Abstract.
West Nile virus (WNV) is an important emerging flavivirus in North America. Experimental studies in animals infer the development of persistent infection in the kidneys. In humans, recent studies suggest the possibility of persistent renal infection and chronic kidney disease. Considering the discrepancies between published studies on viral RNA detection in urine of convalescing WNV-positive patients, we explored the use of electron microscopy (EM) with anti-WNV E protein antibody immunogold labeling to detect virus in the urine sediment from a subset of study participants in the Houston WNV cohort. In 42% of evaluated study participants had visible sediment present in urine after centrifugation; viral particles consistent with the size and morphology of WNV were successfully detected using EM in the urine sediment up to 9 years postinfection. The anti-WNV immunogold labeling bound to virus envelope in the sediment allowed for enhanced detection when compared with PCR and provide a new technique for understanding kidney disease in WNV patients. These results provide further evidence of persistent infection in at least a subset of individuals infected with WNV. These findings present a novel tool to diagnose persistent WNV infection and its possible link with progressive renal pathology.
INTRODUCTION
West Nile virus (WNV) is an important flavivirus that has emerged across the United States, with an estimated 3 million people infected to date.1 Experimental studies in various animal models support the development of persistent infection in the central nervous system and the epithelium of the distal renal tubules.2–6 There is an urgent need to understand the mechanisms and potential pathology related to persistent renal infection in humans. In Houston, we have been prospectively following a cohort of study participants with a history of infection with WNV since 2003. In a prior publication, we reported five of 25 cohort participants were positive for WNV RNA in urine up to 7 years postinfection,7 and in a larger study, we found that 40% of our cohort population had evidence of chronic kidney disease (CKD), with a history of the more severe form of WNV infection, neuroinvasive disease, being identified as an independent risk factor for CKD.8 Of those with stage III-IV CKD, 83% were found to be positive for WNV RNA in urine.
Our current methods of viral RNA detection are not optimal. Serial collections and testing of positive patients only detected viral RNA 39% of the time (unpublished data), and we hypothesize this is either due to intermittent shedding or natural PCR inhibitors present in the urine environment. One study of Colorado failed to detect viral RNA in a group of 40 patients up to 6 years postinfection; however, urine collection, storage, RNA extraction, and polymerase chain reaction (PCR) methods in this study all differed from our published approach.9 Another study conducted in Arizona on 63 patients within 5 months postinfection identified one woman who was positive for viral RNA in urine through transcription-mediated amplification (TMA) and not by PCR.10 The authors speculated that the discrepancy in testing results was a result of low numbers of viral copies that were below the threshold of detection by PCR. Conversely, other studies have been successful in detecting viral RNA in urine, particularly during the acute phase (up to 30 days) after onset of symptoms,11–15 with immunosuppressing conditions and history of neuroinvasive disease enhancing the opportunity for detection. Similar to our observation, one study by Papa et al. 15 described differences in the ability to detect viral RNA if specimens were not properly handled and stored before testing.
Based on the challenges and discrepancies of the current PCR detection method, we explored other methods to detect and potentially diagnose persistent renal infection in patients with a history of WNV infection. This paper presents our methodology and results of using electron microscopy (EM), including the use of labeling immunogold with anti-WNV E protein polyclonal antibody, to detect virus envelope in the sediment of urine.
METHODS
Study population and specimen collection.
Of our cohort of 220 study participants with a history of WNV infection, we first collected urine sediment from one participant (Case 1) who we consistently found to be positive for WNV RNA in urine for evaluation of the sediment for viral particles using EM. After visualizing virus-like particles on EM that were consistent with the size and morphology of WNV, we collected urine samples from other WNV-positive study participants who had visible urine sediments observed after collection and centrifugation (N = 12). As a control, we collected and analyzed urine sediments from patients with chronic kidney disease without a prior WNV infection (N = 3). Sediments were analyzed using EM with anti-WNV E protein polyclonal antibody–labeled immunogold. In addition, one of our WNV participants who we identified as positive on urine sediment using EM had undergone a renal biopsy as part of her evaluation by her physicians for CKD. We obtained banked fixed tissue from her nephrologist for additional EM. This study was reviewed and approved by the University of Texas Health Science Center Committee for the Protection of Human Subjects (HSC-SPH-03-039) and Baylor College of Medicine Institutional Review Board (H-30533). Written informed consent was obtained from all study participants.
