2 × reaction mixture, 1 μL of Bst DNA polymerase, and 2.5 μL of prepared DNA templates was used. To determine the optimal temperature, the mixture was incubated at different temperatures ranging from 57°C to 69°C at 2°C intervals for 60 minutes, followed by enzyme inactivation at 80°C for 5 minutes. In addition, to determine the optimal amplification time, the reaction was conducted at 63°C for 15, 30, 45, 60, 75, and 90 minutes. Positive reaction containing LAMP products was observed by imaging under an ultraviolet (UV) light source after 1 μL SYBR Green (Invitrogen, Shanghai, China) was added to each reaction tube. In addition, amplicons were also evaluated by 2% agarose gel electrophoresis to observe a ladder of products.
In addition, the sensitivity of the LAMP method for T. pallidum detection was compared with an optimized simple PCR. Serial 10-fold dilutions of genomic DNA of T. pallidum Nichols strain were used as template in the amplification system. The reaction was carried out in a 50-μL reaction mixture using a Q5 High-Fidelity 2 × Master Mix kit (New England Biolabs, Beijing, China) with the following final concentrations: 1 × Master Mix, 200 μM each dNTP and 0.5 μM each primer (F3/B3). A total of 2.5 μL of DNA template was added to each reaction. The reaction conditions were as follows: initial hold at 98°C for 30 seconds, followed by 38 cycles at 98°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 5 minutes. The PCR products were electrophoresed on a 2% agarose gel together with a 100-bp DNA ladder (Invitrogen) at 100 V for 60 minutes and were visualized by UV transillumination of the ethidium bromide-stained gel.
A negative control (deionized water), positive control (T. pallidum Nichols strain), B. burgdorferi, T. denticola, L. interrogans, S. aureus, E. faecalis, E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa were included in LAMP assay and simple PCR assays to evaluate the specificity of these methods. DNA extraction, PCR, LAMP, and electrophoresis were carried out in different rooms or biological safety cabinets to prevent cross-contamination.
LAMP and simple PCR for clinical specimens.
Overall, 642 clinically suspected blood samples and 80 samples from blood donors were tested both by LAMP and simple PCR. The reaction conditions and systems of both methods were described as previously mentioned. Triplicate reactions for each test and positive (T. pallidum Nichols strain) and negative (sterile deionized water) controls were used in all experiments.
Statistical analysis.
Data were analyzed using SPSS (version 19.0) software (SPSS Inc., Chicago, IL). Evaluation of the clinical performance of the LAMP assay was performed by determining the sensitivity, specificity, positive predictive value (PPV), negative predictive values (NPV), and kappa value.
RESULTS
Patient characteristics.
The clinical diagnosis of syphilis was determined by combining the patients’ serological assays, disease history, and clinical characteristics. Overall, 603 patients were diagnosed with SS, and 10 SS-suspected patients were determined to have SS at a subsequent visit 1 month later. Eventually, a total of 613 SS patients were diagnosed definitely. Of these, 209 (34.1%, 209/613) were from males with ages ranging between 19.2 and 69 (median 34.5) years and 404 (65.9%, 404/613) were female patients with ages ranging between 16 and 53.2 years, with a median age of 26.7 years. In addition, 15 SS patients were also known to be HIV-positive. Among the 613 SS patients, 327 cases presented maculopapular exanthem, 105 macular exanthem, 81 papular exanthem, 68 condyloma lata, and 32 diffuse exanthem. Otherwise, of 642 suspected cases, 25 patients were diagnosed with pityriasis rosea, and 4 were diagnosed with psoriasis.
Optimization of reaction conditions for LAMP assay.
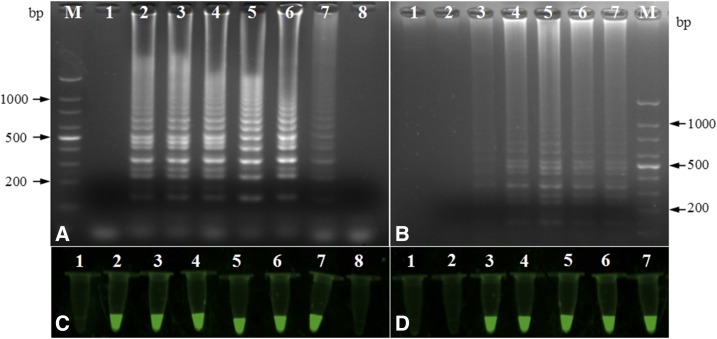
To optimize the conditions for LAMP, reaction temperatures ranging from 57°C to 69°C at 2°C intervals were compared with determine the optimal amplification temperature. The amplification efficiency was highest at 63°C (Figure 1A and C), which was therefore used in subsequent experiments. In addition, different incubation times (15, 30, 45, 60, 75, and 90 minutes) of LAMP reaction were evaluated to determine the optimal amplification time. The amplification efficiency was highest at 60 minutes (Figure 1B and D), which was therefore used in subsequent experiments.