With these participants, 12 mL urine was collected fresh and poured into a 15 mL RNASE/DNASE-free conical tube and then centrifuged at low speed (1,800 × g) at 4°C for 10 minutes. After spinning, urine sediment reached at least the 100 uL line of the 15 mL conical tube. Urine was decanted, and the pellet was resuspended in a fixation solution of 3% formalin/0.15% glutaraldehyde and stored at 4°C for up to 2 weeks. In two cases (Case 1 and Case 2), we also examined the samples directly by applying a small portion of each pellet to a formvar/carbon nickel grid, heat fixed, and then immunogold-labeled without staining.
PCR of urine samples.
PCR analysis of urine samples was performed as described previously.7 Briefly, urine was aliquoted into tubes containing Protector RNase Inhibitor (Roche Diagnostics, Indianapolis, IN), and RNA was extracted with QIAamp MinElute Virus Spin kit (Qiagen, Frederick, MD) following the manufacturer’s protocol. WNV RNA was detected in samples by end-point reverse transcription-PCR using One-Step RT-PCR kit (Qiagen), followed by a nested round with HotStarTaq DNA Polymerase (Qiagen). Primer sequences and PCR conditions were previously described.7,16
Immunogold labeling and election microscopy.
Pellets were partially dehydrated, embedded in LR-White resin, polymerized and thin sectioned to 100 nm and placed on nickel grids. The free aldehyde groups were blocked in 50 mM glycine in a phosphate buffer solution (PBS) for 15 minutes, then blocked in a blocking solution (Aurion) for goat anti-rabbit IgG gold conjugate for 30 minutes. The specimens were then washed in BSA-c buffer (PBS+0.1% BSA-c, pH 7.4) three times for 5 minutes and incubated in anti-WNV rabbit polyclonal antibody corresponding to the 14 C-terminal amino acids of WNV E protein (ab25886, Abcam Inc., Cambridge, MA) and diluted (1–5 μg/mL) in a BSA-c buffer overnight. Controls were generated by leaving the grids in BSA-c a buffer overnight without the primary antibody to estimate the level of nonspecific binding. After incubation, the specimens were washed in a BSA-c buffer six times for 5 minutes, then incubated in goat anti-rabbit Ultra-Small gold (Aurion) diluted 1/400 in a BSA-c buffer for 2 hours. The specimens were then washed in a BSA-c buffer six times for 5 minutes, then washed in PBS three times for 5 minutes. Specimens then were post fixed in 2% glutaraldehyde in PBS for 5 minutes, washed in PBS for 5 minutes, then washed thoroughly in distilled water five times for 2 minutes. Sections were then heavy metal stained (uranyl acetate followed by lead citrate) and air-dried for EM. Grids were imaged using a JEOL 1200 TEM at 60 kv and captured with a 2 k × 2 k Gatan Orius 830 CCD camera. Images were considered positive if the 10 nm immunogold particles bound to viral particles matching the size (45–55 nm) and morphology of WNV.
To evaluate the labeling quality of the antibody binding to infectious virus, Vero cells 3 days postinfection with WNV strain 2004Hou3 (GenBank KC928260.1) were spun down and fixed in 3% formalin/0.15% gluteraldehyde then processed and immunogold-labeled as previously described. The WNV strain 2004Hou3 used for infection of Vero cells was isolated from naturally infected squirrel by single passage of tissue homogenate on Vero cells. All imaging was carried out using a JEOL 1200EX Transmission Electron Microscope and a Gatan 792 Bioscan CCD Camera. In addition, we used noninfected Vero cells as a negative control to assess any nonspecific binding of primary and secondary antibodies used for immunogold staining.
A renal core biopsy section from Case 2 was fixed in Millonigs buffered 3% glutaraldehyde (Electron Microscopy Sciences [EMS], Hatfield, PA) for 24 hours. Fixed samples were placed in 2% osmium for 1 hour at 4°C. The specimens were then dehydrated through a series of graded ethanols (50% ethanol for 5 minutes, 70% for 10 minutes, 95% for 10 minutes and, finally, 100% three times for 10 minutes) followed by three 10-minute changes of propylene oxide (EMS). Using LX-112 resin (LADD Research, Williston, VT) the sample was infiltrated for 2 hours at room temperature in a 50/50 solution of LX-112 resin and propylene oxide. The biopsy section was further infiltrated in 100% LX-112 resin for an additional 2 hours then embedded and polymerized overnight at 70°C. Semi-thin sections (500 nm) were first cut, heat fixed on a glass slide stained with toluidine blue, and a representative block was selected for ultrathin sectioning. Ultrathin sections (120 nm) were then collected on 150 mesh copper grids and stained with uranyl acetate (EMS) for 10 minutes followed by lead citrate (EMS) for 5 minutes. The grids were rinsed with distilled/deionized water, air-dried, and screened using a JEOL 1200EX Transmission Electron Microscope. Images were captured using a Gatan 792 Bioscan CCD Camera.
RESULTS
Study participants ranged from 1 to 9 years postinfection at the time of urine collection. Four study participants had unknown dates of infection as they were asymptomatic and reported no symptoms or illnesses consistent with disease from WNV infection. Additional details regarding the clinical presentation, comorbidities, and testing results of each case can be found in Table 1.
Table 1.
Case demographics
| Case | Age at infection, gender | Race/Ethnicity | Year of onset | Clinical presentation | Comorbidities | Lowest recorded eGFR (mL/min/1.73 m2) | Urinalysis | YPI: Urine collection for EM | Urine pellet | EM results | Urine PCR YPI (PCR results) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 36F | White, non-Hispanic | 2003 | WNND | None | 67 | Normal | 8 | Large white | + | 6 (pos, neg, pos) |
| 7 (pos, pos, neg, neg) | |||||||||||
| 8 (eqv, neg) | |||||||||||
| 2 | 58F | African-American | 2002 | WNND | Diabetes, hypertension | 39 | Proteinuria | 9 | Large white | + | 8 (pos) |
| 9 (pos, pos) | |||||||||||
| 3 | 47F | African-American | UNK | Asymptomatic | None | 88 | Proteinuria | NA | Large white | + | UNK (neg) |
| 4 | 63F | White, non-Hispanic | 2011 | WNND | Hypertension, high cholesterol | 100 | Turbid, proteinuria, hematuria, hyaline cast | 1 | Large white | + | NA |
| 5 | 57M | African-American | 2009 | Asymptomatic | High cholesterol | 59 | Proteinuria | NA | Very small | + | 1 (neg, neg, neg, neg) |
| 2 (neg, neg) | |||||||||||
| 6 | 52M | White, non-Hispanic | 2004 | WNND | None | 71 | Hematuria, turbid | 7 | Small | − | 5 (neg, neg, pos) |
| 6 (eqv, neg, neg, neg) | |||||||||||
| 7 (neg, neg) | |||||||||||
| 7 | 60M | White, non-Hispanic | 2006 | Asymptomatic | Hypertension, high cholesterol | 67 | Proteinuria, turbid | NA | Small white | − | 4 (neg, neg) |
| 8 | 58M | White, non-Hispanic | 2008 | WNND | Cardiovascular disease, hypertension, seizures | 68 | Proteinuria, turbid | 3 | Small | − | 2 (neg, neg, neg) |
| 3 (neg) | |||||||||||
| 9 | 25F | Asian | UNK | Asymptomatic | None | 124 | Hematuria, hyaline cast | NA | Medium | − | UNK (neg) |
| 10 | 45F | Hispanic | 2012 | WNND | High cholesterol | 100 | Hematuria, proteinuria, turbid | 1 | Large brown/red | − | NA |
| 11 | 31M | Hispanic | 2012 | WNND | None | NA | Normal | 1 | Very small | − | NA |
| 12 | 52F | White, non-Hispanic | 2005 | WNF | Hepatitis | 62 | Proteinuria, hematuria, turbid | 6 | Large white | − | 5 (neg, neg) |
YPI = years post infection; WNND = West Nile Neuroinvasive Disease (meningitis, encephalitis, and acute flaccid paralysis); WNF = West Nile Fever; neg = negative/not detected; pos = positive/WNV detected; eqv = equivocal; NA = not available; UNK = unknown. In the event that more than one urine sample and subsequent PCR test was performed in a given year post infection, all test results are listed beside the corresponding year.