Figure 1.
Agarose gel electrophoretic and fluorescence analysis of the loop-mediated isothermal amplification products at different temperature and time. (A and C) 57°C (lane 2), 59°C (lane 3), 61°C (lane 4), 63°C (lane 5), 65°C (lane 6), 67°C (lane 7) and 69°C (lane 8); negative control (lane 1). (B and D): 15 minutes (lane 2), 30 minutes (lane 3), 45 minutes (lane 4), 60 minutes (lane 5), 75 minutes (lane 6) and 90 minutes (lane 7); negative control (lane 1). The DNA size markers are indicated on the sides. This figure appears in color at www.ajtmh.org.
Specificity of LAMP.
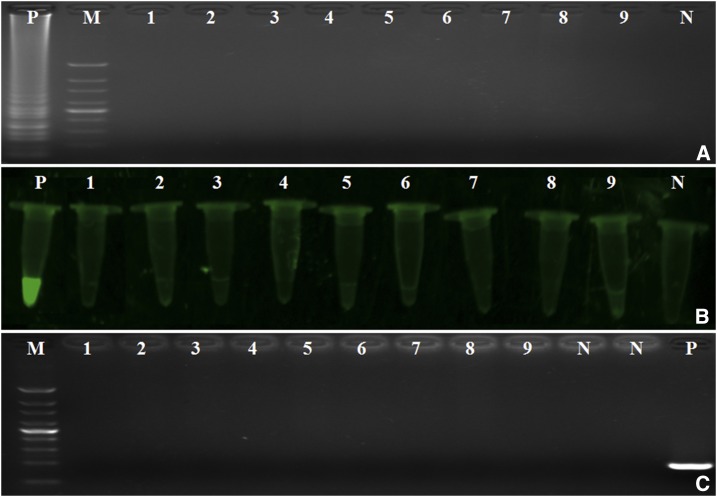
To determine the specificity of the LAMP assay, B. burgdorferi, L. interrogans, T. denticola, S. aureus, E. coli, K. pneumoniae, E faecalis, A. baumannii, and P. aeruginosa were tested for amplification. Our data demonstrate that only the T. pallidum Nichols strain was successfully amplified by LAMP and simple PCR assays without causing cross-reactivity with all other non-T. pallidum strains used in this study. These results indicate that the LAMP assay was specific for T. pallidum (Figure 2).
Figure 2.
The specificity of loop-mediated isothermal amplification (LAMP) and simple polymerase chain reaction (PCR). (A) Agarose gel electrophoretic analysis. (B) The fluorescence of LAMP reaction. (C) Agarose gel electrophoretic analysis of simple PCR of various bacteria strains. Lane 1 = Borrelia burgdorferi; lane 2 = Leptospira interrogans; lane 3 = Treponema denticola; lane 4 = Staphylococcus aureus; lane 5 = Enterococcus faecalis; lane 6 = Escherichia coli; lane 7 = Klebsiella pneumoniae; lane 8 = Acinetobacter baumannii; lane 9 = Pseudomonas aeruginosa; N = negative control (deionized water); P = positive control (T. pallidum Nichols strain); M = 100-bp DNA ladder. This figure appears in color at www.ajtmh.org.
Sensitivity of LAMP.
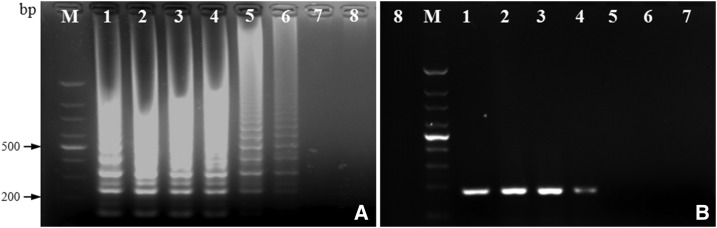
Using 2.5 μL of 10-fold serial dilutions of T. pallidum Nichols DNA as the template for LAMP and simple PCR, the limits of detection of LAMP and simple PCR for the bmp gene were 5.4 × 10−6 and 5.4 × 10−4 ng/μL, respectively (Figure 3). Thus, the sensitivity of LAMP assay is 100 higher than simple PCR for detecting T. pallidum.