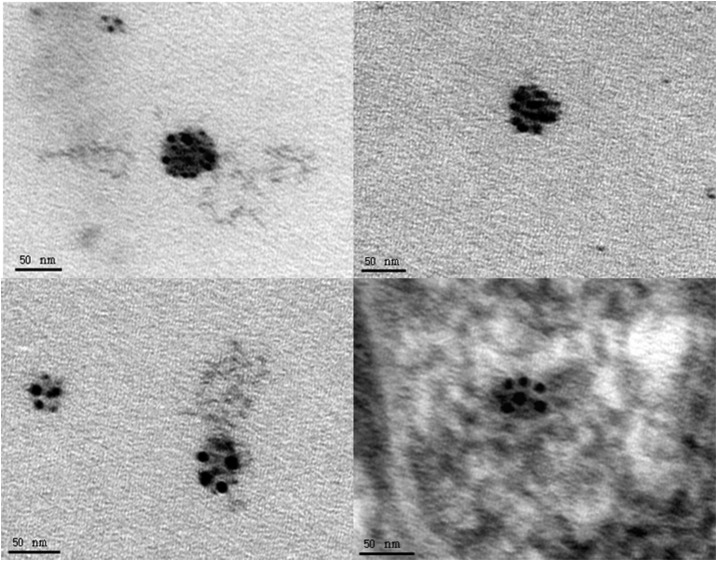
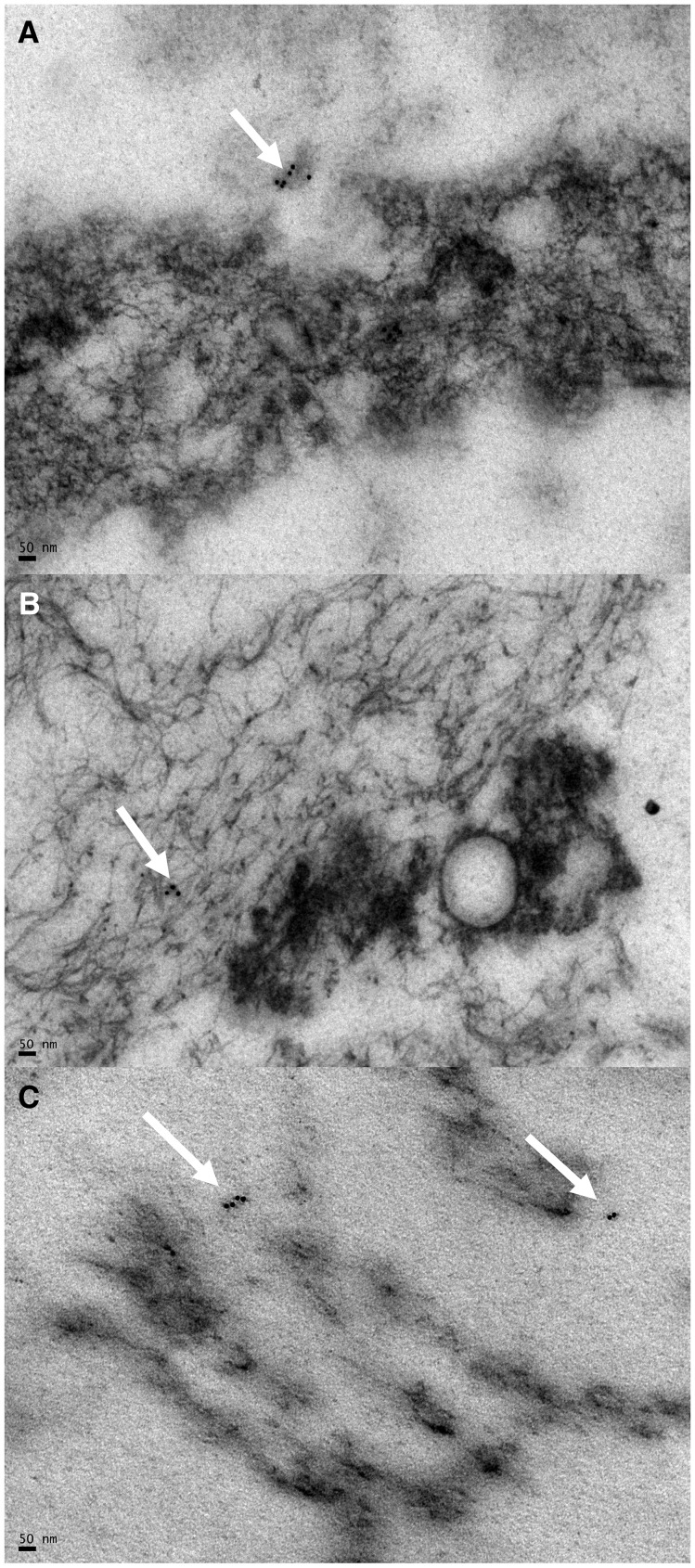
As a positive control measure of our methods, the WNV strain 2004Hou3 infected Vero cells that were immunogold-labeled 3 days postinfection demonstrated specifically labeled particles consistent in size and morphology to WNV (Figure 1). The uninfected (negative) Vero cell control did not display any specific binding (images not shown). Negative control grids without the primary antibody did not show any specific binding, validating our findings (images not shown). In addition to negative control grids, we also processed urine sediment from three WNV-negative controls with chronic kidney disease and found no virus or immunogold labeling visible in any of the sections (Figure 2). Only minor, nonspecific binding was detected as expected.
Figure 1.
Anti-WNV E-protein immunogold labeled 2004Hou3 from 3 day postinfected Vero cells. The bottom right image is immunogold labeled and stained with uranyl acetate and lead citrate; other images are immunogold labeled only.
Figure 2.
Immunogold labeling of urine sediment from three WNV-negative CKD control patients. (A–C) All images are immunogold labeled and stained with uranyl acetate and lead citrate. Arrows indicate nonspecific staining. All bars shown are the width of 50 nm.
Viral particles consistent with the size and morphology of WNV were successfully detected in the urine sediment of five of 12 (42%) participants using EM (Figures 3 and 4). Of the five EM-positive patients, four had large, white sediment pellets, whereas one had a very small sediment pellet.
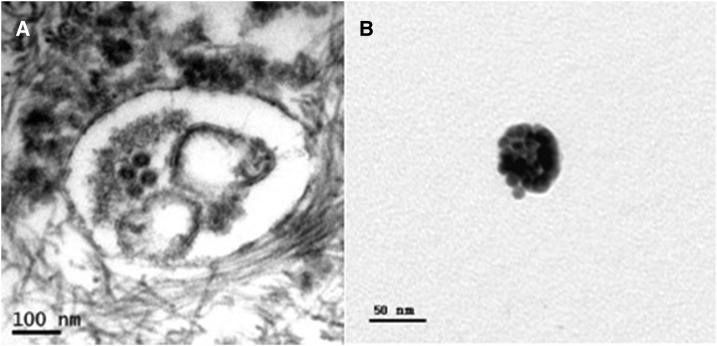
Figure 3.
Identification of WNV particles. Case 1: (A) Electron microscopy of urine sediment identified four possible viral particles consistent with the size, shape, and morphology of WNV stained with uranyl acetate and lead citrate and no immunogold labeling. (B) WNV immunogold labeled with no additional staining.
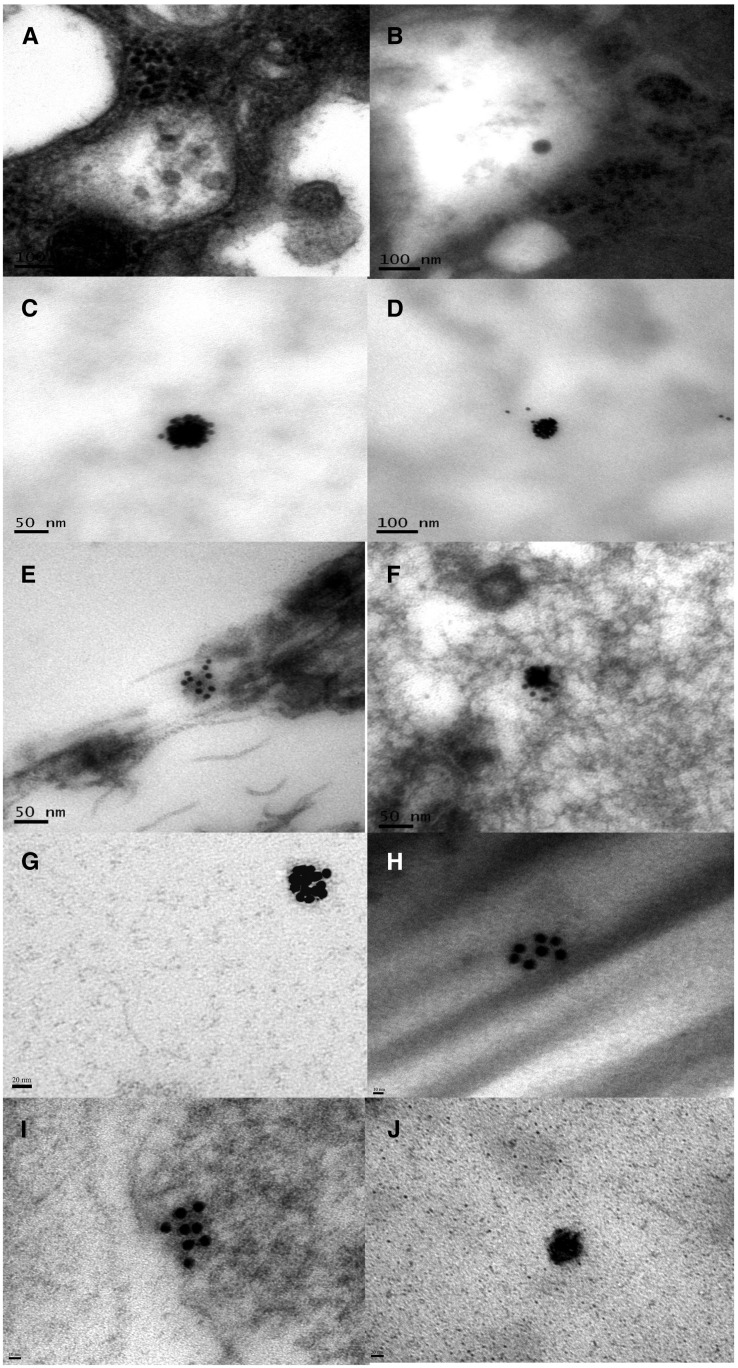
Figure 4.
Cases 2, 3, 4, and 5: Electron microscopy and WNV immunogold images. (A and B) Virion-like structures in the lumen of the renal tubules on sections from renal biopsy tissue from Case 2, stained with uranyl acetate and lead citrate. (C and D) WNV Immunogold-positive viral particles in the urine sediment from Case 2, immunogold labeled only. (E and F) WNV Immunogold-positive viral particles in the urine sediment from Case 3, immunogold labeled and stained with uranyl acetate and lead citrate. (G and H) WNV Immunogold-positive viral particles in the urine sediments from Case 4, immunogold labeled only. (I and J) WNV Immunogold-positive viral particles in the urine sediments from Case 5, immunogold labeled and stained with uranyl acetate and lead citrate. Bars in images (A, B, and D) are 100 nm. Bars in (C, E, and F) are 50 nm. Bars in (G and J) are 20 nm. Bars in (H and I) are 10 nm.