Figure 3.
Agarose gels showing the detection sensitivity of loop-mediated isothermal amplification (LAMP) and polymerase chain reaction (PCR) for detecting Treponema pallidum Nichols DNA. (A) 2.0% agarose gel of LAMP products. (B) 2.0% agarose gel of simple PCR amplification products. Lane 1 = 5.4 × 10−1 ng/μL; lane 2 = 5.4 × 10−2 ng/μL; lane 3 = 5.4 × 10−3 ng/μL; lane 4 = 5.4 × 10−4 ng/μL; lane 5 = 5.4 × 10−5 ng/μL; lane 6 = 5.4 × 10−6 ng/μL and lane 7 = 5.4 × 10−7 ng/μL; lane 8 = negative control.
Examination of clinical samples.
Of the 613 specimens from SS patients, 503 cases were positive by LAMP (82.1%, 503/613) compared with 270 by simple PCR (44.0%, 270/613). The positive rate of LAMP assay was significantly higher than that of simple PCR (χ2 = 103.358, P < 0.001). While 80 blood specimens from healthy donors and 29 from non-syphilis patients (25 cases with pityriasis rosea and 4 with psoriasis) were negative by LAMP or simple PCR. The sensitivity, specificity, PPV, NPV, and corresponding kappa value of LAMP in the SS and non-SS groups were 82.1% (95% confidence interval [CI] = 78.8–84.9), 100.0% (95% CI = 96.6–100.0), 100.0% (95% CI = 99.3–100.0), 49.8% (95% CI = 43.0–56.6), and 0.58, respectively (Table 2). In addition, the results were also compared with those of the TPPA, which is generally accepted as the confirmatory treponemal test. The sensitivity, specificity, PPV, NPV, and kappa value of LAMP were 81.8%, 91.6%, 98.0%, 49.8%, and 0.54, respectively. By contrast, those indexes for simple PCR were 43.4%, 93.3%, 97.0%, 24.6%, and 0.17, respectively (Table 3). Among 613 samples from SS patients with various symptoms, LAMP was positive in 82.9% (271/327) of patients with maculopapular exanthem, 80.0% (84/105) of patients with macular exanthem, 79.0% (64/81) of patients with papular exanthem, 85.3 (58/68) of patients with condyloma lata, and 81.3% (26/32) of patients with diffuse exanthem (Table 4). The detection rates of LAMP assay did not vary during the various symptomatic phases (χ2 = 1.458, P > 0.05).
Table 2.
Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for loop-mediated isothermal amplification (LAMP) and simple polymerase chain reaction (PCR) in secondary syphilis (SS) and non-SS groups
| Group | Number of SS cases (N = 613) | Number of non-SS cases (N = 109) | %Sensitivity (95% CI) | %Specificity (95% CI) | %PPV (95% CI) | %NPV (95% CI) | κ value | Agreement | |
|---|---|---|---|---|---|---|---|---|---|
| LAMP | + | 503 | 0 | 82.1 (78.8–84.9) | 100.0 (96.6–100.0) | 100.0 (99.3–100.0) | 49.8 (43.0–56.6) | 0.58 | 84.8 (82.4–87.1) |
| − | 110 | 109 | – | – | – | – | – | – | |
| PCR | + | 270 | 0 | 44.0 (40.2–48.0) | 100.0 (96.6–100.0) | 100.0 (98.6–100.0) | 24.1 (20.2–28.3) | 0.19 | 52.5 (49.4–55.6) |
| − | 343 | 109 | – | – | – | – | – | – |
CI = confidence interval.
Table 3.