A renal biopsy was collected in 2012 for Case 2. On histopathology, six of 17 glomeruli were globally sclerosed with evidence of hyalin changes. The nonsclerosed glomeruli showed a moderate increase in mesangial matrix and formation of mesangial nodules. Approximately 40% of the renal cortical tissue showed interstitial fibrosis and tubular atrophy with a mild mononuclear infiltrate observed. Diagnosis based on biopsy was diabetic nodular glomerulosclerosis and mild interstitial infiltrate. The pathology report identified segmental positive IgM and C3-positive reactions in the glomerular mesangium. Linear nonspecific reactions with albumin, IgG, and kappa and lambda light chains were observed along the glomerular and tubular basement membranes. C3 was focally positive in the tubules, and a few tubular casts were positive for IgA. Immunogold-positive viral particles consistent with the size and shape of WNV can be seen in the renal biopsy tissue (Figure 4A and B).
DISCUSSION
Our study demonstrated WNV in the urine sediment of study participants with a history of WNV infection, including patients with multiple negative tests through traditional PCR methods (Table 1). These results, for the first time, visualized whole viral particles in urine sediment and kidney biopsies using EM and immunogold staining as well as provide further evidence of persistent renal infection in at least a subset of study participants in Houston. Considering the high prevalence of CKD in our population, with 83% of those with stage III-IV CKD being positive for viral RNA in urine,8 these findings are a critical component to link the chronic renal infection with progressive renal pathology. Interestingly, one case was in an otherwise healthy woman with asymptomatic WNV infection, which raises concerns over the actual prevalence of persistent infection of the kidneys among all infected individuals and not just those with the more severe clinical presentation.
Immunogold labeling enhanced the identification of the envelope of WNV in the urine sediment, providing us with the specificity we needed, that we and other groups have been unable to achieve with various PCR methods. We were concerned about cross-reactivity among North American flaviviruses,17,18 particularly in Houston where St. Louis encephalitis virus (SLEV) has been endemic since 1964.19 We attempted to minimize the possibility of cross-reaction to other flaviviruses during the immunogold labeling procedure by choosing an anti-WNV antibody that corresponded specifically to the 14 amino acids near the C-terminus (domain III) of the WNV E protein. This particular target corresponds to a virus-specific epitope known for its utility in discriminating WNV from other Japanese encephalitis group viruses, including SLEV.20 In parallel, we followed the same immunogold labeling procedures using the WNV-specific H5-46 monoclonal antibody and found the monoclonal was vastly inferior to the polyclonal in binding to virus envelope on our positive control infected Vero cells. Since some nonspecific staining can still occur, it necessary to ensure that positive staining corresponds with the known size (45–55 nm) of WNV.
Rates of recovery for viral RNA from human urine are low, and given the low sensitivity of detecting viral RNA by PCR, the methods presented in this paper allow for definitive identification of WNV in urine sediment; however, there are several limitations worth mentioning. First, using this method we are only able to positively identify binding to the envelope protein of WNV. In some images in both patients and infected Vero cells, viral particles appeared slightly smaller than 45 nm (i.e., Figure 3A where the diameter measures 43 nm). It is possible that these represent empty subviral particles, which can be smaller and have an incomplete or missing genome. This does not detract from the persistence of WNV in the kidneys of some patients as it still represents replication; however, it could provide one explanation for lack of concordance with WNV RNA detection.
Second, EM is a “needle in the haystack” approach, in that it takes hours to examine many sections from a single patient before identifying positive viral particles, thereby preventing it from being a high throughput method for detection of WNV-related renal infection. The number of viral particles present appears to be low in number, which supports the previously reported theory that the number of viral copies present is below the threshold for RT-PCR.10 Second, the cost of EM with immunogold labeling could be somewhat prohibitive in a broad clinical diagnostic setting, with the total research cost in our laboratory being $1,050 per patient. However, this method can be valuable for detecting WNV in the urine and kidney of known WNV cases, especially those that present with kidney disease years after infection. In the future, we hope to extend this method of testing to a larger portion of the cohort and compare the sensitivity of this method with standard RT-PCR, silver enhancement on a glass slide, and other novel detection methods as they become available. The positive urine samples and infected Vero cells as established by immunogold EM will provide us with positive controls for further development and optimization of other more cost-effective methods.
In conclusion, this study successfully detected WNV in urine using EM with immunogold labeling for the first time, providing a method that could be more sensitive than traditional PCR, but requires additional testing with a larger sample size. In addition, the study provides further evidence supporting viral persistence after WNV infection in humans and highlights the urgent need to understand the true prevalence of chronic renal infection in those infected and whether severity of disease or immunosuppression at the time of acute infection places a person at higher risk for chronic infection. Also, we need to understand the mechanisms underlying the establishment of persistent infection and progression of renal pathology so that treatment options can be established. With more than 3 million people now estimated across the United States to have been infected with the virus, the public health cost related to persistent renal infection could be substantial.
Acknowledgment:
We thank the study participants for their generous time and commitment to the Houston WNV cohort study.