Comparison of loop-mediated isothermal amplification (LAMP) and simple polymerase chain reaction (PCR) to the Treponema pallidum particle agglutination (TPPA) using blood from secondary syphilis and non-syphilis groups
| Group | TPPA positive | TPPA negative | %Sensitivity (95% CI) | %Specificity (95% CI) | %PPV (95% CI) | %NPV (95% CI) | κ value | Agreement |
|---|---|---|---|---|---|---|---|---|
| LAMP positive | 493 | 10 | 81.8 (78.5–84.6) | 91.6 (85.2–95.4) | 98.0 (96.4–99.0) | 49.8 (43.0–56.6) | 0.55 | 83.4 (80.9–85.9) |
| LAMP negative | 110 | 109 | ||||||
| PCR positive | 262 | 8 | 43.4 (39.5–47.4) | 93.3 (87.3–96.6) | 97.0 (94.3–98.7) | 24.6 (20.7–28.8) | 0.17 | 51.7 (48.6–54.8) |
| PCR negative | 341 | 111 |
CI = confidence interval; PPV = positive predictive value; NPV = negative predictive values.
Table 4.
Comparison of 613 samples from secondary syphilis patients with various symptoms by loop-mediated isothermal amplification (LAMP) and simple polymerase chain reaction (PCR)
| Symptoms | Number of samples | LAMP | Simple PCR | ||
|---|---|---|---|---|---|
| Positive (%) | Negative (%) | Positive (%) | Negative (%) | ||
| Maculopapular | 327 | 271 (82.9) | 56 (17.1) | 142 (43.4) | 185 (56.6) |
| Macular | 105 | 84 (80.0) | 21 (20.0) | 42 (40.8) | 61 (59.2) |
| Papular | 81 | 64 (79.0) | 17 (21.0) | 38 (46.9) | 43 (53.1) |
| Condyloma lata | 68 | 58 (85.3) | 10 (14.7) | 35 (51.5) | 33 (48.5) |
| Diffuse exanthem | 32 | 26 (81.3) | 6 (18.7) | 14 (43.8) | 18 (56.2) |
DISCUSSION
Occurring within 4–10 weeks of the initial infection, SS is characterized by a variety of dermal manifestations and is difficult to distinguish from other dermatoses because of the lack of specific clinical symptoms. Rapid and accurate laboratory diagnostic methods are essential in controlling the spread of syphilis. However, the most reliable diagnostic approaches for SS at present largely depend on the results of laboratory tests, including nontreponemal and treponemal serological assays, which greatly enhanced the diagnostic rate with the application of reverse algorithm to the clinical laboratory.2,17 However, it is well known that the antigen-based methods have limitations, of which, the most important is their inability to distinguish between recent and remote syphilis, or between treated and untreated infected patients. Moreover, PCR-based methods demonstrate a satisfactory sensitivity when used in chancre and cutaneous lesion specimens,4–6,18,19 but low detection rates in blood samples.3–6 Therefore, together with the need for specialized and costly instrumentation, the low sensitivity of T. pallidum-PCR assays has hindered their further application in routine laboratory.
A previous in vitro study demonstrated that the three common genes, polA, tpp47, and bmp, allowed positive detection when used as target genes to amplify the T. pallidum DNA in blood specimens from syphilis patients.16 Designing LAMP primers for polA and tpp47 might be unadvisable because the former is rich in cysteine codons20 while the latter encodes a penicillin-binding protein that can eventually develop mutants under antibiotics selective pressure.21 Therefore, the bmp gene was selected as the target gene for the LAMP assay in the present study. This gene encodes a basic outer membrane protein that has hydrophilic property and can induce a humoral immune response in the host at different stages of disease progression.22 In addition, whole blood specimens were used because of better detection of spirochetes than in serum.4,6,23,24
The LAMP assay is a potentially useful alternative to the existing molecular method, which is time consuming and requires sophisticated equipment.25 In this study, LAMP tests reacted specifically with T. pallidum Nichols strain but showed no cross-reactivity with other non-T. pallidum bacteria, demonstrating the specificity of LAMP assay. Furthermore, comparison of the sensitivity of LAMP and simple PCR using 10-fold serial dilutions of DNA template from T. pallidum Nichols strain showed that LAMP assay was 100 times more sensitive than simple PCR. To validate the LAMP methodology for the diagnosis of SS, LAMP targeting bmp gene, as well as simple PCR, was performed on clinical samples. The results suggested that LAMP was better than simple PCR in diagnosing SS. Despite the relative low NPV (49.8%), the κ value of LAMP reached 0.58, which showed a moderate agreement. In addition, ten specimens from SS patients were positive by LAMP, but negative by serological tests. Therefore, we hypothesized that these patients may have been in the very early stage of the disease. Moreover, when the 613 patients were stratified into different groups based on the symptoms of SS, the results of LAMP assay showed no significant difference, which indicated that the bacterial load in the blood of SS patients was not associated with the clinical manifestations.
Admittedly, there are some shortcomings in the present study. First, more bacterial strains, including T. pallidum subsp. pertenue, T. pallidum subsp. endemicum, and T. paraluiscuniculi, must be evaluated to determine the specificity of LAMP assay. Second, additional work need to be performed to quantify the sensitivity of the LAMP assay, i.e., to detect the lower limit of detection as expresses as copies per reaction. Third, the advantage of the present assay in monitoring the therapeutic effect was not examined because no effective monitoring of the patients could be ensured. Most importantly, more extensive testing of a larger number of suitable blood samples collected from individuals infected with other pathogens, and individuals with SS and at other stages of the disease need to be performed to further validate the clinical utility of LAMP assay. Moreover, on account of the diagnostic challenges for diseases with low pathogen burden, the strategies for template preparation also need to be further improved in the future. Although the LAMP assay showed only a moderate agreement with TPPA and cannot replace serological testing at this time, the LAMP assay could play an important role for the management of patients with SS combined with serological assays, especially in those high-prevalence, resource-limited regions because of its rapidity, simplicity, high sensitivity, specificity, and especially no need for specialized equipment such as a thermal cycler.
Acknowledgments:
We are grateful to Tianci Yang of the Zhongshan Hospital, Medical College of Xiamen University, Xiamen, China, for the gift of the T. pallidum Nichols strain. We also thank Kanglin Wan of the Chinese Center for Disease Control and Prevention, Beijing, China, Liang Cai of the Hunan Provincial Center for Disease Control and Prevention, Changsha, China, and Xianfei Liu of the Department of Stomatology, First Affiliated Hospital, Xinjiang Medical University, Urumqi, China, for the gift of DNA template of B. burgdorferi, L. interrogans, and T. denticola, respectively; Yanjun Li of The First Affiliated Hospital, University of South China, Jinhong Xiao of the Hunan Provincial People’s Hospital, Changsha, China, Hongliang Chen of The First People’s Hospital of Chenzhou, Chenzhou, China, Li Zhang of The First People’s Hospital of Changde, Changde, China, Dawei Liu of The First People’s Hospital of Yueyang, Yueyang, China, Hua Wu of the Hengyang Central Hospital, Hengyang, China, Xuefeng Peng of the Changsha Central Hospital, Changsha, China, and Zhijun Jiang of the Yongzhou Central Hospital, Yongzhou, China, for providing SS patients’ blood samples and Rong Duan of Hengyang Central Blood Station for providing healthy blood samples for this study.
REFERENCES
- 1.Balagula Y, Mattei PL, Wisco OJ, Erdag G, Chien AL, 2014. The great imitator revisited: the spectrum of atypical cutaneous manifestations of secondary syphilis. Int J Dermatol 53: 1434–1441. [DOI] [PubMed] [Google Scholar]
- 2.French P, Gomberg M, Janier M, Schmidt B, van Voorst Vader P, Young H, IUST, 2009. IUSTI: 2008 European guidelines on the management of syphilis. Int J STD AIDS 20: 300–309. [DOI] [PubMed] [Google Scholar]
- 3.Marfin AA, Liu H, Sutton MY, Steiner B, Pillay A, Markowitz LE, 2001. Amplification of the DNA polymerase I gene of Treponema pallidum from whole blood of persons with syphilis. Diagn Microbiol Infect Dis 40: 163–166. [DOI] [PubMed] [Google Scholar]
- 4.Grange PA, et al. 2012. Evaluation of a PCR test for detection of Treponema pallidum in swabs and blood. J Clin Microbiol 50: 546–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin IE, Tsang RS, Sutherland K, Tilley P, Read R, Anderson B, Roy C, Singh AE, 2009. Molecular characterization of syphilis in patients in Canada: azithromycin resistance and detection of Treponema pallidum DNA in whole-blood samples versus ulcerative swabs. J Clin Microbiol 47: 1668–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gayet-Ageron A, Lautenschlager S, Ninet B, Perneger TV, Combescure C, 2013. Sensitivity, specificity and likelihood ratios of PCR in the diagnosis of syphilis: a systematic review and meta-analysis. Sex Transm Infect 89: 251–256. [DOI] [PubMed] [Google Scholar]