REFERENCES
- 1.Petersen LR, Carson PJ, Biggerstaff BJ, Custer B, Borchardt SM, Busch MP, 2013. Estimated cumulative incidence of West Nile virus infection in US adults, 1999–2010. Epidemiol Infect 141: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appler KK, Brown AN, Stewart BS, Behr MJ, Demarest VL, Wong SJ, Bernard KA, 2010. Persistence of West Nile virus in the central nervous system and periphery of mice. PLoS One 5: e10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pogodina VV, Frolova MP, Malenko GV, Fokina GI, Koreshkova GV, Kiseleva LL, Bochkova NG, Ralph NM, 1983. Study on West Nile virus persistence in monkeys. Arch Virol 75: 71–86. [DOI] [PubMed] [Google Scholar]
- 4.Siddharthan V, Wang H, Motter NE, Hall JO, Skinner RD, Skirpstunas RT, Morrey JD, 2009. Persistent West Nile virus associated with a neurological sequela in hamsters identified by motor unit number estimation. J Virol 83: 4251–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tesh RB, Siirin M, Guzman H, Travassos da Rosa AP, Wu X, Duan T, Lei H, Nunes MR, Xiao SY, 2005. Persistent West Nile virus infection in the golden hamster: studies on its mechanism and possible implications for other flavivirus infections. J Infect Dis 192: 287–295. [DOI] [PubMed] [Google Scholar]
- 6.Tonry JH, Brown CB, Cropp CB, Co JK, Bennett SN, Nerurkar VR, Kuberski T, Gubler DJ, 2005. West Nile virus detection in urine. Emerg Infect Dis 11: 1294–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray K, Walker C, Herrington E, Lewis JA, McCormick J, Beasley DW, Tesh RB, Fisher-Hoch S, 2010. Persistent infection with West Nile virus years after initial infection. J Infect Dis 201: 2–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nolan MS, Podoll AS, Hause AM, Akers KM, Finkel KW, Murray KO, 2012. Prevalence of chronic kidney disease and progression of disease over time among patients enrolled in the Houston West Nile virus cohort. PLoS One 7: e40374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibney KB, Lanciotti RS, Sejvar JJ, Nugent CT, Linnen JM, Delorey MJ, Lehman JA, Boswell EN, Staples JE, Fischer M, 2011. West Nile virus RNA not detected in urine of 40 people tested 6 years after acute West Nile virus disease. J Infect Dis 203: 344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baty SA, et al. 2012. Evaluation for West Nile virus (WNV) RNA in urine of patients within 5 months of WNV infection. J Infect Dis 205: 1476–1477. [DOI] [PubMed] [Google Scholar]
- 11.Barzon L, Pacenti M, Franchin E, Pagni S, Martello T, Cattai M, Cusinato R, Palu G, 2013. Excretion of West Nile virus in urine during acute infection. J Infect Dis 208: 1086–1092. [DOI] [PubMed] [Google Scholar]
- 12.Barzon L, Pacenti M, Franchin E, Squarzon L, Sinigaglia A, Ulbert S, Cusinato R, Palu G, 2014. Isolation of West Nile virus from urine samples of patients with acute infection. J Clin Microbiol 52: 3411–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ergunay K, Karagul A, Abudalal A, Hacioglu S, Us D, Erdem Y, Ozkul A, 2015. Prospective investigation of the impact of West Nile virus infections in renal diseases. J Med Virol 87: 1625–1632. [DOI] [PubMed] [Google Scholar]
- 14.Nagy A, Ban E, Nagy O, Ferenczi E, Farkas A, Banyai K, Farkas S, Takacs M, 2016. Detection and sequencing of West Nile virus RNA from human urine and serum samples during the 2014 seasonal period. Arch Virol 161: 1797–1806. [DOI] [PubMed] [Google Scholar]
- 15.Papa A, Testa T, Papadopoulou E, 2014. Detection of West Nile virus lineage 2 in the urine of acute human infections. J Med Virol 86: 2142–2145. [DOI] [PubMed] [Google Scholar]
- 16.Johnson DJ, Ostlund EN, Pedersen DD, Schmitt BJ, 2001. Detection of North American West Nile virus in animal tissue by a reverse transcription-nested polymerase chain reaction assay. Emerg Infect Dis 7: 739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crill WD, Trainor NB, Chang GJ, 2007. A detailed mutagenesis study of flavivirus cross-reactive epitopes using West Nile virus-like particles. J Gen Virol 88: 1169–1174. [DOI] [PubMed] [Google Scholar]
- 18.Martin DA, Biggerstaff BJ, Allen B, Johnson AJ, Lanciotti RS, Roehrig JT, 2002. Use of immunoglobulin m cross-reactions in differential diagnosis of human flaviviral encephalitis infections in the United States. Clin Diagn Lab Immunol 9: 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lillibridge KM, et al. 2004. The 2002 introduction of West Nile virus into Harris County, Texas, an area historically endemic for St. Louis encephalitis. Am J Trop Med Hyg 70: 676–681. [PubMed] [Google Scholar]
- 20.Beasley DW, et al. 2004. Use of a recombinant envelope protein subunit antigen for specific serological diagnosis of West Nile virus infection. J Clin Microbiol 42: 2759–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]