- 7.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T, 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28: E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomita N, Mori Y, Kanda H, Notomi T, 2008. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 3: 877–882. [DOI] [PubMed] [Google Scholar]
- 9.Pöschl B, Waneesorn J, Thekisoe O, Chutipongvivate S, Karanis P, 2010. Comparative diagnosis of malaria infections by microscopy, nested PCR, and LAMP in northern Thailand. Am J Trop Med Hyg 83: 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamburger J, Abbasi I, Kariuki C, Wanjala A, Mzungu E, Mungai P, Muchiri E, King CH, 2013. Evaluation of loop-mediated isothermal amplification suitable for molecular monitoring of schistosome-infected snails in field laboratories. Am J Trop Med Hyg 88: 344–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Q, Tian S, Yu N, Zhang X, Jia X, Zhai H, Sun Q, Han L, 2016. Development and evaluation of a loop-mediated isothermal amplification method for rapid detection of Aspergillus fumigatus. J Clin Microbiol 54: 950–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Souza DK, Quaye C, Mosi L, Addo P, Boakye DA, 2012. A quick and cost effective method for the diagnosis of Mycobacterium ulcerans infection. BMC Infect Dis 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Li X, Mo Z, Jin F, Wang B, Zhao H, Shan X, Shi L, 2012. Rapid identification of Chikungunya and Dengue virus by a real-time reverse transcription-loop-mediated isothermal amplification method. Am J Trop Med Hyg 87: 947–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao X, Li P, Xu J, Zhang M, Ren R, Liu G, Yang X, 2016. Rapid and sensitive detection of Didymella bryoniae by visual loop-mediated isothermal amplification assay. Front Microbiol 7: 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan L, Zhang L, Wang G, Liu Q, 2012. Rapid, simple, and sensitive detection of the ompB gene of spotted fever group rickettsiae by loop-mediated isothermal amplification. BMC Infect Dis 12: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao Y, Liu S, Liu Z, Xie Y, Jiang C, Xu M, Zhao F, Zeng T, Yu J, Wu Y, 2016. Molecular subtyping and surveillance of resistance genes in Treponema pallidum DNA from patients with secondary and latent syphilis in Hunan, China. Sex Transm Dis 43: 310–316. [DOI] [PubMed] [Google Scholar]
- 17.Loeffelholz MJ, Binnicker MJ, 2012. It is time to use Treponema-specific antibody screening tests for diagnosis of syphilis. J Clin Microbiol 50: 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai T, Li K, Lu H, Gu X, Wang Q, Zhou P, 2012. Molecular typing of Treponema pallidum: a 5-year surveillance in Shanghai, China. J Clin Microbiol 50: 3674–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouznetsov AV, Weisenseel P, Trommler P, Multhaup S, Prinz JC, 2005. Detection of the 47-kilodalton membrane immunogen gene of Treponema pallidum in various tissue sources of patients with syphilis. Diagn Microbiol Infect Dis 51: 143–145. [DOI] [PubMed] [Google Scholar]
- 20.Rodes B, Liu H, Johnson S, George R, Steiner B, 2000. Molecular cloning of a gene (poIA) coding for an unusual DNA polymerase I from Treponema pallidum. J Med Microbiol 49: 657–667. [DOI] [PubMed] [Google Scholar]
- 21.Cha JY, Ishiwata A, Mobashery S, 2004. A novel beta-lactamase activity from a penicillin-binding protein of Treponema pallidum and why syphilis is still treatable with penicillin. J Biol Chem 279: 14917–14921. [DOI] [PubMed] [Google Scholar]
- 22.Brinkman MB, McKevitt M, McLoughlin M, Perez C, Howell J, Weinstock GM, Norris SJ, Palzkill T, 2006. Reactivity of antibodies from syphilis patients to a protein array representing the Treponema pallidum proteome. J Clin Microbiol 44: 888–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz AR, et al. 2010. Secondary syphilis in cali, Colombia: new concepts in disease pathogenesis. PLoS Negl Trop Dis 4: e690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salazar JC, Rathi A, Michael NL, Radolf JD, Jagodzinski LL, 2007. Assessment of the kinetics of Treponema pallidum dissemination into blood and tissues in experimental syphilis by real-time quantitative PCR. Infect Immun 75: 2954–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Biswas G, Sakai M, 2014. Loop-mediated isothermal amplification (LAMP) assays for detection and identification of aquaculture pathogens: current state and perspectives. Appl Microbiol Biotechnol 98: 2881–2895. [DOI] [PubMed] [Google Scholar]